Abstract
CD4 T cells play critical roles in mediating adaptive immunity to a variety of pathogens. They are also involved in autoimmunity, asthma, and allergic responses as well as in tumor immunity. During TCR activation in a particular cytokine milieu, naive CD4 T cells may differentiate into one of several lineages of T helper (Th) cells, including Th1, Th2, Th17, and iTreg, as defined by their pattern of cytokine production and function. In this review, we summarize the discovery, functions, and relationships among Th cells; the cytokine and signaling requirements for their development; the networks of transcription factors involved in their differentiation; the epigenetic regulation of their key cytokines and transcription factors; and human diseases involving defective CD4 T cell differentiation.
Keywords: CD4 effector T cells, regulatory T cells, T cell differentiation, cytokines, transcription factors, human diseases
INTRODUCTION
CD4 T cells play central roles in the function of the immune system: They help B cells make antibody, enhance and maintain responses of CD8 T cells, regulate macrophage function, orchestrate immune responses against a wide variety of pathogenic microorganisms, and regulate/suppress immune responses both to control autoimmunity and to adjust the magnitude and persistence of responses. CD4 T cells are important mediators of immunologic memory, and when their numbers are diminished or their functions are lost, the individual becomes susceptible to a wide range of infectious disorders. Indeed, in HIV infection, it is when CD4 T cell numbers in blood fall below 200/mm3 that opportunistic infections are most likely to occur.
The Th1/Th2 Paradigm
These various functions are achieved through the differentiation of naive CD4 T cells as they are stimulated by their cognate antigen presented by competent antigen-presenting cells to become effector and/or memory cells of specialized phenotypes. The initial understanding of the existence of distinctive populations of differentiated CD4 T cells came from the analysis of mouse CD4 T cell clones that were shown by Mosmann & Coffman (1) and slightly later by Bottomly and her colleagues (2) to be divisible into two major groups, designated Th1 and Th2 cells by Mosmann & Coffman. Th1 and Th2 clones could be distinguished mainly by the cytokines produced by the cells, but also through the expression of different patterns of cell surface molecules. With regard to cytokine expression, Th1 cells make IFN-γ as their signature cytokine and also uniquely produce lymphotoxin. Th1 cells tend to be good IL-2 producers, and many make TNF-α as well. By contrast, Th2 cells fail to produce IFN-γ or lymphotoxin. Their signature cytokines are IL-4, IL-5, and IL-13. They also make TNF-α, and some produce IL-9. Although initially thought to be unable to make IL-2, later results indicated that Th2 cells could often produce relatively modest amounts of IL-2.
For some time, investigative concern focused on whether the Th1/Th2 dichotomy was principally applicable in mice but not in humans and on whether it was mainly a property of in vitro–differentiated cells. Indeed, although the study of clones had clearly shown a dichotomy between the two cell types, one could often observe cells obtained directly from mice or humans that produced both IL-4 and IFN-γ. Nonetheless, with growing experience, it has become clear that specialization in patterns of cytokine production and other phenotypic characterstics do occur in vivo in mice and in humans. Still important is to compare, in detail, IFN-γ-producing and IL-4-producing CD4 T cells (i.e., Th1 and Th2 cells) generated in vitro and those that appear in vivo in mice and humans to determine the degree to which in vitro–generated cells truly reflect the biology of responses generated under physiologic conditions.
Within a few years of the description of distinct populations existing among CD4 T cell clones, methods were developed to differentiate naive CD4 T cells into IL-4-producing (Th2) cells in vitro (3–6). Such Th2 differentiation required the activation of naive cells, initially with polyclonal stimuli such as anti-CD3 and anti-CD28, and later with cognate antigen, in the presence of a particular set of cytokines. To obtain Th2 cells, the presence of both IL-2 and IL-4 during the differentiation process was essential. The particularly provocative aspect here was that a major product of the Th2 cell, IL-4, was also a critical inducer. The significance of this finding is now clear in vitro and is discussed in detail below.
It was subsequently shown that Th1 cells could be differentiated in vitro from naive CD4 T cells if IL-4 was neutralized and IL-12 was added to the culture (7). Experiments in which single naive CD4 T cells were primed in vitro indicate that individual CD4 T cells can be made to differentiate into Th1 or Th2 cells (8–10). There was considerable controversy as to whether the inducing cytokines led individual CD4 T cells to adopt a Th1 or Th2 phenotype or whether such adoption was a stochastic event and the added cytokines functioned by selectively promoting the outgrowth of differentiated cells. Several lines of evidence strongly support the notion that the cytokines play a major role in inducing the transcription factors that determine differentiation (11, 12), although there may be elements of selective outgrowth (12, 13).
Th17 Cells and iTregs
That two major cell types differentiate from naive CD4 T cells dominated the field for more than a decade and a half. Other types of CD4 T cells were recognized, such as NKT cells and natural regulatory T cells (nTregs), but these cells were not derived in the periphery from the naive cells that could also give rise to Th1 and Th2 cells. Rather, they were members of lineages that developed in the thymus and that were distinct from the cells undergoing parallel thymic differentiation to become the naive “conventional” CD4 T cells that were progenitors of Th1 and Th2 cells (14, 15).
In 2003, a third major effector population of CD4 T cell that could be derived from naive CD4 T cells was shown to exist (16–18). These cells, designated Th17 cells (19–21), were characterized by the production of IL-17A, IL-17F, and IL-22 as signature cytokines, molecules not produced by Th1 or Th2 cells. Th17 cells also were good producers of IL-21 (22–24), although IL-21 can be made by several Th cell types.
At the same time, it was shown that cells with the characteristics of regulatory T cells could be induced to differentiate in vitro from naive CD4 T cells (25–28). These cells were designated induced Tregs (iTregs) to distinguish them from nTregs. In mice, iTregs show in vitro and in vivo functions similar to those of nTregs (29), but iTregs in humans have thus far failed to demonstrate activity in an in vitro Treg functional assay (30).
Thus, four major T cell populations clearly emerge from naive CD4 T cells. It is virtually certain that the same precursor can be caused to differentiate either into a Th1 or a Th2 cell. Although not yet definitively shown for Th17 cells or iTregs, it is most likely the case for those cells as well.
Are Tfh Cells a Fifth Lineage?
The activation of CD4 T cells in vivo in response to appropriate stimulation results in the generation of effector populations that have the capacity to enter the tissues and to mediate their immune functions at the site of pathogen invasion. In parallel, a population develops of central memory cells that reside principally in the lymph nodes and spleen and, presumably, are available to reconstitute effector cells upon subsequent antigenic challenge. Both central memory and effector (sometimes effector/memory) cells appear to be distinctive states of each of the Th1, Th2, and Th17 populations and possibly also of Tregs.
A major function of CD4 T cells is to help B cells produce antibody in response to T-dependent antigens. CD4 T cells are also important in the induction and control of immunoglobulin class switching and somatic hypermutation. These events occur mainly within germinal centers, and the CD4 T cells that enter the germinal center to mediate their helper function for antibody production are often designated T follicular helper (Tfh) cells (31).
Whether Tfh cells are an independent lineage (essentially parallel to Th1, Th2, and Th17 cells) or a phenotypic state of each of the three effector lineages remains uncertain. Recently, several studies have appeared in which Tfh cells were analyzed in germinal centers of mice infected with different parasites that induce responses typically associated with Th2 cell development and with immunoglobulin class switching to IgE, an Ig isotype for which IL-4 is essential (32–34). In each instance, cells with the Tfh phenotype produced IL-4, and these cells could be shown to reside within germinal centers and to form contacts with responding B cells (32). This finding implies that individual Tfh cells mediate both help and class switching. In other experiments, individual Tfh cells directly interacting with B cells in the germinal center produced either IL-4 or IFN-γ, depending upon how they had been primed (31). This implies that the issue of Tfh lineage is not limited to the IL-4 pathway but is general.
The demonstration that Tfh cells produce IL-4 or IFN-γ, depending on how they are primed, does not distinguish the possibility that a cell of the Tfh lineage subsequently acquires the capacity to produce IL-4 or IFN-γ from the possibility that Th2 or Th1 cells acquire the capacity to act as Tfh cells. Pearce and colleagues (33) harvested antigen-responsive non-Tfh cells that were competent to produce IL-4 but that were not actually producing the cytokine from lymph nodes of mice infected with Schistosoma mansoni. When these cells were transferred into mice also infected with S. mansoni, some of the transferred cells acquired the Tfh phenotype, consistent with the concept that Th2 cells can become Tfh cells. However, one cannot rule out the possibility that among the cells harvested from the lymph nodes, there were subpopulations already determined to become conventional Th2 CD4 T cells and others committed to become Tfh cells. It has recently been reported that the bulk of memory cells that participate in affinity maturation reside within the bone marrow, raising the possibility that such T cells are specialized as Tfh memory cells (35).
It is not yet clear whether Tfh cells are a distinct lineage. Further studies are awaited to clarify this interesting and important question. Detailed analysis of the pattern of gene expression or of genome-wide chromatin accessibility might well allow one to determine whether IL-4- and IFN-γ-producing Tfh cells are more closely related to one another than are IL-4-producing Th2 and Tfh cells or vice versa. Such data would provide a reasonable basis upon which to reach a conclusion as to whether Tfh cells are an independent lineage.
More on Lineage Relationships
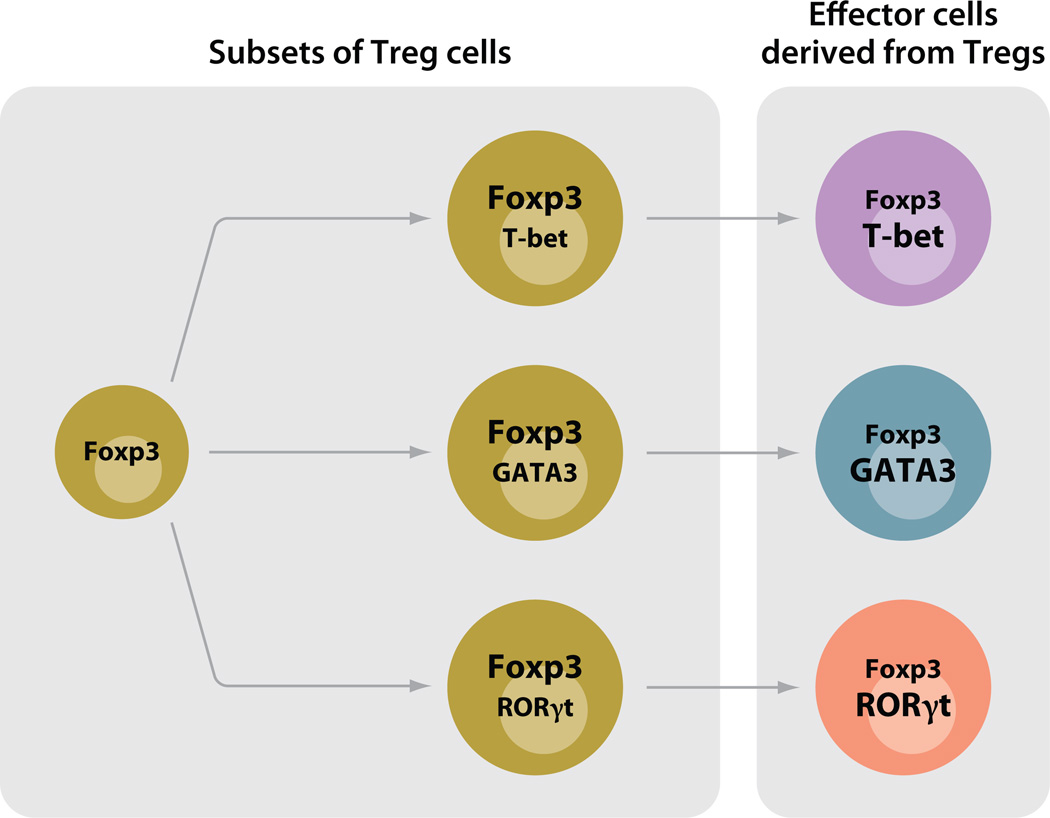
The discussion as to whether Tfhs should be regarded as a distinctive differentiated state of the major effector lineages (i.e., Th1, Th2, or Th17 cells that acquire Tfh character) or an independent lineage of cells, parallel to Th1, Th2, and Th17, that can subsequently acquire the capacity to produce the distinct regulatory cytokines that control switching to different immunoglobulin isotypes raises the possibility that other lineage relationships may be more complex than previously thought. Principal among these are Tregs, where recent work suggests that different Treg populations may be specialized to control mainly the function of particular subsets of CD4 T cells.
Campbell and colleagues (36) have reported that in response to IFN-γ, Foxp3+ Tregs upregulated T-bet and that T-bet-expressing Tregs accumulated at sites of Th1 cell–mediated inflammation. Furthermore, T-bet− Tregs proliferated less well than T-bet+ Tregs, and when T-bet was lacking from Tregs, they were relatively ineffective in controlling the expansion of T-bet+ conventional CD4 T cells, whereas they seemed as effective as conventional Tregs in controlling Th2 and Th17 cells.
Rudensky and colleagues (37) reported that conditionally deleting Irf4 in Tregs selectively allowed the uncontrolled expansion of Th2 cells, suggesting that IFN regulatory factor 4 (IRF4) expression, important in Th2 differentiation, also played a role in differentiation of those Tregs that could control Th2 cells. Similarly, STAT3 expression in Tregs seems to be essential for the ability of Tregs to suppress immune pathology mediated by Th17 cells, whose differentiation requires STAT3 (38). Thus, iTregs should not necessarily be considered as one of a set of distinct fates of CD4 T cells, equivalent to Th1, Th2, and Th17 cells, but possibly as a lineage parallel to the effector CD4 T cells (Th1, Th2, and Th17) as a whole and capable of differentiating into specialized cells that show distinctiveness in their regulatory targets. The finding that transcription factors associated with a particular Th fate differentiation are also important in the specialization of Tregs suggests that the priming conditions that lead conventional cells to adopt one of their possible fates may be the same conditions that call forth Tregs specialized to control these very Th cells.
THE CYTOKINE ENVIRONMENT PLAYS A CENTRAL ROLE IN FATE DETERMINATION AND EFFECTOR FUNCTION
The distinctive differentiated states of the various CD4 effector/regulatory subpopulations are determined largely by the set of transcription factors they express and the genes they transcribe. The induction of the distinctive patterns of gene expression may be achievable in several ways, but in vitro the major determinants of the differentiated state of the cell are the set of cytokines present during the T cell receptor (TCR)-mediated activation process. Our understanding of this process has evolved over an extended period and is described in detail below.
As discussed above, it was first demonstrated that naive CD4 T cells could differentiate into IL-4-producing CD4 T cells if the cytokines IL-4 and IL-2 were present at the time of stimulation by cognate antigen (3–6). That one of the key inducing cytokines is also a major product was a striking finding; this has proven not to be unique for Th2 differentiation. For Th1 differentiation, it was first shown that IL-12 (7) played a central role and only somewhat later was it appreciated that IFN-γ also played an important role in the induction of Th1 cells (39), of which IFN-γ is a signature cytokine. Indeed, in vitro neutralization of IFN-γ will often markedly diminish Th1 development.
Understanding of Th17 differentiation went through a complex evolution, beginning with the recognition of the existence of an IL-12 congener (IL-23) that shared one chain with IL-12 (p40) but expressed a unique chain (p19), distinct from IL-12 p35 (40). This led to the recognition that, in much research that had relied on deleting p40 to block Th1 differentiation, the development/maintenance of both Th1 and Th17 cells were blocked and that IL-23 played an important role in the development and/or maintenance of Th17 cells. However, it was soon appreciated that IL-23 did not act on naive CD4 T cells, but rather was more important later in the Th17 priming process or in the maintenance of the Th17 phenotype.
Further analysis revealed that in vitro Th17 differentiation was most efficient when TGF-β and IL-6 were available (21, 41, 42) but that IL-21 could mediate many of the functions of IL-6 (22–24). IL-6, IL-21, and IL-23 can be regarded, at least at one level, as congeners since each mediates its function through the activation of STAT3. The relative efficacy of the three cytokines may be determined, at least in part, by the number of specific receptors that exist at any one time. For example, IL-23 receptors appear not to be expressed until after the naive cell has partially completed its differentiation to becoming a Th17 cell, and consequently IL-23 plays little part in the initial determination of Th17 differentiation (21, 41, 43). In accord with the importance of products of the differentiated cells playing a role in differentiation, Th17 cells produce IL-21, and IL-21 can certainly propagate the Th17 differentiation process, even if it is less effective than IL-6 in initiating differentiation.
The induction of iTregs from naive CD4 T cells relies on T cell activation in the presence of TGF-β and IL-2. Since Tregs are good TGF-β producers, the principle that a major product of the differentiated cell plays a major role in induction is also applicable to iTregs.
Reliable means of developing Tfh cells in vitro are still being uncovered. It has been proposed that the inclusion of IL-6 or IL-21 together with a TCR-mediated stimulation will induce these cells (44, 45), but this idea is still controversial. Some of these difficulties may stem from an uncertainty as to the proper starting cells for such differentiation—be they naive CD4 T cells or already differentiated Th cells— and the possibility that environmental factors from the germinal center or provided by B cells may play an important role in such differentiation (33, 34).
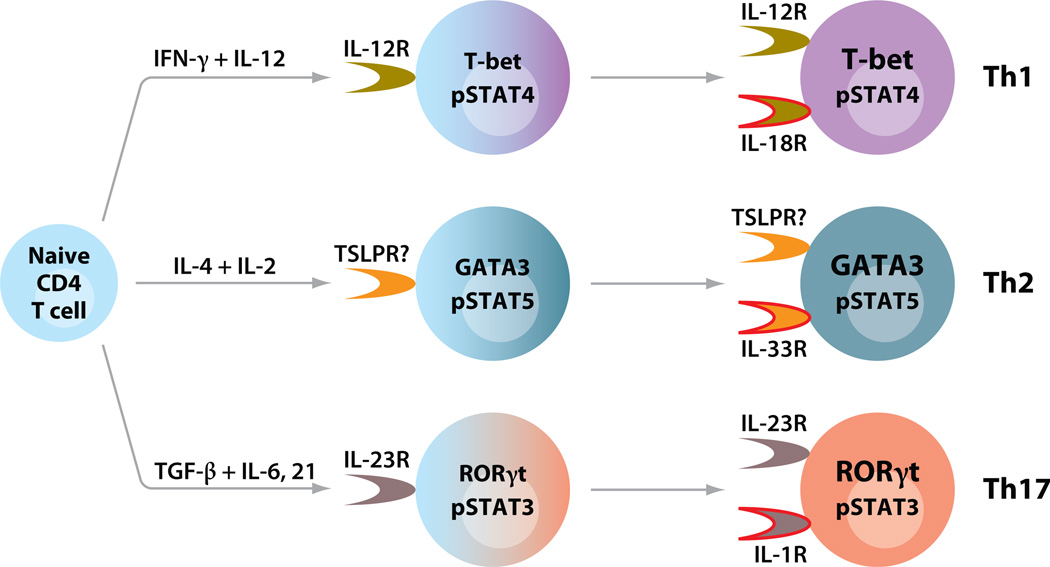
Cytokines may also play a role in effector cytokine production by differentiated Th1, Th2, and Th17 cells (Figure 1). At later stages of Th cell differentiation, a distinct member of the family of IL-1 receptors is selectively upregulated in each lineage. Together with a STAT5 inducer including IL-2, IL-7, or TSLP (thymic stromal lymphopoietin), the IL-1 analog IL-33 causes TCR-independent IL-13 production in a cyclosporine A–independent manner in Th2 cells (46), suggesting an innate-like effector function of Th cells. TCR-independent cytokine production can also be induced in Th1 and Th17 cells by IL-12/IL-18 and IL-23/IL-1, respectively (46–49).
Figure 1.
Cytokines play critical roles in differentiation and effector functions of Th1, Th2, and Th17 cells. Upon TCR activation triggered by antigen-presenting cells, naive CD4 T cells differentiate into distinct Th lineages in the context of combinations of cytokines. The differentiation processes involve upregulation of master transcriptional regulators and activation of STAT proteins (185). Each lineage expresses unique cytokine receptors, which can respond to cytokines produced by accessory cells. At later stages of Th cell differentiation, different Th cells preferentially express an IL-1 family receptor. Together with a STAT activator, an IL-1 family cytokine is capable of inducing effector cytokine production from Th cells in a TCR-independent manner (46–49).
TRANSCRIPTION FACTORS FOR T HELPER CELL FATE DETERMINATION
The master transcription factors and the signaling transducer and activator of transcription (STAT) proteins are indispensable for Th cell fate determination and cytokine production. The activities of the master transcription factors are mainly determined by their expression levels, whereas those of the STATs are regulated by cytokine-mediated posttranslational modification, including tyrosine and/or serine/threonine phosphorylation. Not only do the activated STAT proteins, in collaboration with master transcription factors, regulate the production of the key cytokines by Th cells, they also play important roles in the induction of the master transcription factors. Other transcription factors, either constitutively expressed or induced by TCR and/or cytokine-mediated signaling, are also involved in executing or fine-tuning Th cell functions. These molecules form a sophisticated transcription factor network (Figure 2), which is critical for Th cell fate determination, expansion, and function.
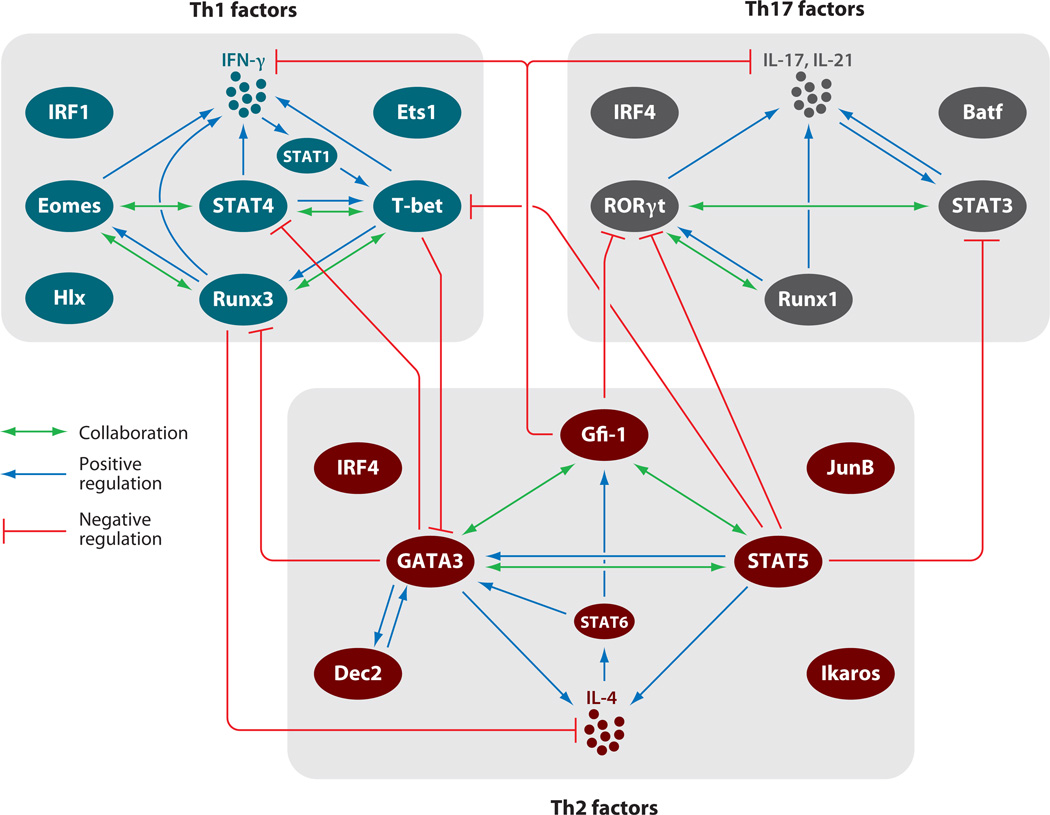
Figure 2.
Networks of transcription factors for Th cell differentiation. Critical transcription factors involved in Th1, Th2, and Th17 differentiation and the relationships among these factors are shown. The cell fate of each Th lineage is determined by many transcription factors, including master regulators and STAT family proteins. There are collaborations and positive regulations among the transcription factors during Th cell differentiation and lineage-specific cytokine production. Some factors negatively regulate expression or function of transcription factors of other Th lineages.
Master Transcription Factors for Each Lineage
GATA3
GATA3, the Th2 master regulator, was the first master regulator to be identified (50, 51). GATA3 is also critical for the development of CD4 T cells, and naive CD4 T cells express it at detectable levels (52). GATA3 expression is upregulated or downregulated during Th2 or Th1 differentiation, respectively (51, 53, 54). Expression of retrovirally encoded GATA3 in Th1 cells makes these cells competent to produce IL-4 and induces endogenous GATA3 production (53, 55). Introducing a dominant-negative (DN) form of GATA3 in T cells reduces Th2 cytokine expression and, in vivo, blocks induction of airway hypersensitivity (56). Th2 differentiation is totally abolished in vitro and in vivo in the absence of GATA3, as shown by the failure of such differentiation in mice in which Gata3 is deleted in peripheral CD4 T cells (57, 58). Deleting Gata3 from fully differentiated Th2 cells by the introduction of retrovirally encoded Cre has only a modest effect on IL-4 production but completely blocks the production of IL-5 and IL-13 (57), consistent with direct GATA3 binding to the IL-5 (59) and IL-13 (60, 61) promoters, but only to IL-4 enhancers (62).
GATA3 promotes Th2 differentiation through instructing Th2 commitment, selectively stimulating the growth of Th2 cells, and suppressing Th1 differentiation (63). GATA3 is also expressed at intermediate levels in NKT cells (64) and Treg cells (J. Zhu and W.E. Paul, unpublished observations). NKT cell development and survival is defective in Gata3 conditional knockout mice (64), but the function of GATA3 in Tregs is not clear.
T-bet/Eomes
T-bet is a major factor for inducing IFN-γ production and Th1 cell differentiation (65). Overexpression of T-bet either during Th2 differentiation or in fully differentiated Th2 cells causes such cells to acquire competence to produce IFN-γ while, at the same time, suppressing their capacity to produce IL-4. T-bet induces IFN-γ partly through remodeling the Ifng gene and by upregulating IL-12Rβ2 expression, thus promoting both IFN-γ expression and selective Th1 cell expansion in response to IL-12 (12, 66). Tbx21−/− (T-bet knockout) cells have severe defects in Th1 cell differentiation both in vitro and in vivo (67). IFN-γ responses to Leishmania major are significantly diminished, although not abolished, in Tbx21−/− mice; these mice show increased IL-4 and IL-5 production (67) in response to L. major infection. Notably, Tbx21+/− cells display a partial phenotype.
T-bet-expressing CD4 T cells are dramatically reduced in human asthmatic airways, and Tbx21−/− mice spontaneously develop airway hypersensitivity (68). Despite the very important role of T-bet in Th1 differentiation and in the acquisition of competence to produce IFN-γ, it has been reported that Tbx21−/− cells can produce normal amounts of IFN-γ in vitro when naive cells are differentiated under Th1 conditions (i.e., when IL-4 was neutralized), suggesting that a main function of T-bet is to inhibit GATA3 expression and that IFN-γ may be redundantly controlled (54). However, others have reported that, although capable of inducing IL-12Rβ2 expression, T-bet fails to suppress GATA3 and Th2 cytokine production (69). The controversy among these studies regarding T-bet function has not been resolved. Possibly, differences in the timing and/or culture conditions of the experiments account for the different observations. T-bet deficiency also causes defective IFN-γ production by NK cells. However, IFN-γ production by CD8 T cells from Tbx21−/− mice is relatively normal (67), although it has been reported recently that T-bet contributes to optimal IFN-γ production at early stages of the CD8 response (70).
The differential requirement for T-bet for IFN-γ production by CD4 and CD8 T cells may be explained by the heightened expression of another T-box family member, Eomesodermin (Eomes), in CD8 T cells (71). Indeed, it has recently been reported that CD8 T cells from T-bet/Eomes double knockouts but not single knockouts produce very little IFN-γ and fail to control lymphocytic choriomeningitis virus (LCMV) infection. Rather, such CD8 T cells aberrantly produce IL-17, which results in a wasting disease after LCMV infection (72). IL-21 inhibition of Th1 cell IFN-γ production may be mediated by suppression of Eomes, not T-bet (73), suggesting that Eomes is also upregulated during Th1 differentiation and involved in optimal IFN-γ production by CD4 T cells. Indeed, CD4 T cells from T-bet/Eomes double knockout mice infected with LCMV fail to produce any IFN-γ in response to challenge with the GP61–81 LCMV envelope peptide (72).
T-bet and Eomes are also involved in regulating IL-2Rβ, whose expression is critical for IL-15-mediated CD8 T cell memory (74). Furthermore, T-bet suppresses IL-7Rα expression and thus affects the generation of central memory CD8 T cells (75, 76). A subset of Treg cells also express T-bet, and these Tregs are important for controlling Th1 responses (36). T-bet is also important for the development and/or function of other immune cells, including B cells, NK cells, NKT cells, and dendritic cells (DCs) (77). Therefore, one should consider the multiple functions of T-bet in the immune system while interpreting results obtained from the analysis of T-bet germline knockout mice; conditional deletion of Tbx21 may help in dissecting the functions of T-bet in different cell types.
Foxp3
Scurfy mice and patients with IPEX (immunodeficiency, polyendocrinopathy, and enteropathy, X-linked syndrome) lack detectable nTregs. Both the mutant mice and the patients have mutations in Foxp3 (78–80), which is reported to be the master transcriptional regulator for nTregs (81, 82). Continuous expression of Foxp3 in Tregs is required to maintain the suppressive activity of such cells (83). In addition, conventional T cells transduced with retroviral Foxp3 acquired a Treg phenotype, including the inability to produce cytokines (anergy) and suppressive activity (81). By contrast, limiting Foxp3 expression appears to divert cells that would have differentiated into Tregs to develop into Th2-like cells, implying a close relationship of the Th2 and Treg lineages (84).
Culturing Foxp3− naive CD4 T cells with a TCR stimulus and TGF-β converts these cells into Foxp3+ CD4 T cells, which have been designated iTregs (25). TGF-β is also critical for the development, homeostasis, and function of nTregs (85–88). Smad3 and NFAT, activated by TGF-β and TCR-mediated signaling, respectively, cooperate in Foxp3 gene remodeling and expression (89).
RORγt/RORα
Th17 cells do not express GATA3 or T-bet (19, 20); instead, they express high levels of RORγt (90). RORγt is induced in naive CD4 T cells within 8 h of TCR stimulation in association with TGF-β and IL-6. RORγt is the master regulator of Th17 cells; ~50% of activated cells overexpressing RORγt produce IL-17, and RORγt-deficient cells produce very little IL-17. Furthermore, RORγt-deficient mice are partially resistant to experimental autoimmune encephalomyelitis. The residual IL-17 production in RORγt-deficient cells appears to be dependent on the activity of a related nuclear receptor, RORα, which is also upregulated in Th17 cells (91). Although deleting RORα resulted in minimal reduction of IL-17 expression, deficiency of both RORγt and RORα completely abolished IL-17 production. RORγt is also expressed in double-positive (DP) thymocytes (92) and other cell types including lymphoid tissue inducer (LTi) cells, where IL-17 is also produced (93).
Bcl-6
Bcl-6 is a transcriptional repressor. Bcl-6 germline knockout mice develop Th2 diseases even in the absence of STAT6 (94, 95), possibly due to the derepression of GATA3 in the absence of Bcl-6 (96). Bcl-6 is frequently translocated and hypermutated in diffuse large B cell lymphoma and is critical for germinal center B cell differentiation and thus germinal center formation (94). In addition, Bcl-6 expression is greater in CD25+ germinal center B cells than in CD25− B cells in the germinal center (97). Unlike plasma cells, the CD25+ B cells, representing memory B cells, express lower levels of Blimp-1. STAT5 activation, critical for the self-renewal of these cells, directly induces Bcl-6 expression. Bcl-6 is also expressed in Tfh cells (98) as is CXCR5, a critical chemokine receptor that allows cells to home to B cell follicles (99). Indeed, three recent reports showed that Bcl-6 is critical for Tfh cell differentiation (100–102). Bcl-6 is necessary and sufficient to induce Tfh-related molecules, including CXCR5, PD-1, IL-6R, and IL-21R, but has no effect on IL-21 production. Bcl-6 also suppresses the expression of Th1, Th2, and Th17 cytokines. In addition, enforced Bcl-6 expression induces endogenous Bcl-6 transcription (100), and Blimp-1 represses Bcl-6 (101).
Signaling Transducer and Activator of Transcription (STAT) Proteins
As mentioned above, the cytokine milieu present during TCR-mediated activation of naive CD4 T cells is the most important determinant of CD4 T cell fate. The major signaling pathway triggered by cytokines is the activation of the STAT family of proteins. STATs play critical roles in the differentiation and expansion of Th cells. They are important both for the induction of the master regulators and for cytokine production in collaboration with master regulators.
STAT1
Activation of STAT1 by IFN-γ is important for the induction of T-bet during in vitro Th1 differentiation (39, 69). IFN-γ, through STAT1 activation, also induces T-bet expression in monocytes, macrophages, DCs, and B cells. The existence of a positive feedback loop in which IFN-γ, acting through T-bet, induces more IFN-γ indicates that STAT1 serves as a critical mediator for the amplification of in vitro Th1 responses. However, in the acute phase of Toxoplasma gondii infection in mice, the appearance of CD4 T cells capable of producing IFN-γ does not require STAT1 (103). Serum IFN-γ levels in Stat1−/− mice 7 days after T. gondii infection are comparable to those in wild-type mice. Furthermore, Stat1−/− CD4 T cells from T. gondii–infected mice produce amounts of IFN-γ in response to soluble T. gondii antigen comparable to cells from similarly infected wild-type mice, although these knockout CD4 T cells expressed lower levels of T-bet than did wild-type CD4 T cells. Stat1−/− mice have an increased parasite burden and died 7–12 days after infection, most likely owing to the failure of macrophage activation. Thus, to determine whether the IFN-γ/STAT1 autocrine pathway plays an important role in in vivo CD4 T cell differentiation will require a T cell–specific conditional Stat1 knockout mouse. The need for STAT1 in order for CD4 T cells from T. gondii–infected mice to obtain optimal expression of T-bet suggests that the IFN-γ/STAT1 pathway has a role during in vivo Th1 responses and may be particularly important for IFN-γ production in responses that are less robust than those elicited by T. gondii.
STAT2
STAT2 forms a heterodimer with STAT1 in response to type I IFNs. Stat2-deficient mice have increased susceptibility to viral infection owing to defective type I interferon responses (104). Although type I IFNs have been reported to influence CD4 T cell differentiation, such function is largely attributed to the activation of a STAT1 homodimer, and possibly also of STAT4, by type I IFNs.
STAT3
STAT3 is activated by IL-6, IL-21, and IL-23, cytokines that are involved in Th17 cell differentiation, amplification, and maintenance (21–24, 41, 42). Deletion of Stat3 in mice and DN STAT3 mutations in humans result in the loss of IL-17-producing CD4 T cells (22, 23, 105–109). STAT3 binds to Il17 (110) and Il21 (111) and is responsible for the induction of RORγt and the IL-23R (22, 23, 108). In parallel, STAT3 activation by IL-6 is responsible for Foxp3 downregulation in both differentiating and differentiated Tregs (24, 108, 112, 113), accounting for the critical role of IL-6 in determining the balance between Th17 and iTreg induction. In the absence of STAT3, Foxp3 is up-regulated when cells are cultured under Th17 conditions (108). Curiously, IL-10 and IL-27, which are negatively involved in Th17 differentiation, also activate STAT3. IL-6, -21, or -23, through the activation of STAT3, together with IL-1, an NF-κB activator, induce TCR-independent, cyclosporine A–independent IL-17A production (46).
STAT4
STAT4, activated mainly by IL-12, is important for Th1 responses in vitro (114, 115) and in vivo in response to T. gondii infection (116). STAT4 expression is higher in Th1 than in Th2 cells (117). STAT4 expression is likely to be regulated positively by IFN-γ (118) and negatively by IL-4 and GATA3 (117, 118). Activated STAT4 can directly induce IFN-γ production and expression of IL-12Rβ2 and T-bet during Th1 differentiation (54, 117). IL-12, by activating STAT4, together with IL-18, an NF-κB activator, induces TCR-independent IFN-γ production (48, 49). STAT4 also plays an important role in IL-12-mediated activation of NK cells (114, 115).
STAT5
STAT5a and STAT5b, the two isoforms of STAT5, are critical for the signaling of many cytokines that utilize the common γ chain as a subunit of their receptors (119). Deletion of both STAT5a and STAT5b affects many aspects of cellular responses, including cell proliferation (120). Low levels of STAT5 activation are sufficient for cell proliferation and survival; however, strong STAT5 signaling is required for Th2 differentiation (121, 122). Thus, even in the presence of STAT5b activation, STAT5a single knockout cells displayed profound defects in Th2 cell differentiation both in vitro and in vivo (121–124). STAT5 directly binds to the DNase I hypersensitive sites (HS) II and HSIII in the second intron of the Il4 locus in Th2 but not in Th1 cells (122). Such binding may be critical for IL-2-mediated induction and maintenance of accessibility at the HSII site of the Il4 locus. In addition, STAT5a-deficient cells are hyperresponsive to IL-12, which leads to better Th1 differentiation (125).
STAT5 activation by IL-2 is also critical for Treg development (126–128). STAT5 may contribute to Foxp3 induction by binding to its promoter (127, 129). STAT5 activation also regulates the activity of the Bcl6 promoter in B cells (97); in view of the expression of Bcl-6 in Tfh cells, this regulation raises the possibility that such an effect may be important in Tfh cell differentiation. On the other hand, STAT5 suppresses Th17 cell differentiation (107) but is required for the expansion of differentiated Th17 cells (130). It has been reported recently that STAT5 serves as a pioneer factor in regulating the accessibility of the Ifng locus and thus is also involved in Th1 differentiation (131). Therefore, a low level of STAT5 activation is required for cell proliferation and survival, possibly also for Th1 differentiation and Th17 cell expansion. However, enhanced STAT5 activation suppresses Th1 and Th17 differentiation while Th2 and Treg differentiation is promoted, which correlates with higher expression levels of CD25 in Th2 and Treg cells. The quantitative regulation of STAT5 signaling that results in qualitative differences in Th cell differentiation may be explained by the differential affinity of STAT5 binding to the different gene targets in distinct cell types.
STAT6
STAT6 is the major signal transducer in IL-4-mediated Th2 differentiation and expansion (132–134). In vitro, STAT6 activation is necessary and sufficient for inducing high expression levels of the Th2 master regulator gene, GATA3 (135, 136). STAT6 does not regulate IL-4 transcription directly, but it may regulate the activity of the Il4/Il13 locus control region (137). Although STAT6 appears indispensable for Th2 differentiation in vitro, one can induce STAT6-independent Th2 cell differentiation in vivo. Despite their STAT6 independence, these responses are still GATA3-dependent (138–142). However, some in vivo Th2 responses such as those elicited by Trichuris muris (143), as well as the accumulation of Th2 cells in lung tissue in response to Nippostrongylus brasiliensis infection, depend on the IL-4/STAT6 pathway (141). STAT6 may also be important for the amplification of Th2 responses at later stages and/or for the generation of Th2 memory cells in vivo (139).
Other Factors Involved in Fine-Tuning Th Differentiation
Runx family members
Runx3, a transcriptional repressor important for silencing CD4 during CD8 T cell development, is highly expressed in both CD8 and Th1 CD4 T cells (144, 145). In CD8 T cells, Runx3 appears to be responsible for the induction of Eomes, granzyme B, and perforin and for optimal expression of IFN-γ at later stages of responses (70), although such Runx3 functions need to be further verified in Runx3 conditional knockout mice, given that Runx3 germline knockout mice show aberrant CD8 development. Our unpublished data also indicate that enforced expression of Runx3 in Th2 cells induces the capacity to produce IFN-γ independent of T-bet and that such IFN-γ induction is partly due to upregulation of Eomes. Runx3-deficient cells produce less IFN-γ than wild-type Th1 cells (70, 145). Runx3 also represses IL-4 transcription through its binding to the DNase I HSIV region of the Il4 gene (144).
The Runx family includes Runx1, Runx2, and Runx3; only Runx1 and Runx3 are expressed in T cells. Runx1 is required for CD4 T cell development (146) and may be responsible for IL-2 production by naive CD4 T cells (147). In Tregs, Runx1 interacts with Foxp3, an interaction that is required for the function of Tregs (147) and through which Foxp3 suppresses IL-2 production. Recently, Runx1 has been shown to be critical for maintaining Foxp3 expression and suppressive function of Tregs (148, 149). Naive CD4 T cells from mice conditionally lacking CBFβ, the cofactor for Runx protein, express reduced Foxp3 when stimulated by TGF-β, possibly because the induction of Foxp3 expression requires Runx binding to the Foxp3 promoter (150, 151). Runx1 also interacts with RORγt and induces optimal RORγt expression and IL-17 production in Th17 cells (152).
Since Runx1 can interact with Foxp3 and RORγt and Runx3 can interact with T-bet and GATA3 (J. Zhu, R. Yagi, W.E. Paul, unpublished data), Runx family members should be considered as important fine-tuners of the master regulator genes. That Runx3 negatively regulates Runx1 expression adds complexity to this transcriptional regulatory network (70).
IFN regulatory factor family members
IRF4 expression is important for Th2 cell differentiation (153, 154). IRF4-deficient Th2 cells produce diminished amounts of IL-4, but this defect can be rescued by overexpression of GATA3, suggesting that IRF4 is involved in upregulating GATA3 (153). In addition, IRF4 may be important in regulating IL-4 expression by collaborating with NFATc2 and c-Maf (154). IRF4 is also indispensable for Th17 differentiation (155). Irf4−/− T cells fail to produce IL-17, and Irf4−/− mice are resistant to experimental autoimmune encephalomyelitis induction. IRF4 appears to play a role in regulating RORγt expression but not Foxp3 expression. Mice with a conditional Irf4 deletion in Tregs may develop Th2-like diseases, although the number of Foxp3+ cells in these mice appears to be normal (37). IRF4 seems to play an important role in regulating some Foxp3 functions through protein-protein interactions, but the detailed mechanisms for such regulation have not been determined.
IRF1 is an IFN-γ-inducible transcription factor. It was recently reported to regulate IL-12Rα expression in Th1 and Th17 cells. Deletion of Irf1 resulted in decreased levels of IL-12Rα expression, leading to loss of the responsiveness of such cells to IL-12 (156). Although IL-12Rα is also a component of the receptor complex for IL-23, such decreased IL-12Rα expression did not affect the cells’ responsiveness to IL-23. Thus, fine-tuning of IL-12Rα expression by IRF1 may regulate Th1 and Th17 differentiation and expansion in response to IL-12 and IL-23, respectively.
Gfi-1
Growth factor independent 1 (Gfi-1) is a transcriptional repressor. Its locus is the most frequent insertion site in Moloney murine leukemia virus (MoMLV)-induced lymphomas (157). Gfi-1 is involved in many aspects of immune cell functions, including homeostasis of hematopoietic stem cells and development of neutrophils, T cells, and mature DCs, as revealed by the phenotypes of Gfi1 germline knockout mice (158). Loss of Gfi-1 also results in myeloid leukemias. TCR activation transiently induces Gfi-1, and IL-4 prolongs its expression (13). Gfi-1 selects GATA3hi cells for growth by modulating both upstream and downstream IL-2 signaling events, suggesting that it mediates a selective function during Th2 cell differentiation (13, 159).
Gfi-1 also suppresses non-Th2 lineages. IFN-γ and IL-17 production are increased in Gfi1 conditional knockout T cells (159, 160). In addition, loss of Gfi-1 results in increased numbers of CD103+ Tregs (160). Overexpression of Gfi-1 suppresses TGF-β-mediated functions, and TGF-β downregulates Gfi-1 expression, suggesting a reciprocal regulation between TGF-β signaling and Gfi-1 expression.
Ikaros family members
Ikaros is critical for the development of T and B lymphocytes and NK cells (161). The Ikaros family consists of five members, Ikaros, Helios, Aiolos, Eos, and Pegasus. Their functions require the formation of homoor heterodimers. Recently, Ikaros was reported to be important for Th2 cell differentiation (162). In the absence of Ikaros, IFN-γ and T-bet are dramatically upregulated even under Th2 polarizing conditions, suggesting that Ikaros plays an important role in suppressing Th1 differentiation. Eos has been reported to play an important role in regulating the repressive activity of Foxp3 in Tregs through interacting with Foxp3 (163). Another family member, Helios, is among the very few genes that are highly expressed in nTregs but not in iTregs, but its function in nTregs is unknown (164).
c-Maf
c-Maf, selectively upregulated in Th2 cells, enhances production of IL-4 but not of other Th2 cytokines (165). c-Maf was also reported to induce CD25 (IL-2Rα) expression (166). Recently, c-Maf has been found to be highly expressed in Tfh cells that are capable of producing IL-17, and IL-17-producing Tfh cells are less frequent in Maf knockout mice (167). However, the function of c-Maf in Tfh cells in general has not been established.
Other transcription factors
Many other transcription factors are involved in the differentiation of at least one Th lineage. Selected examples follow: Hlx, a transcription factor induced by T-bet, enhances T-bet-mediated IFN-γ production (66). Ets-1, a cofactor for T-bet for Th1 differentiation, plays a negative role in Th17 cell differentiation (168, 169). JunB, whose expression is selectively upregulated in Th2 cells, collaborates with c-Maf in inducing IL-4 production through binding to the Il4 promoter (170). Expression of Blimp-1, an important transcription factor for long-lived plasma cells, is induced in Th2 cells where it suppresses IFN-γ and IL-2 production (171–173). Blimp-1 has recently been reported to oppose the expression of Bcl-6 and thus to inhibit Tfh differentiation (101). TIEG1, a TGF-β inducible transcription factor, together with Itch, an E3 ubiquitin ligase, play important roles in Foxp3 induction (174). The aryl hydrocarbon receptor (AhR), induced in Th17 cells independent of RORγt, plays a critical role in the production of Th17 cytokines, particularly IL-22 (175). Cell culture medium IMDM (Iscove’s Modified Dulbecco’s Medium), which contains higher levels of AhR ligands than RPMI medium 1640, supports better Th17 differentiation (176). BATF (basic leucine zipper transcription factor, ATF-like), an AP-1 family transcription factor, plays a critical role in Th17 but not in Th1 and Th2 cell differentiation (177). BATF is required for RORγt induction, but overexpression of RORγtin Batf−/− T cells fails to completely restore IL-17 production. Indeed, BATF binds to the Il17 gene directly. Dec2 is able to induce GATA3 expression, and Dec2 deficiency leads to impaired Th2 responses both in vitro and in vivo (178). GATA3 also regulates Dec2 expression, suggesting that Dec2 and GATA3 form a positive regulatory feedback loop during Th2 differentiation. In addition, Dec2 upregulates IL-2Rα expression, which may be partially responsible for Dec2-mediated enhancement of Th2 responses (179).
COLLABORATION BETWEEN TRANSCRIPTION FACTORS
GATA3 and STAT5
Both IL-4 and IL-2 are required for Th2 differentiation in vitro (3, 121). IL-4 can be either provided exogenously or produced by the cultured T cells. In either case, IL-4-mediated STAT6 activation enhances GATA3 expression (135, 136). GATA3 binds to regions of the Il4/Il13 locus, including DNase I HSVA (62), whereas STAT5 binds to HSII of the Il4 gene (121, 122). Our unpublished data show that GATA3 also binds to HSII. However, GATA3 alone is not sufficient to induce IL-4 production in the absence of STAT5 activation (121), and a constitutively active form of STAT5a loses its ability to induce IL-4 when basal GATA3 expression is eliminated by gene deletion (57). Thus, both GATA3 and STAT5 are required for IL-4 production, and a higher degree of STAT5 activation can lower the GATA3 level required to induce IL-4.
In differentiated Th2 cells, STAT5 activation is critical to maintain the expression of GATA3 (46). We also found that in Th2 cells GATA3 binds to the Cd25 locus and maintains CD25 expression (J. Zhu and W.E. Paul, unpublished observation). In addition, a recent report showed that STAT5 regulates IL-4Rα expression, especially during the initiation of Th2 differentiation (180). These data indicate a positive crosstalk between the IL-2/STAT5 and the IL-4/STAT6/GATA3 pathways. Thus, the collaboration of STAT5 and GATA3 at different regulatory levels accounts for full Th2 differentiation.
Besides the collaborative effect between GATA3 and STAT5, other factors, including NFAT, c-Maf, IRF4, and JunB, are also involved in IL-4 production through formation of a transcriptional complex in the promoter region of Il4 gene, as mentioned above.
T-bet and STAT4
Similar to the collaborative effects between GATA3 and STAT5, T-bet and STAT4 also synergize in the induction of many Th1-specific genes, including IFN-γ, IL-18R1, IL-12Rβ2, and Hlx, although the expression of some Th1 molecules, like CXCR3, is T-bet-dependent but STAT4-independent (181). T-bet expression is partially reduced in Stat4−/− Th1 cells, which cannot be rescued by the addition of exogenous IFN-γ. However, enforced expression of T-bet in Tbx21/Stat4 double knockout failed to restore the defects in the induction of Th1 genes. Both STAT4 and T-bet bind to the IFN-γ promoter; optimal binding of one factor requires the presence of the other. Consistent with this observation, both STAT4 and T-bet are required for the chromatin remodeling at the IFN-γ locus. As mentioned above, Runx3, Hlx, and Ets-1 are also cofactors of T-bet for IFN-γ induction.
RORγt and STAT3
Both RORγt and STAT3 are critical for Th17 cell differentiation (22, 90). Both directly bind to the Il17a/Il17f locus (91, 110, 152). The collaborative effect of these two molecules is indicated by the findings that deletion of either results in almost complete loss of IL-17 production and that enforced expression of RORγt and STAT3C, an active form of STAT3, synergistically induces IL-17 production. STAT3 activation or TGF-β signaling alone induces RORγt expression; however, induction is further enhanced when both are present (22). Even in the absence of TGF-β signaling, STAT3 activation fully induces the expression of two Th17-related molecules, IL-21 and IL-23R (22, 23). IL-21 expression induced by the STAT3 pathway is independent of RORγt, but optimal IL-23R expression requires RORγt. Although induction of RORγt in response to STAT3 activation alone is much lower in the absence than in the presence of TGF-β signaling (22), it is sufficient for IL-23R expression. Importantly, TGF-β-mediated induction of Foxp3, a negative regulator of RORγt function, is suppressed by the IL-6/STAT3 pathway (24, 182), providing another mechanism for the collaboration between STAT3 and RORγt.
CROSS-REGULATION AMONG TRANSCRIPTION FACTORS DURING Th DIFFERENTIATION
Transcriptional Repression of Transcription Factors and Cytokines
During Th cell differentiation toward one lineage, the other lineage fates are usually suppressed. There are several mechanisms for such inhibition. An important cross-regulation during Th differentiation is through repression of transcription factors that are important for lineage determination. For example, GATA3 downregulates expression of STAT4, which is the important factor for mediating IL-12 signaling and Th1 differentiation (117). A constitutively active form of STAT5 inhibits T-bet expression while it also promotes Th2 differentiation (122). On the other hand, GATA3 expression is suppressed by T-bet during Th1 differentiation (54).
The transcription factors expressed in one lineage also suppress the production of cytokines of other lineages. In Tregs, Foxp3 inhibits IL-2 production, possibly by binding NFAT (183) and Runx1 (147). In Th1 cells, Runx3 inhibits IL-4 production through binding the Il4 locus at the HSIV region (144). Gfi-1, which is a regulator for Th2 cell growth, suppresses both IFN-γ (58) and IL-17 production (160). A Th17-specific factor(s) that suppresses Th1 or Th2 cytokines has not been identified.
Cross-Regulation between Transcription Factors through Protein-Protein Interaction
Another interesting mechanism of cross-regulation is suppressive protein-protein interaction between master regulators. Itk-mediated phosphorylation of T-bet at position Y525 induces its interaction with GATA3 and the repression of GATA3 function (184). The Y525F T-bet mutant fails to suppress GATA3-mediated IL-4, IL-5, and IL-13 production while maintaining its ability to suppress IL-2 and to induce IFN-γ. GATA3 may also suppress T-bet function through such interaction.
Our unpublished data indicate that Runx3 induces IFN-γ in the absence of T-bet by up-regulating Eomes expression. GATA3 blocks this Runx3-Eomes-IFN-γ pathway presumably through interaction with Runx3. Therefore, when Gata3 is deleted from Th2 cells, the Runx3-Eomes pathway becomes active and IFN-γ is produced even when IL-12 and IFN-γ are neutralized.
TGF-β is required to differentiate both Th17 and Treg cells. At an intermediate stage of Th17 or Treg differentiation, both RORγt and Foxp3 are induced. Foxp3 interacts with RORγt and, by blocking its function, inhibits IL-17 production (182). A low concentration of TGF-β in combination with a STAT3 activator (IL-6, IL-21, or IL-23) is sufficient to induce RORγt expression; however, Foxp3 induction requires high concentrations of TGF-β. Thus, the relative expression of RORγt and Foxp3, controlled by the amount of TGF-β and of the STAT3-activating proinflammatory cytokines, determines whether the Th17 or Treg fate is adopted.
The mutual exclusivity among master transcription factors, at the transcriptional level, appears to be the major mechanism for cross-regulation during Th cell differentiation. However, suppressive protein-protein interactions play an important role in background “cleanup” during the polarization process by neutralizing the function of any aberrantly expressed transcription factors, most of whose expression may be driven to some extent by TCR activation. Such protein-protein interactions may also be important in maintaining the flexibility of the cells when a final fate decision has not yet been made, such as at early stages of Th differentiation.
TRANSCRIPTION FACTORS EXPRESSED AT LOW LEVELS CAN BE FUNCTIONALLY IMPORTANT
High levels of master regulator gene expression are usually correlated with the phenotype of the appropriate Th lineage. However, some effector functions may not require these factors to be highly expressed. Indeed, the functionality of the factor may depend on the cell context or, more precisely, on the relative amounts of other critical transcription factors. Forced expression of GATA3 in Th2 cells does not generally further enhance Th2 cytokine production; it appears that the endogenous level of GATA3 is already capable of inducing a maximum response. On the other hand, a small amount of GATA3, equivalent to or lower than that expressed in Th1 cells, can be sufficient for inducing IL-4 production given strong STAT5 activation (57). Similarly, although T-bet expression is lower in Stat1−/− CD4 T cells than that in wild-type CD4 T cells in response to T. gondii infection, such cells produced normal levels of IFN-γ (103). Furthermore, low expression of T-bet in a subset of Tregs is sufficient to induce these cells to produce CXCR3 but not IFN-γ (36). RORγt is modestly induced by IL-6 or IL-21 in the absence of TGF-β signaling, but this level of RORγt expression, although not sufficient to induce IL-17 production, can induce IL-23R expression (22). Therefore, despite the absolute requirement for master regulators, their expression level may not be correlated with the degree of a response. Caution is needed when interpreting data obtained from knockout cells, especially when a factor is already expressed at measurable levels before its upregulation occurs. Loss of function proves the importance of a regulator but does not necessarily establish the importance of the induction. Under such circumstances, a knockdown experiment may be more appropriate.
Taking this argument further, some in vivo–generated Th2 cells express lower levels of GATA3 than do in vitro–differentiated Th2 cells (J. Zhu and W.E. Paul, unpublished data), although GATA3 is absolutely required for in vivo Th2 cell differentiation (57). In addition, IL-4 strongly induces GATA3 in vitro and is needed for in vitro Th2 responses but is not required for in vivo Th2 cell differentiation under several conditions. Although it is possible that IL-4-independent upregulation of GATA3 expression is important in inducing in vivo Th2 differentiation, it is equally possible that another pathway such as STAT5 activation, but not the upregulation of GATA3, is the main driving force in inducing Th2 cell differentiation in some in vivo models. That is, the level of GATA3 found in any activated CD4 T cells may be sufficient for Th2 differentiation provided that high levels of STAT5 activation can be achieved and thus no preferential induction of GATA3 is required.
POSITIVE FEEDBACK DURING Th CELL DIFFERENTIATION
Each Th lineage can produce a cytokine with the potential to play a positive feedback role in promoting differentiation, i.e., IFN-γ for Th1, IL-4 for Th2, IL-21 for Th17, and TGF-β for Tregs (185). Furthermore, the master regulators and STAT proteins not only directly regulate lineage-specific cytokine production and induce important positive feedback at differentiation levels (Figure 3), but also regulate genes that are associated either with selective growth of this lineage or with repression of alternative lineage fates. In Th1 cells, T-bet and STAT4 are involved in IFN-γ production (positive feedback), IL-12Rβ2 upregulation (selective growth), and GATA3 downregulation (alternative fate repression). In Th2 cells, GATA3 and STAT5 induce IL-4 production (positive feedback) and CD25 (IL-2Rα) upregulation (both positive feedback and selective growth), and suppress IFN-γ and IL-17 expression (alternative fate repression). Transcription factors that are highly expressed in Th2 cells, including c-Maf and Gfi-1, are also involved in modulating IL-2/STAT5 signaling, which is critical for Th2 cell expansion. In Th17 cells, STAT3 is responsible for the upregulation of IL-21 and IL-23R, so that IL-21 and IL-23 can promote the expansion and terminal differentiation of Th17 cells at later stages (positive feedback and selective growth). Therefore, fully polarized Th cells are generated through lineage commitment, selective growth of committed cells, and active suppression of alternative lineage fates.
Figure 3.
Positive regulatory circuits for Th cell differentiation. Both TCR- and cytokine-mediated signaling, through activation of NFAT/NFκB/AP-1 and STAT proteins, respectively, are critical for early cytokine production and upregulation of a master transcription factor. The master transcription factor induces secondary transcription factors, which collaborate with the master transcription factor to enhance the expression of cytokine and cytokine receptors. In some cases, the master transcription factor also promotes its own expression. Elevated cytokine production and cytokine receptor expression provide powerful positive feedback loops for promoting Th cell fate determination as well as for selective expansion of committed Th cells.
The negative feedback of Th cell differentiation is less well studied. Each lineage can produce IL-10 under certain circumstances, which may serve as an example of negative feedback (186).
SIGNALING PATHWAYS CONTROL THE REGULATION OF TRANSCRIPTION FACTORS AND CYTOKINES
As indicated above, naive CD4 T cells stimulated by their cognate antigen can, when conditions are correct, rapidly express master transcriptional regulators and cytokines important for the differentiation of particular Th phenotypes. This process has been studied intensively for Th2 cells, but the paradigm applies to all three of the effector Th lineages and probably for iTregs and even for Tfh cells.
For Th2 cells, TCR engagement, under the appropriate conditions (discussed in the next section), results in GATA3 expression and IL-2 production/STAT5 activation. Jointly, these result in early IL-4 production, which is IL-4-independent but TCR dependent. This early IL-4 (produced in the induction phase of Th2 polarization) then acts through the IL-4 receptor and STAT6 to further enhance GATA3 expression and, together with continued STAT5 phosphorylation in response to endogenous IL-2 or possibly other STAT5 activators such as IL-7 or TSLP, leads to marked enhancement of IL-4 production and completion of the Th2 polarization process. This latter phase can be thought of as the polarization phase.
Analysis of how various signaling pathways impinge on Th differentiation is best understood in the context of this two-phase process and in terms of the need for early induction of a master transcription factor and early activation of a critical STAT. As noted above, the pairs are as follows: Th2, GATA3/STAT5; Th1, T-bet/STAT4; Th17, RORγt/STAT3; iTregs, Foxp3/STAT5. Recent studies suggest that Tfh cells may also fit the paradigm, with the factors being Bcl6/STAT3. In many instances, the STAT involved also plays a role in the induction of the master transcriptional regulator.
Pathways Downstream of TCR Signaling
The strength of TCR signaling during in vitro differentiation regulates Th1/Th2 polarization. In general, weak signaling favors Th2 differentiation and stronger signaling leads to Th1 differentiation (187). Priming of TCR-transgenic T cells, specific for moth cytochrome c (MCC), with an altered peptide ligand (K99R), preferentially induces Th2 differentiation (188). Jorritsma et al. (189) showed that K99R stimulates weak and transient activation of extracellular signal-regulated kinase (ERK), compared with the “cognate” MCC peptide. The reduced ERK activation by K99R is associated with early IL-4 production by naive CD4 T cells and with a distinct pattern of DNA-binding activity of AP-1 to the Il4 promoter, dominated by a JunB homodimer (189). This finding is consistent with a previous report showing that JunB, when directly bound to the Il4 promoter, synergizes with c-Maf to activate an Il4 luciferase reporter gene (170).
When naive TCR-transgenic CD4 T cells are stimulated with low concentrations of cognate peptide, ERK activation is weak and transient (190). The “low concentration–stimulated” T cells rapidly produce GATA3 and activate STAT5 in response to endogenously produced IL-2. GATA3 and STAT5 synergize to result in TCR-dependent, IL-4-independent early IL-4 transcription (induction). These T cells go on to complete their differentiation into Th2 cells by responding to the endogenously produced IL-4 and continued STAT5 activation (polarization) (190). By contrast, stimulating TCR-transgenic naive CD4 T cells with high concentrations of cognate peptide results in failure of Th2 differentiation. TCR-dependent IL-4-independent early GATA3 expression is suppressed and IL-2R-mediated STAT5 activation is transiently blocked, resulting in failure of early IL-4 production. Under these stimulation conditions, strong and prolonged ERK activation is observed. Blockade of the ERK pathway with an inhibitor of MAPK/ERK kinase (MEK) allows T cells stimulated with high peptide concentrations to express early GATA3 and to respond to endogenously produced IL-2, leading to the restoration of early IL-4 production, completion of the induction phase, and subsequent completion of the Th2 polarization process. These results imply that strong ERK activation prevents early GATA3 production and “de-sensitizes” the IL-2 receptor, thus blocking the Th2 induction phase.
However, cells expressing a DN Lck trans-gene or a DN H-Ras transgene under the control of the Lck-proximal promoter show diminished in vitro Th2 differentiation. This result has been interpreted by the authors to indicate that TCR-mediated Ras/ERK activation is required for Th2 differentiation, possibly by enhancing tyrosine phosphorylation of STAT6 in response to IL-4 (191) and/or preventing ubiquitin/proteasome-mediated degradation of GATA3 in developing Th2 cells by inhibiting the activity of Mdm2, an E3 ubiquitin ligase for GATA3 (192).
The difference in these views of the role of ERK function in Th2 differentiation needs to be considered in terms of the two-phase model of T cell polarization. Indeed, naive CD4 T cells from the mice expressing DN Lck actually produce significantly more IL-4 than do those from the littermate control mice during the early induction phase (193), implying that the Lck/Ras/ERK cascade inhibits early IL-4 production and the Th2 induction phase. However, IL-2 production by naive CD4 T cells from mice expressing DN Lck is significantly decreased compared with that by control cells (193). Diminished IL-2 production could account for diminished Th2 differentiation since STAT5 activation is essential for both the induction and polarization phase of Th2 differentiation, and the degree of activation may have fallen below the threshold level during the polarization phase. Indeed, this suggestion is consistent with our data showing that blockade of the ERK pathway leads to a substantial diminution in IL-2 production by CD4 T cells stimulated with low peptide concentrations (190) and that, in the presence of a MEK inhibitor, low-peptide-concentration Th2 differentiation requires exogenous IL-2.
TCR-Proximal Src Family Tyrosine Kinases: Lck and Fyn
Lck and Fyn belong to the Src family of tyrosine kinases and are involved in optimal T cell activation. Lck−/− mice exhibit a prominent but incomplete developmental arrest at the transition from the DN to the DP stage during thymocyte development. The few peripheral T cells that do develop have markedly impaired responses to TCR stimulation (194). By contrast, Fyn−/− mice show virtually no abnormality in thymocyte development and in the compartment of peripheral T cells (195, 196). Naive CD4 T cells from Fyn−/− mice on a C57BL/6 background display enhanced Th2 polarization upon TCR/CD28 stimulation under neutral conditions (197). Similarly, Fyn−/− DO11.10 TCR-transgenic CD4 T cells, on a BALB/c background, are more ready to differentiate into IL-4-producing cells than are wild-type DO11.10 TCR-transgenic cells when activated by cognate peptide in vitro (198). Our unpublished data suggest that activated Fyn−/− CD4 T cells have increased STAT6 phosphorylation in response to limited amounts of IL-4, implying that Fyn negatively regulates Th2 differentiation by attenuating IL-4R signaling and thus diminishes the polarization phase of Th2 differentiation. The effects of Fyn on IL-4-mediated STAT6 phosphorylation may be explained by a functional association between Fyn and PTP1B phosphatase (199, 200), which could result in desensitization of activated CD4 T cells to IL-4 (201).
Tec Family Kinases: Itk and Rlk
T cells express three different Tec family kinases, Itk, Rlk (also known as Txk), and Tec. The role of Itk during T cell activation and Th1/Th2 differentiation is well established. Following TCR ligation, Itk is recruited to LAT/SLP-76/PLC-γ1 complex and activates PLC-γ1, which induces hydrolysis of PIP3 to IP3 and DAG, resulting in Ca2+ release from the endoplasmic reticulum and PKC activation, respectively (202). Itk−/− mice exhibit diminished in vivo Th2 responses to Leishmania major, and CD4 T cells from these mice show impaired Th2 differentiation in vitro due to a deficit in the Ca2+/NFATc1 pathway (203), which presumably results in a decrease in early IL-2 and/or IL-4 production. Berg and colleagues (204) reported that naive Itk−/− TCR-transgenic CD4 T cells have aberrant expression of T-bet and polarize toward the Th1 phenotype in response to a weak TCR signal that would normally have induced Th2 differentiation. Recently, Schwartzberg and colleagues (205) found that a defect in the Ca2+/NFAT pathway in Itk−/− CD4 T cells results in diminished IL-17A but not IL-17F production when naive Itk−/− CD4 T cells are stimulated under Th17-polarizing conditions. Rlk has been reported to be involved in Th1 responses (206–208).
Itk−/−Rlk−/− mice show enhanced Th2 responses to Schistosoma mansoni eggs in vivo, although Itk−/− mice have a defect in Th2 responses to the same organism, and Rlk−/− mice behave just as wild-type mice do (209). Consistent with these data, our unpublished data indicate that naive CD4 T cells from Itk−/−Rlk−/− TCR transgenic mice undergo Th2 differentiation in response to high concentrations of cognate peptide that would have caused Th1 polarization in wild-type cells. The Itk−/−Rlk−/− cells show a marked diminution in ERK activation at high peptide concentration, presumably allowing them to express early GATA3 and to respond to endogenously produced IL-2, implying the involvement of Itk and Rlk in TCR-mediated signal strength and thus in inhibition of the induction phase of Th2 differentiation in response to high peptide concentrations.
NF-κB Pathway
Nfkb1−/− CD4 T cells have been reported to undergo diminished Th2 differentiation in vitro and in vivo due to impaired GATA3 expression in the nucleus (210). Because NF-κB1 has no transactivation domain, it must form a complex with other protein(s) to activate NF-κB1-dependent gene expression. Boothby and colleagues (211) identified Bcl-3, which belongs to the IκB family and possesses a transactivation domain, as the partner of NF-κB1 for binding to the κB-like consensus sequence located at 310 to 301 bp upstream of the Gata3 transcriptional initiation site, implying the potential importance of NF-κB1 and Bcl-3 in regulating TCR-driven GATA3 expression (212). However, because the NF-κB pathway also plays a critical role in the expression of IL-2 and CD25 (213), entities essential for Th2 differentiation, a careful study is needed to determine the relative importance of NF-κB-mediated GATA3 upregulation versus enhanced STAT5 phosphorylation during Th2 cell differentiation induced by this pathway.
Ca2+/NFAT Pathway
The NFAT family consists of five members, of which four, NFAT1 (also known as NFATp, NFATc2), NFAT2 (NFATc, NFATc1), NFAT3 (NFATc4), and NFAT4 (NFATx, NFATc3), are regulated by calcium and the fifth member, NFAT5/TonEBP (tonicity-responsive enhancer-binding protein), by osmotic shock (214). T cells express NFAT1, NFAT2, and NFAT4. TCR ligation increases intracellular Ca2+ concentration. Ca2+ binds to calmodulin, which in turn triggers the activation of calcineurin. Activated calcineurin dephosphorylates NFAT proteins, which results in the translocation of NFAT to the nucleus and the subsequent induction of NFAT-dependent gene transcription (214).
NFAT proteins are indispensable for effector cytokine production upon TCR activation in already differentiated Th cells and also play important roles in regulating Th differentiation. Here, we focus on the functions of NFAT1, NFAT2, and NFAT4 during the differentiation of Th1/Th2 cells.
The role of NFAT1 in regulating Th1/Th2 differentiation has been the subject of controversy. Naive CD4 T cells from Nfat1−/− mice are biased toward Th2 differentiation owing to an increased production of IL-4 in response to anti-CD3 stimulation (215). They exhibit diminished IFN-γ production and subsequent Th1 differentiation through mechanisms independent of IL-4, GATA3, and c-Maf (216). However, NFAT1 binds to the Il4 promoter in cooperation with IRF4 and c-Maf and enhances Il4 transcription (154). NFAT1 also binds to the 3′ enhancer of the Il4 gene (HSVA), whose activity is Ca2+-dependent (62). In agreement with these reports, the enhancement of Th2 differentiation by IL-6 is mediated through induction of NFAT1 and preferential accumulation of NFAT1 in the nucleus (217).
Nfat4−/− mice have impaired development of CD4 and CD8 SP thymocytes due to diminished Bcl-2 expression, which leads to increased apoptosis (218). Peripheral T cells from these mice exhibit an increased frequency of CD4 T cells of an activated/memory phenotype and become hyperactive in response to anti-CD3 stimulation, although skewing toward either Th1 or Th2 lineage is not observed (218). A combined deficiency in NFAT1 and NFAT4 results in greater Th2 responses with increased expression of Th2 cytokines and increased serum IgG1 and IgE levels (219, 220).
T cells from mice lacking NFAT2 in the lymphoid system generated by blastocyst complementation show impaired Th2 cytokine production, and the sera from these mice display reduced IgG1 and IgE levels (221, 222), implying that NFAT2 is required for Th2 differentiation. Consistent with these results, Th2 differentiation of naive MCC-specific TCR-transgenic CD4 T cells induced by K99R is accompanied by greater expression of nuclear NFAT2 than that of NFAT1 (223). Moreover, the inducible costimulator (ICOS) molecule substantially upregulates Nfat2 gene transcription, leading to enhanced TCR/CD28-driven early IL-4 production and IL-4-dependent c-Maf expression (224).
Notch Signaling
The Notch pathway plays a crucial role in the development of the central nervous system and vascular system, among others. The mammalian Notch family has four members, Notch1, 2, 3, and 4. They are expressed on the cell surface following various posttranslational modifications, such as fucosylation by Pofut, glucosylation by Fringe, and S1 cleavage by a Furin-like protease. In mammals, there are five Notch ligands: Jagged (Jag) 1 and 2 and Delta-like (Dll) 1, 3, and 4. When a Notch interacts with a Notch ligand, the γ-secretase complex proteolytically releases the Notch intracellular domain (NICD). NICD translocates into the nucleus, where it displaces a corepressor complex from CBF1/Su(H)/Lag-1 (CSL, also known as RBP-J) and recruits a coactivator complex, leading to Notch-dependent gene transcription (225).
In the immune system, Notch is important in thymic T cell differentiation and in the development of marginal zone B cells (226). Notch also plays a role in regulating differentiation of naive CD4 T cells into distinct Th lineages. It was first reported that the Notch3-Dll1 interaction results in Th1 differentiation (227). Skokos & Nussenzweig (228) found that LPS induces MyD88-dependent Dll4 expression on CD8α− DCs and that these DCs then direct Th1 differentiation in an IL-12-independent, Notch-dependent manner. Enforced expression of Dll1 and Dll4 on IL-12 p40−/− bone marrow–derived DCs has been reported to promote Th1 differentiation in a T-bet-dependent manner and to suppress Th2 development (229). Osborne and colleagues (230, 231) found that Notch1 ICD (N1ICD) can form a complex with NF-κB1 and c-Rel, allowing these NF-κB isoforms to be retained in the nucleus and, with the binding of the N1ICD/NF-κB complex to the IFN-γ promoter, to activate IFN-γ expression. Also reported is that the N1ICD/CSL complex binds to the T-bet promoter and that T-bet gene expression is directly regulated by the Notch1 pathway (232).
Notch has also been reported to be important in directing Th2 lineage commitment. CD4 T cells from mice with a conditional deletion of Rbpj (which specifies CSL) by CD4-Cre fail to undergo Th2 differentiation under non-polarizing conditions (233, 234). Amsen et al. (233) reported that Jag1/Notch interaction directs Th2 differentiation, whereas Dll1/Notch interaction leads to Th1 polarization. They identified a CSL-binding site in the HSV site of the Il4 gene and found that the binding of the N1ICD/CSL complex to this site upregulates IL-4 gene expression. The N1ICD/CSL complex is also reported to directly regulate Gata3 gene transcription in a STAT6-independent manner through its binding to an alternative Gata3 promoter located ~10 kb upstream of the conventional Gata3 promoter (235). Mice that overexpress a DN form of Mastermind-like 1, one of the coactivator components required for Notch-dependent gene expression, also have a defect in Th2 responses (236, 237).
The Notch pathway also regulates naive CD4 T cell differentiation into iTreg and Th17 cells. Pretreatment of CD4 T cells with a γ-secretase inhibitor diminishes the frequency of Foxp3+ cells induced by TGF-β1 and reduces the binding of the N1ICD/CSL complex to the Foxp3 promoter (238). γ-secretase inhibitor treatment also blocks the recruitment of Smad proteins to the Foxp3 promoter (238), consistent with a previous finding that N1ICD associates with Smad3 to integrate Notch and TGF-β signals in myogenic cells (239). The interaction of Notch with Dll4 has recently been reported to cause a substantial increase in expression of Th17-related genes in CD4 T cells stimulated under Th17-polarizing conditions (240). Binding of CSL to the RORγt and IL-17A promoter regions is greatly enhanced by Notch ligation by Dll4 but abrogated by treatment with a γ-secretase inhibitor.
Collectively, the Notch pathway appears to govern differentiation of naive CD4 T cells into each of the Th lineages by directly controlling expression of lineage-specific transcription factors and cytokines. However, given the common machinery to activate the Notch pathway upon interaction with any Notch ligand, it is difficult to provide a reasonable explanation for how different Notch ligands instruct naive CD4 T cells to undergo such a diverse set of Th differentiation outcomes.
An alternative view is that the Notch pathway does not instruct Th1/Th2 fate determination but rather regulates the cellular expansion and cytokine production of differentiated cells (241). It was recently reported that neither Dll1-nor Jag1-expressing artificial antigen-presenting cells instruct naive DO11.10 CD4 T cells to differentiate into Th1 or Th2 cells under nonpolarizing conditions (241). Rather, conditional deletion of Rbpj or of presenilin, one component of the γ-secretase complex, in mature CD4 T cells does not affect T-bet or GATA3 expression at day 6 of priming under Th1-and Th2-polarizing conditions, respectively. However, loss of RBP-J or presenilin reduces the capacity of differentiated cells to secrete effector cytokines upon challenge, which is associated with decreased T cell proliferation during the priming period. These results are consistent with those in earlier reports suggesting that Notch signaling controls T cell activation. Osborne and colleagues (231) showed that inhibition of Notch activation dramatically decreases the division of both CD4 and CD8 T cells in response to TCR stimulation. γ-secretase inhibition causes a decrease in IL-2 production and CD25 expression, resulting in diminished proliferation of activated CD4 T cells (242). These data suggest an important costimulatory role of Notch signaling in controlling optimal T cell activation.
Our unpublished results indicate that Notch is required for Th2 differentiation induced by weak TCR signaling under nonpolarizing conditions. An analysis of the kinetics of early expression of genes essential for Th2 commitment reveals that IL-4-independent early induction of GATA3 and IL-4 by a weak TCR signal is intact in CD4 T cells deprived of Notch signal, but that IL-2 gene expression is greatly reduced. Consistent with our previous reports demonstrating the central role of IL-2 in Th2 differentiation (121, 190), Notch signal–deprived CD4 T cells fail to complete the polarization phase of Th2 differentiation because of diminished IL-4-dependent late Gata3 and Il4 gene expression resulting from limited STAT5 signaling. Exogenous IL-2 corrects the defect in late expression of Gata3 and Il4 and thus fully restores Th2 differentiation in Notch signal–deprived CD4 T cells. These results imply that during Th2 differentiation, Gata3 and Il4 expression do not require direct binding of Notch to their genes, but rather that, in the absence of Notch signaling, it is the failure to produce sufficient amounts of IL-2 needed to sustain IL-4 production that is responsible for defective Th2 differentiation.
TRANSCRIPTION FACTOR–MEDIATED EPIGENETIC MODIFICATIONS
Th cell differentiation involves epigenetic modification and chromatin remodeling at specific loci (243–245). Epigenetic regulation includes modification of both DNA and histones, including DNA CpG methylation, histone methylation and acetylation, as well as DNase I HS induction. Epigenetic modification and chromatin remodeling play critical roles in determining specific gene expression induced by common transcription factors such as NFAT. Indeed, NFAT binding to the Il4 promoter increases, whereas its binding to the Ifng promoter decreases during Th2 differentiation, possibly owing to opposite epigenetic modifications at these two loci during Th2 differentiation (246).
Rao and colleagues (247) identified Th cell lineage–specific HSs in the Il4 and Ifng loci. A series of specific HSs in different Th cells identified at these loci have proven to be critical regulatory elements. Most of these HSs, located in the promoter and enhancer regions, exert positive function in cytokine production, but some, such as HSIV within the Il4 gene, serve as silencers. Deletion of HSIV results in IL-4 production from Th1 cells both in vitro and in vivo (246). The appearance of the lineage-specific HSs may depend on the induction of master regulator genes. Indeed, many studies show that GATA3 and T-bet are directly responsible for chromatin remodeling at cytokine loci (12, 55, 248, 249). DNA CpG methylation plays an important role in gene silencing. Binding of a DNA methyltransferase, Dnmt-1, to the Il4 locus is dramatically reduced during Th2 differentiation, and Dnmt-1 deficiency results in abnormal IL-4 expression in CD8 T cells without GATA3 upregulation (250). Deficiency in a methyl CpG-binding domain protein-2 (MBD2) also results in heritable, aberrant IL-4 production in Th1 cells without induction of GATA3 expression (251). GATA3 can block the binding of MBD2 to methyl CpG, suggesting an interesting mechanism through which GATA3 induces IL-4 production.
Recently, genome-wide histone modification patterns have been profiled in human and mouse T cells using next-generation high-throughput DNA sequencing (164, 252, 253). High-throughput DNA sequencing has also been used in genome-wide mapping of DNase I HS sites in Th cells (254).
There are at least 20 different histone modifications involving multiple positions in individual histones. Among these, histone H3 trimethylation at lysine position 4 (H3K4me3) is strongly associated with active chromatin, whereas H3K27me3 is associated with repressed chromatin. Genome-wide profiling of these two histone modifications in different Th lineages, including nTregs, iTregs, Th1, Th2, and Th17 cells, confirms the specificity of epigenetic modification at cytokine loci. For example, the Il4/Il13 locus displays H3K4me3 in Th2 cells but H3K27me3 in Th1 cells (164). The molecule responsible for H3K4 methylation at the Il4/Il13 locus during Th2 differentiation does not appear to be the histone H3K4 methyltransferase MLL, which is required for maintaining H3K4 modification at the Gata3 and Il4/Il13 loci in memory Th2 cells (255). EZH2, a H3K27 methyltransferase, binds to the Il4/Il13 locus and may be responsible for suppressive H3K27me3 Il4/Il13 modification in Th1 cells (256). The genome-wide profiling of H3K4 and H3K27 modifications also revealed the flexibility of master regulator expression in different lineages, as indicated by bivalent H3K4me3 and H3K27me3 modifications, particularly for the Tbx21 locus (164). Indeed, T-bet can be induced in differentiated Th17 cells and even in nTregs. Strikingly, T-bet associates with RbBp5, a core component of H3K4 methyltransferase complexes, and JMJD3, a H3K27 demethylase (257). These data suggest that the flexibility of T-bet expression underlies the plasticity of Th cells to produce IFN-γ, possibly by reversing the repressive epigenetic modifications at the Ifng locus. Differential epigenetic modification at the Foxp3 locus between nTregs and iTregs has been observed (164, 258).
Through transcription factor binding and epigenetic modification studies, many critical regulatory elements have been identified at each signature cytokine gene locus (Figure 4).
Figure 4.
Important cis-regulatory elements at the Ifng, Il4/Il13, and Il17a/Il17f loci and binding of transcription factors to these sites. (a) Within ~140 kb flanking the Ifng gene, many conserved noncoding sequences and their epigenetic modifications have been studied (259). CNS–22 has been shown to be critical for IFN-γ production (261). Many transcription factors, including T-bet, STAT4, Runx3, STAT5, and CTCF, bind to different regions of the Ifng gene (65, 131, 144, 181, 243, 261, 262). Our unpublished data suggest that Runx3 also binds to other regulatory elements in addition to the Ifng promoter. (b) Within ~70 kb flanking the Il4/Il13 genes, several important regulatory elements, including the locus control region (LCR), CNS1, Il4 HSII, Il4 HSIV, and Il4 HSVA/V, have been identified (62, 246, 263–267). Many transcription factors, including GATA3, c-Maf, STAT5, STAT6, Runx3, NFAT, and Notch/CSL, directly bind to different regions of the Il4/Il13 locus (61, 62, 122, 137, 144, 165, 233). Our unpublished data obtained from anti-GATA3 ChIPseq showed six GATA3-binding sites across this ~70-kb region in Th2 cells, with three located in LCR at RHS4, 5, and 6 and the other three at ~1.6 kb upstream of Il13, Il4 HSII, and Il4 HSVA. (c) Epigenetic modifications of the CNSs across ~160 kb of Il17a/Il17f have been reported (268). Some transcription factors critical for regulation of IL-17 expression, including RORγt, STAT3, BATF, Runx1, NFAT, and Gfi-1/LSD1, directly bind to the CNS or promoter regions of Il17a/Il17f (90, 110, 152, 160, 177, 205).
IFN-γ
Using a combination of computational analysis, epigenetic profiling of histone H3 modification, DNA CpG methylation and DNase I HS and functional assays, Wilson and colleagues (259) identified several important conserved noncoding sequence (CNS) sites within 100 kb of the Ifng gene, representing enhancers and insulators. IfngCNS–6, located 6 kb upstream of the Ifng transcription start site, has enhancer activity consistent with an earlier report (260). However, this site was not shown to be hypersensitive to DNase I in primary Th1 cells in this study (259). IfngCNS+29, located 29 kb downstream of the transcription start site, is also an enhancer; it displays DNase I hypersensitivity in Th1 but not in Th2 cells. IfngCNS–22 and IfngCNS–34 contain T-bet responsive elements. These two CNS sites show strong H3K4 methylation and CpG DNA demethylation in Th2 cells, both usually associated with active chromatin, which suggests that they may be the silencers of IFN-γ in Th2 cells or that they are critical elements in maintaining plasticity. However, deletion of CNS–22 from a BAC transgene reduces expression of the IFN-γ reporter in T cells and in NK cells (261). Finally, IfngCNS–54 and IfngCNS+46, serving as insulators, may be the boundaries of the IFN-γ transcription unit (259).
IL-4/IL-13
The Il4/Il13 Th2 cytokine locus is the most extensively studied among those of the Th “signature” cytokines. GATA3 has been reported to bind to the Il13 but not to the Il4 promoter. In addition to the promoters, there are several important regulatory elements within the locus including CNS1 (263), HSVa (264), HSII (265), the locus control region (LCR) (266, 267), and HSIV (246). All these elements except HSII have been shown by genetic deletion studies to be important in regulating IL-4 or IL-13 expression.
IL-17A/IL-17F
The Il17a/Il17f locus is less well studied, partly because of the relatively short time since Th17 cells were recognized. Nevertheless, STAT3-binding sites have been identified in the Il17a and Il17f promoters (110). Chen and colleagues (268) studied chromatin remodeling of Il17a/Il17f in Th17 cells and found eight CNS sites, all associated with histone 3 acetylation. These modifications are specific for Th17 cells at all but the CNS5 site, which is located in the intergenic region between Il17a and Il17f. In addition, functional ROR response elements have been identified within the CNS2 region (also called CNS-5), located ~5 kb upstream of Il17a and within 2 kb of the Il17a promoter region (91, 152). Both RORγt and Runx1 bind to these two regions and play important roles in regulating IL-17 production (152). We have recently identified a Gfi-1/LSD1 complex binding element located ~10 kb downstream of the Il17a transcription start site (CNS3), which may negatively regulate IL-17 production (160). BATF, an AP-1 factor critical for IL-17 production, also strongly binds to this region (177), suggesting that CNS3 contains both positive and negative regulatory elements for IL-17 transcription.
microRNA AND T CELL DIFFERENTIATION
By targeting the 3′ UTR of a transcript, miRNAs can repress translation and/or reduce mRNA stability. Approximately 400 miRNAs have been identified in humans, and more than half of mammalian transcripts are predicted to be targets of miRNAs. Not surprisingly, many reports indicate that miRNAs play important roles in regulating virtually all biological processes including hematopoiesis and the functions of immune cells, including T cells (269).
Dicer and Drosha are the RNase III enzymes critical for generating mature miRNA (270). Deletion of Dicer at an early stage of T cell development by Lckcre greatly reduces the cellularity of the thymi owing to the increased death of CD4/CD8 DP cells (271). When Dicer is deleted at DP stages, by CD4-cre, the total number of thymocytes is normal (272), but the frequency of CD8 T cells is dramatically reduced, especially in the periphery. CD4 T cell proliferation in response to TCR stimulation is also impaired, correlated with poor IL-2 production by these cells. In addition, Dicer-deficient Th2 cells produce a substantial amount of IFN-γ, suggesting that miRNAs play a negative role in Th1 differentiation and/or IFN-γ production (272). The specific miRNA(s) responsible for these defects remains to be identified. miRNAs are also involved in the generation and expansion of Tregs. Dicer deletion by CD4-cre results in reduced numbers of Tregs and affects the induction of Foxp3+ cells from CD4+CD25− T cells by TGF-β (273).
miRNA-155-deficient mice have reduced numbers of Tregs. miRNA-155, whose expression is regulated by Foxp3, directly suppresses SOCS-1 translation and promotes sensitivity of Tregs to IL-2, thereby enhancing Treg proliferation (274). However, the expression of miRNA-155 is not restricted to Tregs. T cell activation by anti-CD3/anti-CD28 dramatically induces miRNA-155 expression within 5 h (275), suggesting that miRNA-155 also plays an important role in conventional T cells. Indeed, miRNA-155-deficient T cells show a Th2 differentiation bias with increased IL-4 and decreased IFN-γ production possibly due to the increased expression of c-Maf, which is a direct target of miRNA-155 (276, 277).
Given the critical importance of both the master transcription factors and miRNAs, study of the regulation of miRNA expression by master transcription factors and of the involvement of miRNAs in modulating master transcription factors promises to be of great importance in understanding Th cell differentiation.
HETEROGENEITY OF THE Th CELLS
Th cells are very diverse owing to the existence of different lineages and heterogeneity within each lineage. As discussed above, Th cells can be classified in at least four lineages, Th1, Th2, Th17, and Treg cells. Some labs have also described TGF-β-producing Th3 cells (278), IL-10-producing TR1 (279) cells, IL-9-producing Th9 cells (280, 281), and T follicular helper Tfh cells (31, 44, 45) as separate lineages. However, Tregs also produce TGF-β, and all Th cells are capable of producing IL-10 under certain circumstances, whereas IL-9 was originally described as a Th2 cytokine and Th9 cells are developmentally related to Th2 cells. Therefore, the Th3, TR1, and Th9 cells may not be members of lineages that are distinct from Th1, Th2, Th17, and Treg cells but that rather represent diversity within Th lineages. Tfh cells may be members of a separate lineage, although this is still a matter of debate. As discussed above, Tfh cells generated in vivo in the course of Th1, Th2, or Th17 responses may express Th1, Th2, or Th17 signature cytokines in addition to IL-21 and CXCR5 (32–34, 167). Are these further differentiated Tfh cells or do each of the effector lineages have the potential to express the molecules that allow them to mediate Tfh function? In the gut, Tfh cells appear to originate mainly from Tregs (282). A possible relationship among conventional Th2, IL-4-producing Tfh, and Th9 cells is illustrated in Figure 5.
Figure 5.
Possible relationships among classic Th2, IL-4-producing Tfh, and Th9 cells. Depending on the cytokine milieu, naive CD4 T cells may differentiate directly into Th2, Tfh, and Th9 cells once they are activated (33, 121, 280, 281). Classic Th2 cells may become IL-4-producing Tfh cells upon receiving IL-6/IL-21 and ICOSL stimulation or IL-9-producing cells upon receiving TGF-β signaling. Whether Th9 cells or IL-4-producing Tfh cells can become classic Th2 cells is not known. The transcription factor directly responsible for IL-9 production has not been identified.
Heterogeneity exists within each cell lineage. Some of the heterogeneity results from the pattern of cytokine production. Each lineage is able to express a set of cytokines. For example, Th2 cells express IL-4, IL-5, IL-10, and IL-13. But some cells produce more of one cytokine than of the others, although there are many double or triple expressors. Even for a given cytokine, the probability of expression from the two alleles may differ, which is largely determined by different epigenetic modifications at each allele (283, 284). Heterogeneity of cytokine production is also true for Th1 and Th17 cells. For example, the IFN-γ+TNF-α+IL-2+ multifunctional Th1 cells, but not the single cytokine producers, are likely to be the long-lasting memory cells (285). In addition to differential epigenetic modification and stochastic events during T cell signaling, variable expression of some transcription factors may also be responsible for heterogeneity in cytokine secretory capacity. For example, PU.1 is selectively expressed in IL-4-nonproducing Th2 cells; such cells are capable of producing more CCL22 (286). Within Th17 cells, differential expression of IL-17 and IL-22 may be due to the induction and activation of the aryl hydrocarbon receptor (AhR) (175). Although both RORγt and AhR have been shown to be highly expressed in Th17 cells, whether they are coexpressed in all Th17 cells or the AhR is preferentially expressed in IL-22-producing Th17 cells has not been determined.
Recent reports suggest that the Tregs are more complex than originally thought. Cells coexpressing RORγt and Foxp3 exist in vivo. Although such cells were thought to represent an intermediate stage in the process of Th17 or iTreg differentiation (182), it has now been shown that treatment of differentiated nTregs with IL-6 will induce them to express RORγt and IL-17 (112, 113). Furthermore, T-bet-expressing Tregs exist in normal mice, and loss of T-bet from Tregs diminishes both the capacity of these Th1-like Tregs to proliferate and their capacity to suppress Th1-related autoimmune diseases (36). Our unpublished data indicate that GATA3 is expressed in some Tregs and may contribute to certain Treg functions. Indeed, Tregs may consist of Th1-, Th2-, and Th17-like populations in which differentiation is accompanied by the upregulation of Th1, Th2, and Th17 master regulators; under certain circumstances, these Tregs may become effector cells (287) (Figure 6).
Figure 6.
Complexity of Tregs. Foxp3 is the master regulator for Tregs. Recent reports and our unpublished data show that the transcription factors for Th1, Th2, and Th17 cells, T-bet, GATA3, and RORγt, respectively, can also be coexpressed in some Tregs (36, 113, 182). A relatively high expression ratio of Foxp3 over another master transcription factor prevents the induction of effector cytokines. Different combinations of transcription factors subdivide the Tregs with possible distinct regulatory functions. In some cases, the level of Foxp3 expression may fall, leading to expression of effector cytokines controlled by the other master regulator expressed by the cell. Effector cells derived from Tregs, because of their unique antigen specificity, may participate in normal immune responses to infections or contribute to autoimmune diseases (287).
PLASTICITY OF T HELPER CELLS
A multipotential precursor cell can differentiate into distinct lineages, a process that was generally thought to move toward a fixed fate. However, an increasing number of studies have shown that the differentiated cells retain the flexibility to dedifferentiate or redifferentiate. Even terminally differentiated fibroblasts can be reprogrammed to become inducible pluripotent stem cells by the transduction of the transcription factors Oct4/Klf4/Sox2/Myc (288, 289). Likewise, Pax5 deletion in B cells causes them to lose B cell identity and to gain the potential to develop into T cells (290). Deleting Gata3 from Th2 cells allows the production of IFN-γ (57); reduction of Foxp3 in Tregs renders them able to gain a Th2 phenotype (84); Gfi1 deletion from Th2 cells results in active epigenetic modifications at Th17- and iTreg-related gene loci, including Rorc, Il23r, and Cd103 (160).
Not only can artificial genetic modification cause the reversal of differentiation, so too can physiologic stimuli. For example, Th2 cells can be induced by IL-12 to produce IFN-γ; Th17 cells isolated using an IL-17F-Thy1.1 reporter, upon transfer, lose IL-17-producing capacity while gaining competence to produce IFN-γ, where IFN-γ production is dependent on STAT4 and T-bet (291). Tregs cultured under Th1 conditions gain the capacity to produce IFN-γ (164). These examples imply that although IFN-γ is the Th1 signature cytokine, each of the other lineages retains the capacity to produce it. Such flexibility may be determined by the bivalent H3K4 and H3K27 modification of the Tbx21 gene as well as by the accessibility of IfngCNS-22 in non-Th1 cells (164, 259). Interestingly, by using mice with indicators for Foxp3 and IL-17F (Foxp3-GFP/IL-17F-RFP mice), Dong and colleagues (113) reported the existence of GFP+RFP+ cells and showed that Tregs can produce IL-17 when they are treated with IL-6. Acquisition of IL-6-producing capacity correlated with upregulation of RORγt. Strober’s group had earlier reported a similar observation (112). Thus, differentiated Th cells are somewhat plastic and can be reprogrammed into other lineages given appropriate stimulation. Understanding the detailed mechanisms through which switching of one Th cell type to the other is achieved has important implications for immune intervention.
IN VITRO/IN VIVO
Another important point is to draw a comparison between effector/regulatory CD4 T cell populations differentiated in vitro and those that occur in vivo in response to infectious agents or other naturally occurring stimulants. Substantial evidence suggests that the in vivo cells may have undergone pathways of differentiation that differ from those established in vitro, and hence the cell types could be quite different from one another. In turn, this finding could imply that the highly polarized cells seen in vitro may either represent more extreme cases than those that occur in vivo or only occur under unusual circumstances. Indeed, partial states of differentiation, in which components of individual phenotypes are coexpressed, may dominate physiologic effector/memory CD4 cell populations. Analysis and comparison of the in vitro and in vivo populations would now seem essential. A particularly cogent example of this potential difference is exemplified in the Th2 lineage. In vitro, IL-4, acting through the IL-4 receptor/STAT6/GATA3 pathway, is virtually essential for Th2 differentiation. In vivo, however, although GATA3 remains critical (57), differentiation can often be obtained in mice that lack the IL-4 receptor and/or STAT6 (142).
MONOGENIC HUMAN DISEASES EFFECTING Th CELL DIFFERENTIATION OR FUNCTION
One of the most convincing lines of evidence establishing the importance of a given differentiation process for human physiology is the existence of monogenic abnormalities in particular differentiation processes or effector functions resulting in disease. A growing set of human disorders in the Th differentiation/function pathways has been identified, with a concomitant proliferation of data establishing the importance of these differentiation processes in humans.
Abnormalities in Th1 Differentiation or Function
To our knowledge, there are no defects that uniquely block the Th1 differentiation of CD4 T cells. However, there is a set of monogenic disorders each with a relatively similar phenotype consistent with the absence of Th1 function. These disorders include deficiencies in IFNGR1, IFNGR2, STAT1, IL12RB1, IL12B, and nuclear factor-kappaB-essential modulator (NEMO) (292). In each case, the dominant phenotype is susceptibility to infection with mycobacterial species, including poorly infectious agents such as BCG and atypical mycobacteria, as well as to infection with Mycobacterium tuberculosis. In general, other infectious agents are not a problem for these patients, with the exception of Salmonella.
Of the mutations described, three block the IFN-γ-signaling pathway and two inhibit the IL-12/23 pathway. IFN-γ is not uniquely produced by Th1 cells; in particular, NK cells are an excellent source of IFN-γ. Defects in IL-12RB1 and IL-12B should block or diminish both Th1 and Th17 differentiation, because these receptor and cytokine chains are important in assembling both the IL-12 and IL-23 receptors and the IL-12 and IL-23 proteins (40, 293). Mutations in IL-12RB1 and IL-12B do indeed impair the capacity of patients’ CD4 T cells to develop into Th17 cells (294). However, patients with a defect in Th17 development do not show susceptibility to mycobacterial infection. Taken together, these observations strongly suggest a critical role of human Th1 cells in resistance to infection with various mycobacteria. In mice, although M. tuberculosis infection induces both a Th1 and Th17 response (295), the protective response is dependent mainly upon Th1 cells. Human Th1 cells play a dominant role in protection against mycobacterial infection and surprisingly few other intracellular infectious agents.
Abnormalities in Th2 Function
GATA3 mutations have been detected in human populations, and in the heterozygous form they account for the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome (296). Although haploinsufficiency of GATA3 also causes reduced Th2 differentiation (297), it does not result in any major immunologic abnormalities. By contrast, there is a human disorder resulting from mutations that cause diminished TCR repertoire complexity. As noted above, low peptide concentration and low affinity of the TCR for the peptide/MHC complex are associated with an increased likelihood of Th2 differentiation in experimental systems. Similarly, mutations in key elements of TCR signaling are also associated with Th2 differentiation and often with elevated IgE. Does the same hold for human conditions?
Omenn syndrome, an autosomal recessive disease of neonates characterized by erythroderma, chronic diarrhea, hepatosplenogaly, and lymphadenopathy (298), is fatal unless treated with bone marrow transplantation. Patients display elevated numbers of Th2 cells and overproduction of IL-4 and IL-5 as well as eosinophilia and elevated serum IgE. The disease results from mutations that significantly impair T cell development in the thymus. Most cases are associated with hypomorphic mutations in RAG1 or RAG2, leading to the development of relatively limited numbers of T cells and B cells (298). As a result, the cells that reach the periphery have a very limited repertoire, although they expand greatly in number in the periphery. Thus, while large numbers of CD4 T cells exist in patients, they represent expansions from relatively small numbers of thymic emigrants, and the individuals have a very limited peripheral CD4 T cell repertoire. In one report, a patient with what appears to be a major but not complete absence of ZAP70 presented with eczematous skin lesions simulating atopic dermatitis and with eosinophilia and elevated IgE, suggesting that a thymic signaling defect may result in a limited TCR repertoire (299). Alternatively, this patient’s abnormalities may have resulted from signaling defects among the peripheral CD4 T cells.
Why a limited repertoire should lead to a Th2-mediated disease is not entirely clear. However, we have argued that low repertoire complexity may favor Th2 differentiation because the likelihood of a newly introduced antigen binding T cells that recognize it with high affinity is small, and thus low-affinity responses will predominate, which should favor Th2 differentiation (300). Mouse models of Omenn syndrome with mutations in RAG genes have a phenotype similar to that of the human Omenn syndrome (301, 302), and, as already noted, intentionally providing mice with a limited T cell repertoire by transferring a small number of CD4 T cells to Rag2−/− recipients results in a fulminant, although delayed, Th2 inflammatory disease (300).
Abnormalities in Th17 Differentiation
As noted above, humans with a mutation in IL-12B and IL-12RB1 may display some defects in IL-17 differentiation, but their principal abnormalities are more likely explained by deficiencies in Th1 induction. A very striking example exists of a human deficiency in the development of Th17 cells in patients with mutations in STAT3 that account for Job syndrome, also known as hyper-IgE syndrome (303). The mutations in Job syndrome, which occur mainly in the SH2 domain or the DNA-binding domain of STAT3 (304, 305), have been shown to act as dominant negatives, consistent with the autosomal dominant pattern of inheritance of the syndrome. The striking abnormalities of affected individuals are heightened susceptibility to infection with Staphylococcus aureus and Streptococcus pneumoniae as well as to Candida albicans and other fungi. Blood of such patients lacks CD4 T cells capable of producing IL-17, and their purified naive CD4 T cells fail to differentiate into Th17 cells under conditions in which MHC-matched normal cells differentiate quite efficiently (109).
With the growing body of data indicating an important role for IL-1β in Th17 differentiation or stabilization (47, 306), there will be an interest in mutations affecting IL-1β production, such as those in the inflammasome pathway or in caspsase 1. Thus far, activating mutations have been described that result in several autoinflammatory disorders (307).
Abnormalities in Treg Differentiation
Immunodeficiency, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) results from mutations in FOXP3 (80, 308). Because FOXP3 is on the X chromosome, the affected population is mainly boys. The syndrome often includes insulin-dependent diabetes and elevated serum IgE with aspects of both eczema and psoriasis. Other autoimmune symptoms include hypothyroidism, anemia, thrombocytopenia, and neutropenia. Patients often show an excess production of Th2 cytokines. IPEX-like syndromes have been reported in patients with IL2RA mutations (309).
Significance of Evidence for Differentiation
There is no doubt that the effector cytokines of various Th cell types have a critical role in humans. For both Th17 cells and Tregs, mutations of key molecules in the differentiation pathway lead both to the failure of the cells of that type to appear and to severe abnormalities, very much as expected from the mouse studies. For Th1 and Th2 cells, the human results as to the critical role of the differentiation process are less certain as mutations in the key factors associated with differentiation have not been observed. For Th1 cells, the importance of the differentiated cells is inferred either from the similarity of the phenotype in individuals with mutations that block a key Th1 product or from mutations that block Th1 differentiation but also control another lineage. For Th2 cells, the support from human disease lies in the consequence of overactivity of the cell population, particularly where the TCR repertoire is limited.
Although space does not permit an extended discussion of the role of various genes that regulate either signature cytokines or transcription factors in susceptibility to complex human disorders, there is a very large body of data on this point. Genome-wide association studies have in Th differentiation as being associated with implicated many genes known to play a role specific disorders.
SUMMARY POINTS.
There are at least four types of Th cells, Th1, Th2, Th17, and iTregs. Th1, Th2, and Th17 cells are important for eradicating intracellular pathogens, helminth, and extracellular bacteria/fungi, respectively. Th1 and Th17 cells are also involved in many types of autoimmune diseases, whereas Th2 cells contribute to allergic responses (185). iTreg as well as nTreg cells are critical in maintaining self-tolerance and in modulating immune responses to infections (310).
Cytokines play critical roles in determining Th cell differentiation. A combination of cytokines is required for the differentiation of each lineage: IL-12 and IFN-γ for Th1, IL-4 and IL-2/IL-7/TSLP for Th2, TGF-β and IL-6/IL-21/IL-23 for Th17, and TGF-β and IL-2 for iTreg. One of the effector cytokines produced by Th cells further promotes the differentiation process, providing a powerful positive amplification loop. IL-1 family cytokines may also participate in inducing TCR-independent effector cytokine production by Th cells: IL-18 for Th1, IL-33 for Th2, and IL-1 for Th17.
Transcription factors involved in Th cell differentiation form a sophisticated network involving collaboration and positive and negative regulation. The master regulators and STAT family members collaborate in T cell differentiation and expansion: T-bet and STAT4 for Th1, GATA3 and STAT5 for Th2, RORγt and STAT3 for Th17, Foxp3 and STAT5 for iTreg. Other transcription factors are either secondary to master regulators and STAT proteins or responsible for the induction of master regulators. Mutual repression between transcription factors of different lineages also exists. Therefore, Th cell differentiation involves lineage commitment, selective growth of committed cells, and active suppression of alternative lineage fates.
Most transcription factors involved in Th cell differentiation have been shown to directly bind to the effector cytokine genes (Ifng, Il4/Il13, and Il17a/Il17f) at promoters, enhancers, insulators, and locus control regions (LCR). These genetic elements are generally found at conserved noncoding sequence (CNS) and DNase I hypersensitivity (HS) regions. The binding of transcription factors to the different sites induces gene activation or repression as well as epigenetic modification.
Our knowledge of the Th cells has expanded greatly owing to massive Th cell heterogeneity and their plasticity. T cell heterogeneity and plasticity open new opportunities for targeting or redirecting specific subsets in autoimmune and allergic diseases. These targets may become clinically feasible when we fully understand the regulation of Th cell subsets and their relationships.
Many signaling molecules and transcription factors shown to be critical in Th cell differentiation in a mouse model are also defective in some human diseases related to abnormal Th cell differentiation. Linking animal models with clinical studies should provide greater insight into the details of Th cell differentiation.
FUTURE ISSUES.
Identification of Th cell subsets in vivo and comparison of in vitro and in vivo differentiated Th cells remain important issues for future research.
We need to understand the cellular and molecular mechanisms underlying in vivo Th differentiation decisions.
Specific signaling pathways for regulating master transcription factors of each Th cell lineage have yet to be identified.
The component of transcription factor complexes mediating gene activation and repression is also an area requiring further research.
Genome-wide profiling of transcription factor binding and epigenetic modifications in each Th lineage are needed.
Involvement of specific miRNAs in each Th lineage must be elucidated.
Researchers must discover the relationships between classic Th1/Th2 and newly defined Th17/iTreg/Tfh/Th9 cells and between effector cells and regulatory T cells.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
This is a work of the U.S. Government and is not subject to copyright protection in the United States.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Jinfang Zhu, Email: jfzhu@niaid.nih.gov.
Hidehiro Yamane, Email: hyamane@niaid.nih.gov.
William E. Paul, Email: wpaul@niaid.nih.gov.
LITERATURE CITED
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 2.Killar L, MacDonald G, West J, Woods A, Bottomly K. Cloned, Ia-restricted T cells that do not produce interleukin 4(IL 4)/B cell stimulatory factor 1(BSF-1) fail to help antigen-specific B cells. J. Immunol. 1987;138:1674–1679. [PubMed] [Google Scholar]
- 3.Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J. Exp. Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J. Immunol. 1990;145:3796–3806. [PubMed] [Google Scholar]
- 5.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh CS, Heimberger AB, Gold JS, O’Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an αβ T-cell-receptor transgenic system. Proc. Natl. Acad. Sci. USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 8.Rocken M, Saurat JH, Hauser C. A common precursor for CD4+ T cells producing IL-2 or IL-4. J. Immunol. 1992;148:1031–1036. [PubMed] [Google Scholar]
- 9.Sad S, Mosmann TR. Single IL-2-secreting precursor CD4 T cell can develop into either Th1 or Th2 cytokine secretion phenotype. J. Immunol. 1994;153:3514–3522. [PubMed] [Google Scholar]
- 10.Noben-Trauth N, Hu-Li J, Paul WE. IL-4 secreted from individual naive CD4+ T cells acts in an autocrine manner to induce Th2 differentiation. Eur. J. Immunol. 2002;32:1428–1433. doi: 10.1002/1521-4141(200205)32:5<1428::AID-IMMU1428>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 12.Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, Guo L, Min B, Watson CJ, Hu-Li J, et al. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16:733–744. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 14.Bendelac A, Killeen N, Littman DR, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor α-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 16.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 18.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 19.Park H, Li Z, Yang XO, Chang SH, Nurieva R, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 21.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, et al. IL-6 programs TH -17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 23.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 24.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, et al. IL-21 initiates an alternative pathway to induce proinflammatory TH 17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Jin W, Hardegen N, Lei KJ, Li L, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu S, Zhang N, Yopp AC, Chen D, Mao M, et al. TGF-β induces Foxp3+ T-regulatory cells from CD4+ CD25–precursors. Am. J. Transplant. 2004;4:1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- 27.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-β induces a regulatory phenotype in CD4+CD25− T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 28.Zheng SG, Wang JH, Gray JD, Soucier H, Horwitz DA. Natural and induced CD4+CD25+ cells educate CD4+CD25− cells to develop suppressive activity: the role of IL-2, TGF-β, and IL-10. J. Immunol. 2004;172:5213–5221. doi: 10.4049/jimmunol.172.9.5213. [DOI] [PubMed] [Google Scholar]
- 29.DiPaolo RJ, Brinster C, Davidson TS, Andersson J, Glass D, Shevach EM. Autoantigen-specific TGFβ-induced Foxp3+ regulatory T cells prevent autoimmunity by inhibiting dendritic cells from activating autoreactive T cells. J. Immunol. 2007;179:4685–4693. doi: 10.4049/jimmunol.179.7.4685. [DOI] [PubMed] [Google Scholar]
- 30.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-β dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu. Rev. Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 32.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaretsky AG, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J. Exp. Med. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J. Exp. Med. 2009;206:1001–1007. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokoyoda K, Zehentmeier S, Hegazy AN, Albrecht I, Grun JR, et al. Professional memory CD4+ T lymphocytes preferentially reside and rest in the bone marrow. Immunity. 2009;30:721–730. doi: 10.1016/j.immuni.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, et al. T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 41.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 42.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, et al. Transforming growth factor-β induces development of the TH17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 43.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 44.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Guo L, Wei G, Zhu J, Liao W, Leonard WJ, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. USA. 2009;106:13463–13468. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Zhu H, Murphy TL, Ouyang W, Murphy KM. IL-18-stimulated GADD45 beta required in cytokine-induced, but not TCR-induced, IFN-γ production. Nat. Immunol. 2001;2:157–164. doi: 10.1038/84264. [DOI] [PubMed] [Google Scholar]
- 49.Robinson D, Shibuya K, Mui A, Zonin F, Murphy E, et al. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-γ production and activates IRAK and NFκB. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- 50.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 52.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 54.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J. Exp. Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473–482. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 57.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, et al. Conditional deletion of Gata3 shows its essential function in TH1–TH2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 58.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siegel MD, Zhang DH, Ray P, Ray A. Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J. Biol. Chem. 1995;270:24548–24555. doi: 10.1074/jbc.270.41.24548. [DOI] [PubMed] [Google Scholar]
- 60.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J. Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 61.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, et al. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J. Biol. Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 62.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 63.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 64.Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 65.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 66.Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, et al. Hlx is induced by and genetically interacts with T-bet to promote heritable TH 1 gene induction. Nat. Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- 67.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 68.Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 69.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 70.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin . Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 72.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and Eomesodermin . Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-γ production in developing Th1 cells through the repression of Eomesodermin expression. J. Immunol. 2006;177:3721–3727. doi: 10.4049/jimmunol.177.6.3721. [DOI] [PubMed] [Google Scholar]
- 74.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 75.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glimcher LH. Trawling for treasure: tales of T-bet. Nat. Immunol. 2007;8:448–450. doi: 10.1038/ni0507-448. [DOI] [PubMed] [Google Scholar]
- 78.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 79.Patel DD. Escape from tolerance in the human X-linked autoimmunity-allergic disregulation syndrome and the Scurfy mouse. J. Clin. Investig. 2001;107:155–157. doi: 10.1172/JCI11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse Scurfy. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 81.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 82.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 83.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 84.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 85.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 87.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-β receptor. Immunity. 2006;25:441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-β signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 89.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 90.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 91.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 93.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 95.Dent AL, Doherty TM, Paul WE, Sher A, Staudt LM. BCL-6-deficient mice reveal an IL-4-independent, STAT6-dependent pathway that controls susceptibility to infection by Leishmania major. J. Immunol. 1999;163:2098–2103. [PubMed] [Google Scholar]
- 96.Kusam S, Toney LM, Sato H, Dent AL. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J. Immunol. 2003;170:2435–2441. doi: 10.4049/jimmunol.170.5.2435. [DOI] [PubMed] [Google Scholar]
- 97.Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls BCL-6 expression. Nat. Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 98.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 99.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu D, Rao S, Tsai LM, Lee SK, He Y, et al. The transcriptional repressor BCL-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 103.Lieberman LA, Banica M, Reiner SL, Hunter CA. STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J. Immunol. 2004;172:457–463. doi: 10.4049/jimmunol.172.1.457. [DOI] [PubMed] [Google Scholar]
- 104.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 105.Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 106.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 107.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 108.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 109.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl. Acad. Sci. USA. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: Regulatory T cells induce CD4+CD25−Foxp3− T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-β. J. Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 113.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 115.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 116.Cai G, Radzanowski T, Villegas EN, Kastelein R, Hunter CA. Identification of STAT4-dependent and independent mechanisms of resistance to Toxoplasma gondii. J. Immunol. 2000;165:2619–2627. doi: 10.4049/jimmunol.165.5.2619. [DOI] [PubMed] [Google Scholar]
- 117.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by down-regulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 118.Frucht DM, Aringer M, Galon J, Danning C, Brown M, et al. Stat4 is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages at sites of Th1-mediated inflammation. J. Immunol. 2000;164:4659–4664. doi: 10.4049/jimmunol.164.9.4659. [DOI] [PubMed] [Google Scholar]
- 119.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 120.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 121.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, et al. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 123.Kagami S, Nakajima H, Suto A, Hirose K, Suzuki K, et al. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood. 2001;97:2358–2365. doi: 10.1182/blood.v97.8.2358. [DOI] [PubMed] [Google Scholar]
- 124.Kagami S, Nakajima H, Kumano K, Suzuki K, Suto A, et al. Both stat5a and stat5b are required for antigen-induced eosinophil and T-cell recruitment into the tissue. Blood. 2000;95:1370–1377. [PubMed] [Google Scholar]
- 125.Takatori H, Nakajima H, Kagami S, Hirose K, Suto A, et al. Stat5a inhibits IL-12-induced Th1 cell differentiation through the induction of suppressor of cytokine signaling 3 expression. J. Immunol. 2005;174:4105–4112. doi: 10.4049/jimmunol.174.7.4105. [DOI] [PubMed] [Google Scholar]
- 126.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-β-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 127.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 128.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Amadi-Obi A, Yu CR, Liu X, Mahdi RM, Clarke GL, et al. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat. Med. 2007;13:711–718. doi: 10.1038/nm1585. [DOI] [PubMed] [Google Scholar]
- 131.Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–773. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 133.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat 6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 134.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, et al. Essential role of Stat6 in IL-4 signaling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 135.Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 136.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4’s role in Th2 differentiation and cell expansion. J. Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 137.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc. Natl. Acad. Sci. USA. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J. Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- 139.Finkelman FD, Morris SC, Orekhova T, Mori M, Donaldson D, et al. Stat6 regulation of in vivo IL-4 responses. J. Immunol. 2000;164:2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- 140.Min B, Prout M, Hu-Li J, Zhu J, Jankovic D, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J. Exp. Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 142.van Panhuys N, Tang SC, Prout M, Camberis M, Scarlett D, et al. In vivo studies fail to reveal a role for IL-4 or STAT6 signaling in Th2 lymphocyte differentiation. Proc. Natl. Acad. Sci. USA. 2008;105:12423–12428. doi: 10.1073/pnas.0806372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Else KJ, Finkelman FD, Maliszewski CR, Grencis RK. Cytokine-mediated regulation of chronic intestinal helminth infection. J. Exp. Med. 1994;179:347–351. doi: 10.1084/jem.179.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 145.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 148.Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, et al. Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 149.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, et al. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 156.Kano S, Sato K, Morishita Y, Vollstedt S, Kim S, et al. The contribution of transcription factor IRF1 to the interferon-γ-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol. 2008;9:34–41. doi: 10.1038/ni1538. [DOI] [PubMed] [Google Scholar]
- 157.Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duan Z, Horwitz M. Gfi-1 takes center stage in hematopoietic stem cells. Trends Mol. Med. 2005;11:49–52. doi: 10.1016/j.molmed.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 159.Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc. Natl. Acad. Sci. USA. 2006;103:18214–18219. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhu J, Davidson TS, Wei G, Jankovic D, Cui K, et al. Down-regulation of Gfi-1 expression by TGF-β is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J. Exp. Med. 2009;206:329–341. doi: 10.1084/jem.20081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 162.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J. Immunol. 2009;182:741–745. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pan F, Yu H, Dang EV, Barbi J, Pan X, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–751. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 166.Hwang ES, White IA, Ho IC. An IL-4-independent and CD25-mediated function of c-maf in promoting the production of Th2 cytokines. Proc. Natl. Acad. Sci. USA. 2002;99:13026–13030. doi: 10.1073/pnas.202474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005;201:615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 172.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 173.Wang L, van Panhuys N, Hu-Li J, Kim S, Le Gros G, Min B. Blimp-1 induced by IL-4 plays a critical role in suppressing IL-2 production in activated CD4 T cells. J. Immunol. 2008;181:5249–5256. doi: 10.4049/jimmunol.181.8.5249. [DOI] [PubMed] [Google Scholar]
- 174.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 176.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J. Exp. Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat. Immunol. 2009;10:1260–1266. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu Z, Li Z, Mao K, Zou J, Wang Y, et al. Dec2 promotes Th2 cell differentiation by enhancing IL-2R signaling. J. Immunol. 2009;183:6320–6329. doi: 10.4049/jimmunol.0900975. [DOI] [PubMed] [Google Scholar]
- 180.Liao W, Schones DE, Oh J, Cui Y, Cui K, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 184.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 185.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Garra A, Vieira P. TH1 cells control themselves by producing interleukin-10. Nat. Rev. Immunol. 2007;7:425–428. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 187.Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu. Rev. Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 188.Tao X, Grant C, Constant S, Bottomly K. Induction of IL-4-producing CD4+ T cells by antigenic peptides altered for TCR binding. J. Immunol. 1997;158:4237–4244. [PubMed] [Google Scholar]
- 189.Jorritsma PJ, Brogdon JL, Bottomly K. Role of TCR-induced extracellular signal-regulated kinase activation in the regulation of early IL-4 expression in naive CD4+ T cells. J. Immunol. 2003;170:2427–2434. doi: 10.4049/jimmunol.170.5.2427. [DOI] [PubMed] [Google Scholar]
- 190.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J. Exp. Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yamashita M, Kimura M, Kubo M, Shimizu C, Tada T, et al. T cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T cell differentiation. Proc. Natl. Acad. Sci. USA. 1999;96:1024–1029. doi: 10.1073/pnas.96.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yamashita M, Shinnakasu R, Asou H, Kimura M, Hasegawa A, et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- 193.Yamashita M, Hashimoto K, Kimura M, Kubo M, Tada T, Nakayama T. Requirement for p56lck tyrosine kinase activation in Th subset differentiation. Int. Immunol. 1998;10:577–591. doi: 10.1093/intimm/10.5.577. [DOI] [PubMed] [Google Scholar]
- 194.Molina TJ, Kishihara K, Siderovski DP, van Ewijk W, Narendran A, et al. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 195.Stein PL, Lee HM, Rich S, Soriano P. pp59fyn mutant mice display differential signaling in thymocytes and peripheral T cells. Cell. 1992;70:741–750. doi: 10.1016/0092-8674(92)90308-y. [DOI] [PubMed] [Google Scholar]
- 196.Appleby MW, Gross JA, Cooke MP, Levin SD, Qian X, Perlmutter RM. Defective T cell receptor signaling in mice lacking the thymic isoform of p59fyn. Cell. 1992;70:751–763. doi: 10.1016/0092-8674(92)90309-z. [DOI] [PubMed] [Google Scholar]
- 197.Tamura T, Igarashi O, Hino A, Yamane H, Aizawa S, et al. Impairment in the expression and activity of Fyn during differentiation of naive CD4+ T cells into the Th2 subset. J. Immunol. 2001;167:1962–1969. doi: 10.4049/jimmunol.167.4.1962. [DOI] [PubMed] [Google Scholar]
- 198.Mamchak AA, Sullivan BM, Hou B, Lee LM, Gilden JK, et al. Normal development and activation but altered cytokine production of Fyn-deficient CD4+ T cells. J. Immunol. 2008;181:5374–5385. doi: 10.4049/jimmunol.181.8.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sharma P, Chakraborty R, Wang L, Min B, Tremblay ML, et al. Redox regulation of interleukin-4 signaling. Immunity. 2008;29:551–564. doi: 10.1016/j.immuni.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, et al. PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood. 2008;112:4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhu J, Huang H, Guo L, Stonehouse T, Watson CJ, et al. Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J. Exp. Med. 2000;192:1125–1134. doi: 10.1084/jem.192.8.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: regulators of T-helper-cell differentiation. Nat. Rev. Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 203.Fowell DJ, Shinkai K, Liao XC, Beebe AM, Coffman RL, et al. Impaired NFATc translocation and failure of Th2 development in Itk-deficient CD4+ T cells. Immunity. 1999;11:399–409. doi: 10.1016/s1074-7613(00)80115-6. [DOI] [PubMed] [Google Scholar]
- 204.Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. 2004;21:67–80. doi: 10.1016/j.immuni.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 205.Gomez-Rodriguez J, Sahu N, Handon R, Davidson TS, Anderson SM, et al. Differential expression of IL-17A and IL-17F is coupled to TCR signaling via Itk-mediated regulation of NFATc1. Immunity. 2009;31:587–597. doi: 10.1016/j.immuni.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kashiwakura J, Suzuki N, Nagafuchi H, Takeno M, Takeba Y, et al. Txk, a nonreceptor tyrosine kinase of the Tec family, is expressed in T helper type 1 cells and regulates interferon γ production in human T lymphocytes. J. Exp. Med. 1999;190:1147–1154. doi: 10.1084/jem.190.8.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Takeba Y, Nagafuchi H, Takeno M, Kashiwakura J, Suzuki N. Txk, a member of nonreceptor tyrosine kinase of Tec family, acts as a Th1 cell-specific transcription factor and regulates IFN-γ gene transcription. J. Immunol. 2002;168:2365–2370. doi: 10.4049/jimmunol.168.5.2365. [DOI] [PubMed] [Google Scholar]
- 208.Sahu N, Venegas AM, Jankovic D, Mitzner W, Gomez-Rodriguez J, et al. Selective expression rather than specific function of Txk and Itk regulate Th1 and Th2 responses. J. Immunol. 2008;181:6125–6131. doi: 10.4049/jimmunol.181.9.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schaeffer EM, Yap GS, Lewis CM, Czar MJ, McVicar DW, et al. Mutation of Tec family kinases alters T helper cell differentiation. Nat. Immunol. 2001;2:1183–1188. doi: 10.1038/ni734. [DOI] [PubMed] [Google Scholar]
- 210.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-κB in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat. Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 211.Nolan GP, Fujita T, Bhatia K, Huppi C, Liou HC, et al. The BCL-3 proto-oncogene encodes a nuclear IκB-like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol. 1993;13:3557–3566. doi: 10.1128/mcb.13.6.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J. Immunol. 2005;175:2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- 213.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 214.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 215.Kiani A, Viola JP, Lichtman AH, Rao A. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1. Immunity. 1997;7:849–860. doi: 10.1016/s1074-7613(00)80403-3. [DOI] [PubMed] [Google Scholar]
- 216.Kiani A, Garcia-Cozar FJ, Habermann I, Laforsch S, Aebischer T, et al. Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood. 2001;98:1480–1488. doi: 10.1182/blood.v98.5.1480. [DOI] [PubMed] [Google Scholar]
- 217.Diehl S, Chow CW, Weiss L, Palmetshofer A, Twardzik T, et al. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J. Exp. Med. 2002;196:39–49. doi: 10.1084/jem.20020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Oukka M, Ho IC, de la Brousse FC, Hoey T, Grusby MJ, Glimcher LH. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity. 1998;9:295–304. doi: 10.1016/s1074-7613(00)80612-3. [DOI] [PubMed] [Google Scholar]
- 219.Ranger AM, Oukka M, Rengarajan J, Glimcher LH. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity. 1998;9:627–635. doi: 10.1016/s1074-7613(00)80660-3. [DOI] [PubMed] [Google Scholar]
- 220.Rengarajan J, Tang B, Glimcher LH. NFATc2 and NFATc3 regulate TH2 differentiation and modulate TCR-responsiveness of naive TH cells. Nat. Immunol. 2002;3:48–54. doi: 10.1038/ni744. [DOI] [PubMed] [Google Scholar]
- 221.Yoshida H, Nishina H, Takimoto H, Marengere LE, Wakeham AC, et al. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity. 1998;8:115–124. doi: 10.1016/s1074-7613(00)80464-1. [DOI] [PubMed] [Google Scholar]
- 222.Ranger AM, Hodge MR, Gravallese EM, Oukka M, Davidson L, et al. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity. 1998;8:125–134. doi: 10.1016/s1074-7613(00)80465-3. [DOI] [PubMed] [Google Scholar]
- 223.Brogdon JL, Leitenberg D, Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J. Immunol. 2002;168:3825–3832. doi: 10.4049/jimmunol.168.8.3825. [DOI] [PubMed] [Google Scholar]
- 224.Nurieva RI, Duong J, Kishikawa H, Dianzani U, Rojo JM, et al. Transcriptional regulation of Th2 differentiation by inducible costimulator. Immunity. 2003;18:801–811. doi: 10.1016/s1074-7613(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 225.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Stanley P, Guidos CJ. Regulation of Notch signaling during T- and B-cell development by O-fucose glycans. Immunol. Rev. 2009;230:201–215. doi: 10.1111/j.1600-065X.2009.00791.x. [DOI] [PubMed] [Google Scholar]
- 227.Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, et al. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19:549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 228.Skokos D, Nussenzweig MC. CD8- DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J. Exp. Med. 2007;204:1525–1531. doi: 10.1084/jem.20062305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sun J, Krawczyk CJ, Pearce EJ. Suppression of Th2 cell development by Notch ligands Delta1 and Delta4. J. Immunol. 2008;180:1655–1661. doi: 10.4049/jimmunol.180.3.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, et al. Notch1 augments NF-κB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-γ production in peripheral T cells. J. Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- 232.Minter LM, Turley DM, Das P, Shin HM, Joshi I, et al. Inhibitors of γ-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 233.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different Notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 234.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 235.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tu L, Fang TC, Artis D, Shestova O, Pross SE, et al. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, et al. Notch1 and TGFβ1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 2008;112:1813–1821. doi: 10.1182/blood-2008-03-144980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, et al. Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J. Cell Biol. 2003;163:723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J. Immunol. 2009;182:7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS One. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, et al. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 243.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 244.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 245.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 246.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 247.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 248.Seki N, Miyazaki M, Suzuki W, Hayashi K, Arima K, et al. IL-4-induced GATA-3 expression is a time-restricted instruction switch for Th2 cell differentiation. J. Immunol. 2004;172:6158–6166. doi: 10.4049/jimmunol.172.10.6158. [DOI] [PubMed] [Google Scholar]
- 249.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat. Immunol. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 251.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 252.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 253.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 256.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 257.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Floess S, Freyer J, Siewert C, Baron U, Olek S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nat. Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-γ (IFN-γ) locus revealed by genome sequence comparison. J. Biol. Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 261.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 262.Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, et al. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 264.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 265.Hural JA, Kwan M, Henkel G, Hock MB, Brown MA. An intron transcriptional enhancer element regulates IL-4 gene locus accessibility in mast cells. J. Immunol. 2000;165:3239–3249. doi: 10.4049/jimmunol.165.6.3239. [DOI] [PubMed] [Google Scholar]
- 266.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 267.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 268.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 269.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 270.Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 271.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, et al. A role for Dicer in immune regulation. J. Exp. Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 276.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 277.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chen Y, Kuchroo VK, Inobe J, Hafler DA, Weiner HL. Regulatory T cell clones induced by oral tolerance: suppression of autoimmune encephalomyelitis. Science. 1994;265:1237–1240. doi: 10.1126/science.7520605. [DOI] [PubMed] [Google Scholar]
- 279.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 280.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, et al. Transforming growth factor-β ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 281.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells, and together with TGF-β, generates IL-9+IL-10+ Foxp3− effector T cells. Nat. Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 283.Guo L, Hu-Li J, Zhu J, Watson CJ, Difilippantonio MJ, et al. In TH2 cells the Il4 gene has a series of accessibility states associated with distinctive probabilities of IL-4 production. Proc. Natl. Acad. Sci. USA. 2002;99:10623–10628. doi: 10.1073/pnas.162360199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Guo L, Hu-Li J, Paul WE. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity. 2005;23:89–99. doi: 10.1016/j.immuni.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 285.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 286.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 287.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 290.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 291.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Filipe-Santos O, Bustamante J, Chapgier A, Vogt G, de Beaucoudrey L, et al. Inborn errors of IL-12/23- and IFN-γ-mediated immunity: molecular, cellular, and clinical features. Semin. Immunol. 2006;18:347–361. doi: 10.1016/j.smim.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 293.Trinchieri G, Pflanz S, Kastelein RA. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity. 2003;19:641–644. doi: 10.1016/s1074-7613(03)00296-6. [DOI] [PubMed] [Google Scholar]
- 294.de Beaucoudrey L, Puel A, Filipe-Santos O, Cobat A, Ghandil P, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J. Exp. Med. 2008;205:1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Khader SA, Cooper AM. IL-23 and IL-17 in tuberculosis. Cytokine. 2008;41:79–83. doi: 10.1016/j.cyto.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, et al. GATA3 haploinsufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- 297.Skapenko A, Leipe J, Niesner U, Devriendt K, Beetz R, et al. GATA-3 in human T cell helper type 2 development. J. Exp. Med. 2004;199:423–428. doi: 10.1084/jem.20031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Marrella V, Poliani PL, Sobacchi C, Grassi F, Villa A. Of Omenn and mice. Trends Immunol. 2008;29:133–140. doi: 10.1016/j.it.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 299.Turul T, Tezcan I, Artac H, de Bruin-Versteeg S, Barendregt BH, et al. Clinical heterogeneity can hamper the diagnosis of patients with ZAP70 deficiency. Eur J Pediatr. 2009;168:87–93. doi: 10.1007/s00431-008-0718-x. [DOI] [PubMed] [Google Scholar]
- 300.Milner JD, Ward JM, Keane-Myers A, Paul WE. Lymphopenic mice reconstituted with limited repertoire T cells develop severe, multiorgan, Th2-associated inflammatory disease. Proc. Natl. Acad. Sci. USA. 2007;104:576–581. doi: 10.1073/pnas.0610289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Khiong K, Murakami M, Kitabayashi C, Ueda N, Sawa S, et al. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J. Clin. Investig. 2007;117:1270–1281. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Marrella V, Poliani PL, Casati A, Rucci F, Frascoli L, et al. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J. Clin. Investig. 2007;117:1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Buckley RH. The hyper-IgE syndrome. Clin. Rev. Allergy Immunol. 2001;20:139–154. doi: 10.1385/CRIAI:20:1:139. [DOI] [PubMed] [Google Scholar]
- 304.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 305.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 306.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 309.Caudy AA, Reddy ST, Chatila T, Atkinson JP, Verbsky JW. CD25 deficiency causes an immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like syndrome, and defective IL-10 expression from CD4 lymphocytes. J. Allergy Clin. Immunol. 2007;119:482–487. doi: 10.1016/j.jaci.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 310.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions. Annu. Rev. Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]