Abstract
Pyrazinamide (PZA) is a first-line tuberculosis drug that plays a unique role in shortening the duration of tuberculosis chemotherapy. PZA is hydrolyzed intracellularly to pyrazinoic acid (POA) by pyrazinamidase (PZase), an enzyme frequently lost in PZA-resistant strains, but the downstream target of POA in Mycobacterium tuberculosis (Mtb) has remained elusive. Here we identify a new target of POA as the ribosomal protein S1 (RpsA), a vital protein involved in protein translation and the ribosome-sparing process of trans-translation. Affinity chromatography using immobilized POA selectively retained RpsA and a PZA-resistant clinical isolate without pncA mutation harbored an alanine deletion in its C-terminus. RpsA overexpression conferred increased PZA resistance and we confirmed biochemically that POA bound to RpsA (but not the ΔAla mutant) and inhibited trans-translation rather than canonical translation. Trans-translation is essential for freeing scarce ribosomes in non-replicating organisms and its inhibition may explain the unique ability of PZA to eradicate persisting organisms.
Pyrazinamide (PZA) is a critical first-line TB drug used in combination with isoniazid, ethambutol and rifampin for the treatment of drug susceptible TB and is also frequently used to treat multi-drug resistant TB (MDR-TB) (1). PZA plays a unique role in shortening TB treatment from the 9–12 months required prior to its introduction to the current standard of 6 months, often referred to as short course chemotherapy (2). PZA is an intriguing, unconventional, and paradoxical TB drug. Despite its powerful in vivo sterilizing activity it has no apparent activity for actively growing TB bacilli under normal culture conditions at neutral pH (3). It is instead preferentially active against non-replicating persister bacteria with low metabolism at acid pH in vitro (4) or in vivo during active inflammation (5). Although several new drug candidates are currently in clinical development (6, 7), none can replace PZA. All of the drug candidates, including the highly potent TMC207, will have to be used together with PZA since any drug combination without PZA has been found to be inferior (8–11).
Despite its important role in shortening TB therapy, the mechanism of action of PZA is the least understood of all the current TB drugs (12). Structurally, PZA is an analog of nicotinamide, which, like INH (13), is a prodrug, requiring activation to its active form pyrazinoic acid (POA) by the bacterial pyrazinamidase (PZase) (14). Mutation in the pncA gene encoding the PZase (14) is the major mechanism for PZA resistance in Mtb (14–16). PZA is believed to enter Mtb by passive diffusion, where it is converted to POA by the PZase. POA is an acid with a pKa of 2.9 and is therefore trapped within the cell as the carboxylate anion where it is possibly excreted by a weak efflux pump and passive diffusion (17). Small amounts of protonated POA capable of diffusion across the membrane have been proposed to collapse the proton gradient, reducing membrane potential and affecting membrane transport (18). The observations that energy inhibitors such as DCCD (an F1F0 ATPase inhibitor) (18) and also the new drug candidate TMC207 synergize with PZA in vitro (8, 19) provide some support for this model. However, the real molecular target of PZA is unknown. Although fatty acid synthase-I (Fas-I) was proposed as a target of PZA based on studies with an analog (5-Cl-PZA) (20), a subsequent study showed that Fas-I was not the target of PZA (21).
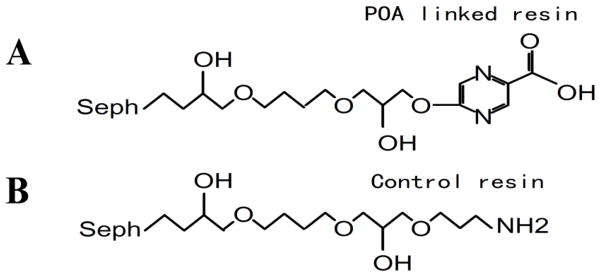
To identify potential targets that bind to POA (2-pyrazinecarboxylic acid) in Mtb we used a proteomic approach to search for proteins that bind to POA by affinity chromatography. The POA analog 5-hydroxyl-2-pyrazinecarboxylic acid (Fig. 1A) was synthesized and covalently coupled to Epoxy Sepharose 6B column. As a control, ethanolamine (Fig. 1B) was also coupled to a separate column. Binding studies with M. tuberculosis cell lysates revealed several proteins that bound to POA (fig. S1). In contrast, no proteins bound to the control column, indicating that the proteins bound specifically to POA. Mass spectrometry analysis and subsequent database searches identified the major POA binding protein as RpsA (table S1), the largest 30S ribosomal protein S1 (Rv1630) from Mtb.
Fig. 1.
Structures of POA derivative (5-hydroxyl-2-pyrazinecarboxylic acid) (A) and the control compound ethanolamine (B) coupled to Sepharose 6B column for the identification of POA binding proteins from M. tuberculosis.
Target overexpression often confers increased drug resistance (22). To confirm that RpsA was indeed a target of PZA, we overexpressed the wild type RpsA in Mtb and measured the PZA sensitivity of these bacilli compared to bacilli carrying only the empty vector control. Overexpression of RpsA caused a 5-fold increase in the minimum inhibitory concentration (MIC) of PZA (MIC=500 μg/ml) compared with the vector control and the parental Mtb strain (MIC=100 μg/ml) at pH 5.5. The susceptibility of the RpsA overexpressing Mtb strain to other drugs, including isoniazid, rifampin, streptomycin, kanamycin, and norfloxacin, remained the same as the parent strain or the vector control strain.
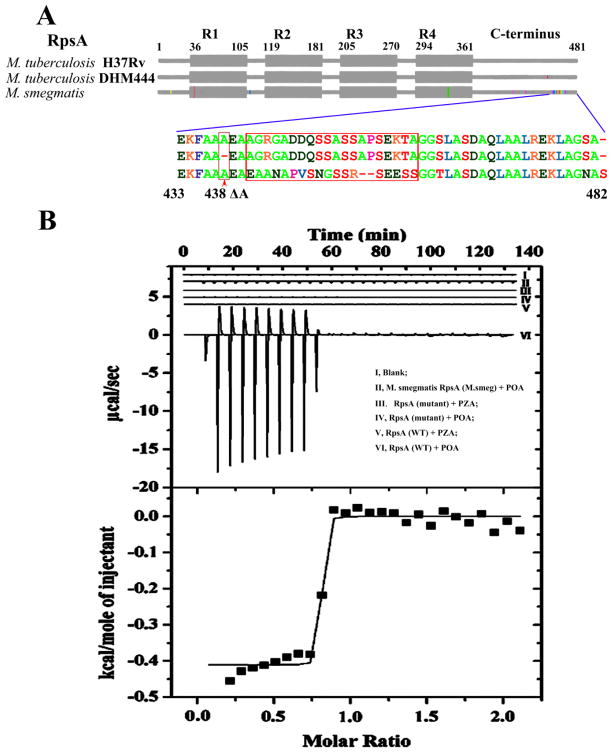
Most PZA-resistant M. tuberculosis strains have mutations in pncA that prevent conversion of PZA to POA (14–16). A small number of PZA-resistant strains, however, have been reported that do not have pncA mutations (15, 16). We previously identified a low level PZA-resistant Mtb clinical isolate DHM444 (MIC=200–300 μg/ml PZA compared with 100 μg/ml in the sensitive control strain H37Rv) that lacked any pncA mutations (15). This suggested that its resistance could be due to alterations in RpsA. We therefore sequenced the rpsA gene from this strain and found that it contained a 3-bp GCC deletion at the nucleotide position 1314 resulting in deletion of an alanine at amino acid 438 (ΔA438) in the C-terminus of RpsA (Fig. 2A), a region that is not considered to be strictly required for protein synthesis in vivo (23).
Fig. 2.
RpsA alignment and isothermal titration calorimetry (ITC) titration of RpsA and POA. (A) Alignment of RpsA from M. tuberculosis H37Rv, M. tuberculosis PZA-resistant strain DHM444 and M. smegmatis. R1 to R4 represent the four homologous RNA-binding domains in RpsA. Colored vertical lines in gray boxes indicate sequence variations in the highly conserved RpsA sequences compared with the wild type M. tuberculosis sequence. The expanded region shows the variability in amino acid sequence in the C-terminus of RpsA among mycobacterial species. The red arrow at position 438 amino acid residue indicates the deletion of alanine in the C-terminal region of the mutant RpsA. ITC binding studies indicate POA bound to the M. tuberculosis H37Rv RpsA (WT) (B, inset VI), but not DHM444 RpsA (Mutant)(Inset, IV), and only weakly with the M. smegmatis RpsA (M. smeg) (Inset II). PZA did not bind to wild type RpsA (Inset V) or mutant RpsA (Inset III). The lower panel of the Fig. 2B shows the typical molar ratio saturation plot of POA with wild type Mtb RpsA.
To determine if the mutant RpsA from the PZA-resistant strain DHM444 has any defect in POA binding, we overexpressed and purified the mutant RpsA (designated RpsAΔA438), the wild type M. tuberculosis RpsA, and the M. smegmatis RpsA, and assessed their ability to bind to POA using isothermal titration calorimetry (ITC). The wild type Mtb RpsA was found to specifically bind to POA (Fig. 2B, VI) with K=(7.53±2.21)×106 M−1, ΔH= −410.9±8.693 Kcal·mol−1, ΔS=27.6 cal·mol−1·K−1 (Fig. 2B, lower panel), but not to the prodrug PZA as expected (Fig. 2B, V). However, the mutant RpsAΔA438 from the PZA-resistant strain DHM444 failed to bind to POA or PZA (Fig. 2B, IV, III), while the RpsA from naturally PZA-resistant M. smegmatis bound to POA only very weakly (Fig. 2B, II). Since the mutant Mtb DHM444 RpsA and the M. smegmatis RpsA had little or no binding to POA (Fig. 2B), it can be inferred that POA binds to the C-terminus of the wild type M. tuberculosis RpsA (Fig. 2A). From the protein sequence alignment of RpsA from different mycobacterial species, the C-terminal region, where the mutation occurs in the PZA-resistant M. tuberculosis strain DHM444, is also the most variable region in the PZA sensitive versus resistant mycobacterial species (Fig. 2A), indicating that changes in this region may alter PZA susceptibility.
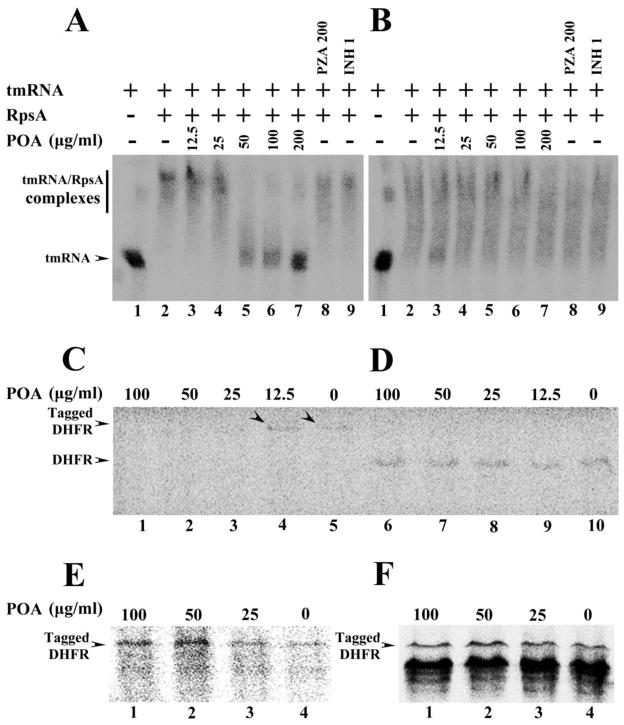
The ribosomal protein S1 (RpsA) is essential for translation, binding directly to both the ribosome and upstream sequences of mRNA (24). In addition to its function in translation, the C-terminus of the RpsA is involved in trans-translation by specifically binding to tmRNA (25–27). Trans-translation is a process involved in rescuing ribosomes that have stalled while in the process of decoding mRNA, tagging the truncated proteins for degradation. Trans-translation has been associated with stress survival, virulence, and recovery from nutrient starvation (28). We therefore evaluated whether the mutant RpsA has any deficiency in tmRNA binding compared with the wild type Mtb RpsA and whether POA affects the interaction between RpsA and tmRNA. Specific binding of RpsA to tmRNA was assessed by changes in gel mobility in the presence of excess tmRNA. The wild type Mtb RpsA and the mutant RpsAΔA438 both bound to the tmRNA in the absence of POA (Fig. 3A, 3B) although the mutant appeared to bind more weakly overall (Fig. 3B). However, when POA was added to the system, it inhibited the wild type Mtb RpsA from binding to tmRNA at its MIC concentration of 50 μg/ml (Fig. 3A), but did not inhibit binding of the mutant RpsAΔA438 (Fig. 3B). The prodrug PZA and the control drug INH had no effect on the binding of wild type or mutant RpsA to the tmRNA (Fig. 3B).
Fig. 3.
(A) Concentration-dependent inhibition of tmRNA binding to wild type M. tuberculosis RpsA by POA (Lanes 2–7). tmRNA from M. tuberculosis was used as RNA alone control (Lane 1). The wild type RpsA interaction with tmRNA was not affected by PZA (200 μg/ml) (Lane 8) or INH (1 μg/ml) (Lane 9). (B) tmRNA had impaired binding to the mutant RpsA (Lane 2), and POA at different concentrations did not inhibit the interaction of the DHM444 mutant RpsA with tmRNA (Lanes 3–7); The mutant RpsA interaction with tmRNA was not affected by PZA (200 μg/ml) (Lane 8) or INH (1 μg/ml) (Lane 9). (C) POA at 100, 50, and 25 μg/ml inhibited trans-translation of the DHFR product in a concentration-dependent manner in the in vitro system that contained ribosomes from M. tuberculosis, tmRNA and recombinant SmpB from M. tuberculosis, template pDHFR-8×AGG rare codons that are required for trans-translation (Lanes 1–5). Arrowheads indicate the trans-translation product DHFR was still present with low concentration of POA at 12.5 μg/ml (Lane 4) or in the absence of POA (Lane 5). POA at different concentrations did not inhibit canonical translation in in vitro translation system using ribosomes from M. tuberculosis, template pDHFR with stop codon (D, Lanes 6–10), nor the trans-translation of DHFR using ribosome from M. smegmatis (E, Lanes 1–4), or using ribosome from E. coli (F, Lanes 1–4) in the trans-translation system that contained tmRNA and recombinant SmpB from M. tuberculosis, template pDHFR-8×AGG rare codons.
RpsA is known to bind to the 3′ terminus of the mRNA-like portion of the tmRNA (25), which has been shown to form a multimeric complex with SmpB, EF-Tu and RpsA for enhanced efficiency of trans-translation (26). Since POA bound to Mtb RpsA (Fig. 2B), and since RpsA is involved in both translation (29) and trans-translation (25–27), we tested whether POA inhibited the translation or the trans-translation function of RpsA. POA had no effect on conventional protein synthesis (Fig. 3D). To examine trans-translation we utilized an in vitro cell-free translation system using the target gene coding for dihydrofolate reductase (DHFR) containing ribosomes from Mtb, M. smegmatis, or E. coli. The DHFR template DNA contained a T7 promoter for transcription and a ribosome-binding site before the DHFR gene with a stop codon or a rare codon cluster for stalled ribosome formation for assessing trans-translation. In the presence of a rare codon cluster in the target DHFR gene, ribosomes stall and translation is blocked (30). When recombinant Mtb SmpB and in vitro transcribed Mtb tmRNA were added a higher molecular weight protein with the tmRNA derived peptide tag was observed for message carrying the rare codon cluster (Fig. 3C, Lane 5), but no such band was observed for the no template control reaction (data not shown). POA inhibited the trans-translation of DHFR with the rare codon cluster at 25 μg/ml or more (Fig. 3C, Lane 3, Lane 2 and Lane 1) in a concentration dependent manner (fig. S2). However, POA did not affect the translation of the normal DHFR gene even at 100 μg/ml with Mtb ribosomes (Fig. 3D) nor did it inhibit the trans-translation of the template bearing the rare codon cluster with either M. smegmatis or E. coli ribosomes (Fig. 3E and 3F). These observations support that POA does not inhibit Mtb SmpB or tmRNA function but instead directly binds to RpsA to cause the inhibition of trans-translation. Under the same conditions, nicotinic acid (50 μg/ml), an analog of POA as a control compound, did not inhibit trans-translation (data not shown).
The Mtb RpsA protein consists of four imperfect repeats of the S1-like domain thought to function directly in binding of RNA, bridging the mRNA (or tmRNA) template and the head of the 30S ribosome terminated at the C-terminus by a 117 amino acid segment. The E. coli RpsA protein contains six repeating S1 domains, with the two N-terminal domains required for ribosome binding. The C-terminal domains have been shown to be specifically involved in trans-translation, thus the deletion of Ala438 observed in our clinical isolate is consistent with POA exerting an effect on ribosome rescue. The deletion occurs within a region homologous (35% identical) to the protein Xrcc4 involved in illegitimate DNA recombination that forms an extensive α-helix (31). Homology modeling of the region suggests that Ala438 lies several turns in to the α-helix connecting a small globular domain to a long helical stalk involved in dimerization. Intriguingly this region has been proposed to be the primary site of nucleic acid interaction in Xrcc4. There are numerous basic residues along the helical face and particularly in the short linker between this helix and the globular domain potentially involved in DNA binding particularly Arg423, Arg424, His 425 and Lys426 that may interact directly with POA disrupting the site of tmRNA interaction.
Trans-translation is dispensable during active growth conditions but becomes important for bacteria in managing stalled ribosomes or damaged mRNA and proteins under stress conditions (32, 33). It is required for stress survival and pathogenesis in some bacteria (28). The levels of RpsA or S1 protein are known to correlate with growth rate, and stress conditions (stationary phase, starvation, acid pH, hypoxia) that halt bacterial growth down-regulate RpsA in bacteria (34). When POA binds to RpsA it prevents the binding of tmRNA to RpsA such that tmRNA cannot function to rescue stalled ribosomes. PZA inhibition of the trans-translation process may therefore be plausibly linked to an interference with survival under stressful, non-replicating conditions in Mtb. The finding that POA binds to RpsA and inhibits the trans-translation process helps to explain how diverse stress conditions such as starvation, acid pH, hypoxia, and energy inhibitors and other drugs could all potentiate PZA activity (18, 35). Trans-translation has been proposed as a good drug target (32). Our finding that PZA inhibits trans-translation qualifies PZA as the first known drug that inhibits the trans-translation process for killing persister bacteria.
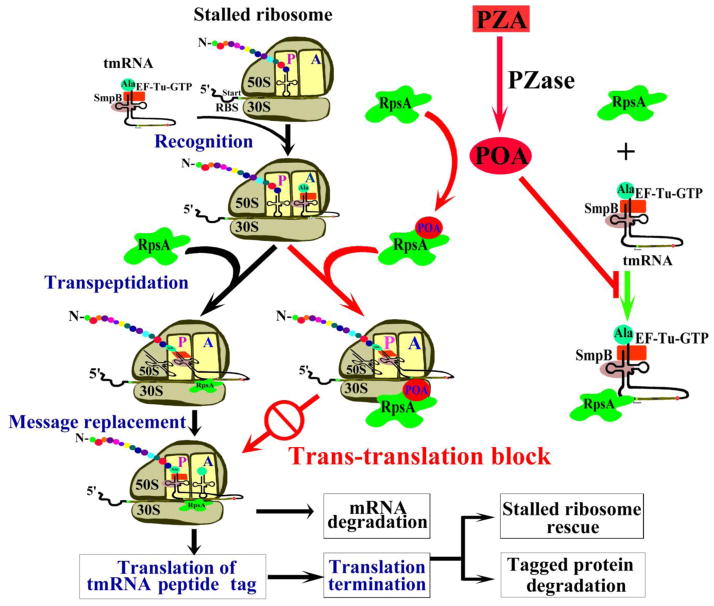
It is worth noting that the conditions that down-regulate RpsA are exactly the stress conditions that also potentiate PZA activity (18, 35). That PZA activity is enhanced by diverse stress conditions (18, 35) has been a mystery. The identification of RpsA as a target of PZA has now afforded a plausible explanation. It is known that under many stress conditions, cells stop growing, translation slows, and levels of RpsA are decreased. Faced with the need to continue to produce proteins required for survival during non-replicating persistence, ribosome rescue becomes even more critical than during periods of rapid growth. POA could therefore saturate the lower levels of RpsA more efficiently and thus show higher activity in metabolically quiescent persister cells than in actively replicating cells. Based on our current and previous studies, we propose a revised new model of the mode of action of PZA that can better explain all the peculiar features of this unique drug (Fig. 4).
Fig. 4.
A new model for the mode of action of PZA. PZA is converted to the active form POA by M. tuberculosis PZase intracellularly and inhibits targets including RpsA. Upon stress, translating ribosomes are stalled and incomplete polypeptides may be toxic to the cell. The bacterial cell resolves this problem by adding tmRNA to the stalled mRNA (28, 36). tmRNA binds to SmpB and EF-Tu, activating the complex for ribosome interaction. The alanyl-tmRNA/SmpB/EF-Tu complex recognizes stalled ribosomes at the 3′ end of an mRNA without stop codon or with rare codons. Translation resumes using tmRNA as a message, resulting in addition of the tmRNA-encoded peptide tag to the C-terminus of the stalled polypeptide. The tagged protein and mRNA are then degraded by proteases and RNases, leading to rescue of stalled ribosomes. POA binding to RpsA interferes with the interaction of RpsA with tmRNA required for trans-translation. POA blockade of the trans-translation pathway leads to a defect in stalled ribosome rescue and depletion of available ribosomes and perhaps increased accumulation of toxic or deleterious proteins, ultimately affecting persister survival under stress conditions.
This study identified RpsA as a new target for PZA in Mtb. PZA is a prototype persister drug that plays an irreplaceable role in any new drug combinations for effective treatment of TB because of its unique ability to shorten the course of chemotherapy. Our study provides a new molecular understanding of the intriguing drug PZA, underscores the importance of trans-translation in persister TB bacteria, and provides a clear pathway for the design of more effective drugs that would further shorten the duration of chemotherapy required to achieve a sterile cure for this devastating disease.
Acknowledgments
This work was supported in part by NIH grant AI44063, in part by the intramural research program of the NIAID, NIH, and by National Key Technologies Research and Development Program of China (2008ZX10003003). We thank Diane E. Griffin and Michael J. Klag for encouragement and support.
References
- 1.WHO. Geneva: 2008. [Google Scholar]
- 2.Mitchison DA. Tubercle. 1985;66:219. doi: 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 3.Tarshis MS, Weed WA., Jr Am Rev Tuberc. 1953 Mar;67:391. doi: 10.1164/art.1953.67.3.391. [DOI] [PubMed] [Google Scholar]
- 4.McDermott W, Tompsett R. Am Rev Tuberc. 1954;70:748. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 5.McCune RM, Feldmann FM, Lambert HP, McDermott W. J Exp Med. 1966 Mar 1;123:445. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yew WW, Cynamon M, Zhang Y. Expert Opin Emerg Drugs. 2010 Sep 26; doi: 10.1517/14728214.2011.521497. [DOI] [PubMed] [Google Scholar]
- 7.Nuermberger EL, Spigelman MK, Yew WW. Respirology. 2010 Jul;15:764. doi: 10.1111/j.1440-1843.2010.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andries K, et al. Science. 2005 Jan 14;307:223. [Google Scholar]
- 9.Rosenthal IM, et al. PLoS Med. 2007 Dec;4:e344. doi: 10.1371/journal.pmed.0040344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasneen R, Tyagi S, Williams K, Grosset J, Nuermberger E. Antimicrob Agents Chemother. 2008 Oct;52:3664. doi: 10.1128/AAC.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuermberger E, et al. Antimicrob Agents Chemother. 2008 Apr;52:1522. doi: 10.1128/AAC.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Mitchison D. Int J Tuberc Lung Dis. 2003;7:6. [PubMed] [Google Scholar]
- 13.Zhang Y, Heym B, Allen B, Young D, Cole S. Nature. 1992 Aug 13;358:591. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 14.Scorpio A, Zhang Y. Nat Med. 1996 Jun;2:662. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 15.Scorpio A, et al. Antimicrobial agents and chemotherapy. 1997 Mar;41:540. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y. Antimicrob Agents Chemother. 2000 Mar;44:528. doi: 10.1128/aac.44.3.528-532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Scorpio A, Nikaido H, Sun Z. J Bacteriol. 1999 Apr;181:2044. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. J Antimicrob Chemother. 2003 Nov;52:790. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim M, et al. Antimicrob Agents Chemother. 2007 Mar;51:1011. doi: 10.1128/AAC.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimhony O, Cox JS, Welch JT, Vilcheze C, Jacobs WR., Jr Nat Med. 2000 Sep;6:1043. doi: 10.1038/79558. [DOI] [PubMed] [Google Scholar]
- 21.Boshoff HI, Mizrahi V, Barry CE., 3rd J Bacteriol. 2002 Apr;184:2167. doi: 10.1128/JB.184.8.2167-2172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Yew WW. Int J Tuberc Lung Dis. 2009 Nov;13:1320. [PubMed] [Google Scholar]
- 23.Boni IV, Artamonova VS, Dreyfus M. J Bacteriol. 2000 Oct;182:5872. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. Cell. 1997 Jan 24;88:235. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 25.Wower IK, Zwieb CW, Guven SA, Wower J. EMBO J. 2000 Dec 1;19:6612. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. J Mol Biol. 2001 Nov 16;314:9. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 27.Saguy M, Gillet R, Skorski P, Hermann-Le Denmat S, Felden B. Nucleic Acids Res. 2007;35:2368. doi: 10.1093/nar/gkm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keiler KC. Annu Rev Microbiol. 2008;62:133. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 29.Suryanarayana T, Subramanian AR. Biochemistry (Mosc) 1983 May 24;22:2715. doi: 10.1021/bi00280a020. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Hirano R, Tagami H, Aiba H. RNA. 2006 Feb;12:248. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junop MS, et al. EMBO J. 2000 Nov 15;19:5962. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thibonnier M, Thiberge JM, De Reuse H. PLoS One. 2008;3:e3810. doi: 10.1371/journal.pone.0003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muto A, et al. Genes Cells. 2000 Aug;5:627. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- 34.Lambert JM, Boileau G, Howe JG, Traut RR. J Bacteriol. 1983 Jun;154:1323. doi: 10.1128/jb.154.3.1323-1328.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Permar S, Sun Z. J Med Microbiol. 2002 Jan;51:42. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- 36.Valle M, et al. Science. 2003 Apr 4;300:127. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]