Abstract
Lysophosphatidic acid (LPA) is a highly potent endogenous lipid mediator that protects and rescues cells from programmed cell death. Earlier work identified the LPA2 G protein-coupled receptor subtype as an important molecular target of LPA mediating antiapoptotic signaling. Here we describe the results of a virtual screen using single-reference similarity searching that yielded compounds 2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid (NSC12404), 2-((3-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)propyl)thio)benzoic acid (GRI977143), 4,5-dichloro-2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid (H2L5547924), and 2-((9,10-dioxo-9,10-dihydroanthracen-2-yl)carbamoyl) benzoic acid (H2L5828102), novel nonlipid and drug-like compounds that are specific for the LPA2 receptor subtype. We characterized the antiapoptotic action of one of these compounds, GRI977143, which was effective in reducing activation of caspases 3, 7, 8, and 9 and inhibited poly(ADP-ribose)polymerase 1 cleavage and DNA fragmentation in different extrinsic and intrinsic models of apoptosis in vitro. Furthermore, GRI977143 promoted carcinoma cell invasion of human umbilical vein endothelial cell monolayers and fibroblast proliferation. The antiapoptotic cellular signaling responses were present selectively in mouse embryonic fibroblast cells derived from LPA1&2 double-knockout mice reconstituted with the LPA2 receptor and were absent in vector-transduced control cells. GRI977143 was an effective stimulator of extracellular signal-regulated kinase 1/2 activation and promoted the assembly of a macromolecular signaling complex consisting of LPA2, Na+-H+ exchange regulatory factor 2, and thyroid receptor interacting protein 6, which has been shown previously to be a required step in LPA-induced antiapoptotic signaling. The present findings indicate that nonlipid LPA2-specific agonists represent an excellent starting point for development of lead compounds with potential therapeutic utility for preventing the programmed cell death involved in many types of degenerative and inflammatory diseases.
Introduction
The growth factor-like lysophospholipids lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) regulate many fundamental cellular responses, ranging from cell survival through cell proliferation to cell motility and migration (Tigyi, 2010). To date, specific inhibitors of the LPA and S1P receptors have taken center stage in drug discovery efforts. The functional antagonist of S1P receptors, fingolimod (Brinkmann et al., 2010), has been recently approved by the Food and Drug Administration for the first-line treatment of multiple sclerosis, and AM152 [(R)-1-phenylethyl-5-(4-biphenyl-4-cyclopropanecarboxylic acid)-3-methylisoxazole-4-yl carbamate sodium salt], an LPA1-selective antagonist, has been granted orphan drug status for the treatment of fibrotic diseases. A decade ago, we had shown that LPA has profound activity in preventing apoptosis and can also rescue apoptotically condemned cells from the progression of the apoptotic cascade (Deng et al., 2002, 2003, 2004, 2007). We developed an LPA mimic, octadecenyl thiophosphate (OTP) (Durgam et al., 2006), which has efficacy superior to that of LPA in vitro and in vivo in rescuing cells and animals from radiation-induced apoptosis (Deng et al., 2007). Development of LPA-based drug candidates has been limited to the discovery of lipid-like ligands, which is understandable because of the hydrophobic environment of the S1P and LPA G protein-coupled receptor (GPCR) ligand binding pockets (Parrill et al., 2000; Wang et al., 2001; Li et al., 2005; Fujiwara et al., 2007; Valentine et al., 2008; Hanson et al., 2012). Only a few LPA receptor ligands break away from lipid-like structural features, among which 3-(4-[4-([1-(2-chlorophenyl)ethoxy]carbonyl amino)-3-methyl-5-isoxazolyl] benzylsulfanyl) propanoic acid (Ki16425), an LPA1/2/3 antagonist (Ohta et al., 2003), and the sodium, (4′-(3-methyl-4-((R)-1-phenyl-ethoxycarbonylamino)-isoxazol-5-yl)-biphenyl-4-yl)-acetate (AM095; Swaney et al., 2011) and AM152 series of LPA1-selective compounds are of importance.
To exploit the potential therapeutic benefits of LPA, discovery and development of drug-like nonlipid compounds might be beneficial. In the present study, we applied virtual screening strategies using similarity searching that we derived from the previously validated molecular models of these receptors, and we limited our searches to chemical libraries with drug-like compounds that satisfy Lipinski's rule of five (Lipinski, 2003). We focused our virtual screen on the discovery of ligands for the LPA2 receptor subtype because of our long-standing interest in developing compounds that can attenuate programmed cell death elicited by radiation and chemotherapy. The choice of this receptor subtype is based on mounting evidence that LPA2 is unique among this class of receptors in its ability to initiate signaling events that promote cell survival and prevent the progression of apoptosis (Deng et al., 2007; Lin et al., 2007; E et al., 2009). This objective was further fueled by our recent successes with OTP (Deng et al., 2007). Our objective in the present study was to identify nonlipid LPA2 agonist scaffolds that can lead to the development of new drug candidates capable of alleviating the side effects of chemotherapy and radiation treatment of cancer patients and potentially function as radiomitigators against lethal levels of radiation injury.
Here we report on the identification of four nonlipid compounds that are specific agonists of LPA2. We selected one of these compounds, 2-((3-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)propyl)thio)benzoic acid (GRI977143) from the Genome Research Institute chemical library, and characterized its cellular, pharmacological, and signaling responses in several assay systems. Our results show that the compound GRI977143 is a specific agonist of LPA2 and does not activate any other known or putative LPA GPCR. We also show that GRI977143 is as effective as LPA and OTP in preventing programmed cell death, although in Ca2+ mobilization and caspase 3 and 7 inhibition assays it has higher EC50 values than the other two ligands. GRI977143 inhibited activation of caspases 3, 7, 8, and 9, B-cell lymphoma 2-associated X protein (Bax) translocation, and poly(ADP-ribose)polymerase 1 (PARP-1) cleavage, leading to reduced DNA fragmentation after activation of the extrinsic or intrinsic apoptotic signaling cascades. We also provide evidence that GRI977143 robustly activates the extracellular signal regulated kinases 1/2 (ERK1/2) survival pathway and leads to the assembly of a macromolecular signalosome consisting of LPA2, thyroid receptor interacting protein 6 (TRIP6), and Na+-H+ exchange regulatory factor 2 (NHERF2), which has been shown to be required for the prosurvival signaling elicited via this receptor subtype. GRI977143 and the three other nonlipid compounds 2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid (NSC12404), 4,5-dichloro-2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid (H2L5547924), and 2-((9,10-dioxo-9,10-dihydroanthracen-2-yl)carbamoyl)benzoic acid (H2L5828102) described in this article represent a good starting point for lead development and optimization, which may yield novel LPA-based drug candidates for therapeutic applications.
Materials and Methods
Materials.
Lysophosphatidic acid (18:1) was purchased from Avanti Polar Lipids (Alabaster, AL). OTP was synthesized and provided by RxBio, Inc. (Johnson City, TN) as described previously (Durgam et al., 2006). The test compounds used in the present study were obtained from the following vendors: Genome Research Institute GRI977143 from the University of Cincinnati Drug Discovery Center (Cincinnati, OH), Hit2Lead (http://www.hit2lead.com) H2L5547924, and H2L5828102 from ChemBridge (San Diego, CA), and NSC12404 from the National Cancer Institute Developmental Therapeutics Program Open Chemical Repository. Stock solutions (10 mM) of GRI977143, H2L5547924, H2L5828102, and NSC12404 were prepared in dimethyl sulfoxide. Stocks of LPA and OTP (1 mM) as an equimolar complex of charcoal-stripped, fatty acid-free bovine serum albumin (Sigma-Aldrich, St. Louis, MO) were prepared just before use in phosphate-buffered saline (PBS). A stock solution of 3.45 mM doxorubicin (Adriamycin) was prepared in distilled water.
Computational Docking.
Compounds were flexibly docked into the activated LPA2 receptor homology model reported by Sardar et al. (2002) using AutoDock Vina (Trott and Olson, 2010). The compounds and receptor homology model were both energy-optimized with the Merck Molecular Force Field 94 (MMFF94) in the Molecular Operating Environment (MOE) software (Chemical Computing Group, 2002) before docking. Docking simulations were performed using a docking box with dimensions of 65 × 63 × 50 Å and a search space of 20 binding modes, and an exhaustive search parameter was set at 5. The best docking pose was chosen on the basis of the lowest energy conformation. Finally, the best pose was further refined using the MMFF94 in MOE.
Ligand-Based Similarity Search.
Similarity searching of NSC12404 was performed using the UC-DCC library database (http://drugdiscovery.uc.edu). The Tanimoto similarity indices for the reference compounds were calculated using ECFC6, FCFP4, and FCFP6 fingerprints in Pipeline Pilot software (Accelerys, Inc., San Diego, CA). The UC-DCC library was screened using Pipeline Pilot fingerprints to identify additional LPA2 ligands. A similarity threshold was set at 80%. Among the 225 returned hits, compounds with similarity >80% were selected by visual inspection, carefully considering the similarity and how closely the structures reflected the reference compound. A total of 27 compounds was selected for evaluation using LPA receptor-activated Ca2+-mobilization assays.
Residue Nomenclature.
Amino acids in the transmembrane (TM) domains were assigned index positions to facilitate comparison between GPCRs with different numbers of amino acids, as described by Ballesteros and Weinstein (1995). An index position is in the format X.YY., where X denotes the TM domain in which the residue appears, and YY indicates the position of that residue relative to the most highly conserved residue in that TM domain, which is arbitrarily assigned position 50.
LPA Receptor-Mediated Ca2+ Mobilization Assay.
Stable cell lines expressing the individual LPA1, LPA2, LPA3, LPA4, and LPA5 established receptor subtypes (Tigyi, 2010), as well as putative LPA receptors GPR87 (Tabata et al., 2007) and P2Y10 (Murakami et al., 2008) or appropriate empty vector-transfected controls, have been previously generated and described (Tabata et al., 2007; Murakami et al., 2008; Williams et al., 2009). Assays for ligand-activated mobilization of intracellular Ca2+ were performed using a FlexStation II robotic fluorescent plate reader (Molecular Devices; Sunnyvale, CA) as described previously (Durgam et al., 2006). The appropriate concentrations of the test compounds were either used alone (for agonist testing) or mixed with the respective ∼EC75 concentration of LPA 18:1 for the LPA receptor being tested (antagonist screen). The cells were loaded with Fura-2-acetoxymethyl ether in Krebs' buffer containing 0.01% pluronic acid for 30 min and rinsed with Krebs' buffer before Ca2+ mobilization was measured. The ratio of peak emissions at 510 nm after 2 min of ligand addition was determined for excitation wavelengths of 340 nm/380 nm. All samples were run in quadruplicate. The inhibition elicited by 10 μM test compound on the EC75 concentration of LPA 18:1 for a given receptor (I10 μM) was interpolated from the dose-response curves. The half-maximal effective concentration (EC50), and inhibitory constant (Ki) values were calculated by fitting a sigmoid function to dose-response data points using Prism 5 software (GraphPad Software, Inc., San Diego, CA).
Cell Culture.
Mouse embryonic fibroblast (MEF) cells were isolated from embryonic day 13.5 LPA1&2 double-knockout embryos (Lai et al., 2007). MEFs were transduced with empty vector or LPA2-containing lentiviruses and selected with 1.5 μg/ml puromycin. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Serum-free medium contained 0.1% (w/v) bovine serum albumin in DMEM. The rat intestinal epithelial cell line 6 (IEC-6) was obtained from the American Type Culture Collection (Manassas, VA) at passage 13; passages 16 to 21 were used in all experiments. IEC-6 cells were maintained in a humidified 37°C incubator in an atmosphere of 90% air and 10% CO2. Growth medium consisted of DMEM supplemented with 5% heat-inactivated FBS, 10 μg/ml insulin, and 50 μg/ml gentamicin. The composition of the serum-starvation medium was the same as that of the full-growth medium except that it contained no FBS. The McArdle rat hepatoma cell line (RH7777) stably expressing LPA2 receptors was a gift from Dr. Fumikazu Okajima (Gunma University, Maebashi, Japan). RH7777 cells stably expressing LPA1 or LPA3 receptors were generated in-house and characterized earlier (Fischer et al., 2001). Wild-type and LPA receptor (LPAR) stably transfected RH7777 cells were grown in DMEM supplemented with 10% FBS and 2 mM l-glutamine in the presence of 250 μg/ml G418. Chinese hamster ovary cells stably expressing either vector or LPA4 receptor were a kind gift from Dr. Takao Shimizu (Tokyo University, Tokyo, Japan). Cells were cultured in Ham's F12 medium containing 10% FBS, 2 mM l-glutamine, and 350 μg/ml G418. Rat neuroblastoma cells (B103) were transduced with the lentivirus harboring wild-type FLAG-LPA5 and selected with puromycin to establish the stable cell lines. The stable cells were maintained in DMEM supplemented with 10% FBS and 0.4 μg/ml puromycin. GPR87- and P2Y10-expressing Chinese hamster ovary cells and vector-transfected control cells were a gift from Dr. Norihisha Fujita (Ritsumeikan University, Shiga, Japan). The highly invasive MM1 rat hepatoma cells (gift from Dr. Michiko Mukai, Osaka University, Osaka, Japan) were grown in suspension in DMEM supplemented with 10% (v/v) FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Human umbilical vein endothelial cells (HUVECs) were purchased from VEC Technologies, Inc. (Rensselaer, NY) and cultured in MCDB-131 complete medium supplemented with 10% (v/v) FBS, 90 μg/ml heparin, 10 ng/ml epidermal growth factor, 1 μg/ml hydrocortisone, 0.2 mg/ml Endo Growth (VEC Technologies, Inc.) supplement, 100 U/ml penicillin G, 100 μg/ml streptomycin, and 25 μg/ml amphotericin B.
Cell Proliferation Assay.
For determination of the effect of the LPA receptor ligands on cell growth, vector- and LPA2-transduced MEF cells (2 × 104) were plated in each well of a 24-well plate in full growth medium. Cells were counted the next day, and the medium was replaced with medium containing 1.5% (v/v) FBS supplemented with or without 1 μM LPA, 1 μM OTP, or 10 μM GRI977143. Media containing LPA, OTP, and GRI977143 were refreshed every 24 h. The growth rate was measured by counting the number of cells as a function of time in triplicate using the Z1 Coulter Particle Counter (Beckman Coulter, Fullerton, CA).
Induction of Apoptosis by Doxorubicin or Serum Withdrawal.
Experiments were performed on vector- and LPA2-transduced MEF cells. To measure caspase 3, 7, 8, or 9 activity and DNA fragmentation, cells were plated in 48-well plates (2 × 104 cells/well). To detect PARP-1 cleavage and Bax translocation, 1.5 × 106 cells were plated in 10-cm dishes and cultured overnight in full growth medium. The next morning, the growth medium was replaced by serum starvation medium, and cells were pretreated for 1 h with LPA (1–10 μM), OTP (1–10 μM), GRI977143 (1–10 μM), or vehicle. Caspase activity, DNA fragmentation, PARP-1 cleavage, and Bax translocation were measured 5 h after incubation with 1.7 μM doxorubicin or 24 h after serum withdrawal.
Induction of Apoptosis by Tumor Necrosis Factor-α in IEC-6 Cells.
Confluent serum-starved IEC-6 cells were treated with or without TNF-α (10 ng/ml)-cycloheximide (CHX) (20 μg/ml) (Deng et al., 2002) in the presence of OTP (10 μM), GRI977143 (10 μM), or LPA (1 μM) for 3 h. Cells were washed twice with PBS, and the quantitative DNA fragmentation assay was carried out as described previously (Valentine et al., 2010).
Caspase Activity Assay.
Caspase-Glo 3/7, Caspase-Glo 8, and Caspase-Glo 9 reagents were purchased from Promega (Madison, WI) and used according to the manufacturer's instructions. In brief, cells were lysed by adding 50 μl of lysis reagent/well, followed by shaking for 30 min at room temperature. Lysate (200 μl) was transferred to a 96-well white-walled plate, and luminescence was measured using a BioTek Instruments (Winooski, VT) plate reader.
DNA Fragmentation Enzyme-Linked Immunosorbent Assay.
Apoptotically challenged cells were washed twice with PBS, and a quantitative DNA fragmentation assay was performed using a Cell Death Detection ELISAPLUS kit (Roche Diagnostics, Penzberg, Germany) and normalized to protein concentration using the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA) as described previously (Valentine et al., 2010). Aliquots of nuclei-free cell lysate were placed in streptavidin-coated wells and incubated with anti-histone-biotin antibody and anti-DNA peroxidase-conjugated antibody for 2 h at room temperature. After the incubation, the sample was removed, and the wells were washed and incubated with 100 μl of 2,2′-azino-di[3-ethylbenzthiazolin]-sulfonate substrate at room temperature before the absorbance was read at 405 nm. Results were expressed as absorbance at 405 nm · min−1 · mg protein−1 as detailed in our previous report (Ray et al., 2011).
MM1 Hepatoma Cell Invasion of Endothelial Monolayer.
HUVECs (1.3 × 105 cells at passages 5–7) were seeded into each well of a 12-well plate precoated with 0.2% gelatin (Sigma-Aldrich). Cells were grown for 2 days until a confluent monolayer was formed. MM1 cells were prelabeled with 2 μg/ml calcein AM (Life Technologies, Grand Island, NY) for 2 h and rinsed twice, and 5 × 104 cells/well were seeded over the HUVEC monolayer. Tumor monolayer cell invasion was performed for 20 h in MCDB-131 complete media containing 1% FBS with or without the addition of 1 μM LPA or 1 to 10 μM GRI977143. Noninvaded tumor cells were removed by repeatedly rinsing the monolayer with PBS (containing Ca2+ and Mg2+), followed by fixation with 10% buffered formalin. Tumor cells that penetrated the monolayer were photographed using a TiU inverted microscope (Nikon, Tokyo, Japan) with phase-contrast and fluorescence illumination. The fluorescent and phase-contrast images were overlaid using Elements BR software (version 3.1x; Nikon). A total of five nonoverlapping fields were imaged per well, and the number of invaded MM1 cells (displaying a flattened morphology underneath the monolayer) was counted.
Immunoblot Analysis.
To detect ligand-induced ERK1/2 activation, vector- and LPA2-transfected MEF cells were serum-starved 3 h before exposure to 1 μM LPA, 1 μM OTP, 10 μM GRI977143, or vehicle for 10 min. For ERK1/2 activation and PARP-1 cleavage measurements, cells were harvested in 1× Laemmli sample buffer and separated using 12% Laemmli SDS-polyacrylamide gels. To assess Bax translocation, cell lysates were separated into cytosolic, mitochondrial, and nuclear fractions using the Cell Fractionation Kit-Standard (MitoSciences, Eugene, OR). Cytosolic fractions were then concentrated by precipitation with 75% trichloroacetic acid, and the pellets were dissolved in 50 mM non-neutralized Tris buffer, pH 10, and 6× Laemmli buffer. Samples were boiled for 5 min and loaded onto 12% SDS-polyacrylamide gels. Western blotting was performed as described previously (Valentine et al., 2010). Primary antibodies against pERK1/2, PARP-1, Bax (Cell Signaling Technology, Danvers, MA), actin (Sigma-Aldrich), and anti-rabbit horseradish peroxidase secondary antibodies (Promega) were used according to the instructions of the manufacturer.
Detection of Ligand-Induced Macromolecular Complex Formation with LPA2.
LPA2 forms a ternary complex with TRIP6 and NHERF2 (Xu et al., 2004; Lin et al., 2007; E et al., 2009). This complex is assembled via multiple protein-protein interactions that include binding of NHERF2 to the C-terminal PSD95/Dlg/ZO-1 domain (PDZ)-binding motif of LPA2, the binding of TRIP6 to the zinc finger-like CxxC motif of LPA2, and binding of NHERF2 to the PDZ-binding motif of TRIP6 (E et al., 2009). To examine ligand-induced macromolecular complex formation, HEK293T cells were transfected with FLAG-LPA2 and enhanced green fluorescent protein (EGFP)-NHERF2, and the cells were exposed to 10 μM GRI977143 for 10 min as described in detail in our previous publication (E et al., 2009). The complex was pulled down using anti-FLAG M2 monoclonal antibody-conjugated agarose beads (Sigma-Aldrich) and processed for Western blotting using anti-EGFP (gift from Dr. A.P. Naren, University of Tennessee Health Science Center, Memphis, TN), anti-FLAG (Sigma-Aldrich), and anti-TRIP6 (Bethyl Laboratories; Montgomery, TX) antibodies.
Statistical Analysis.
Data are expressed as mean ± S.D. or S.E.M. for samples run in triplicate. Each experiment was repeated at least two times. Student's t test was used for comparison between the control and treatment groups. p ≤ 0.05 was considered significant.
Results
Rational Discovery of LPA2 Agonists.
In a virtual screen using a structure-based pharmacophore of LPA1 (Perygin, 2010), we serendipitously identified compound NSC12404, which was a weak agonist of LPA2 (Table 1; Fig. 1). Although this hit was not the intended target of that study, here we returned to this scaffold for the initiation of a virtual homology screen for other nonlipid ligands of LPA2. With the use of this hit, we undertook a database search in the UC-DCC chemical library. The similarity search included the requirement for a fused tricyclic or bicyclic ring system and the presence of an acid moiety linked with a hydrocarbon chain. The similarity fingerprint metrics included 1) extended connectivity fingerprint counts over 6 atoms, 2) functional class connectivity fingerprint counts over 4 atoms, and 3) functional class connectivity fingerprint counts over 6 atoms.
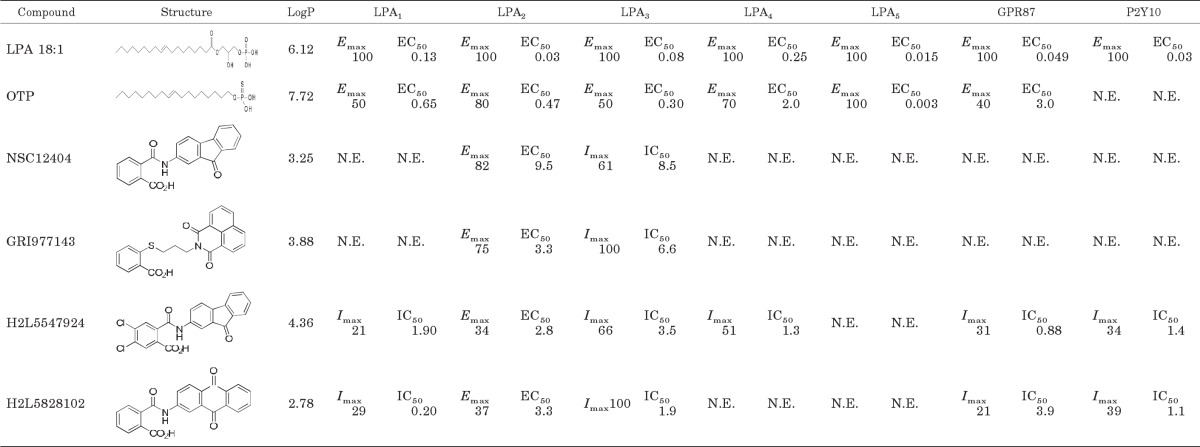
TABLE 1.
LPA receptor-activated Ca2+ mobilization profiles for hit compounds
Every compound was tested on every receptor subtype for agonist activity up to 10 μM and the Emax value listed in the table is normalized to the maximal response of LPA 18:1 at 10 μM. Those compounds that failed to activate a given receptor were tested for inhibition of the LPA 18:1 response. Imax is percentage inhibition of the ∼Emax75 LPA 18:1 response for a given receptor subtype using a 10 μM concentration of the antagonist. EC50 and IC50 concentrations are given as micromolar concentrations for dose-response curves covering the 30 nM to 10 μM range (Fig. 1). For determination of IC50 values, dose-response curves were generated using the ∼Emax 75 concentration of LPA 18:1 for a given LPA receptor subtype, and the ligand was coapplied in concentrations ranging from 30 nM to 10 μM.
LogP, log partition coefficient; N.E., no effect up to 10 μM of the ligand, the maximal concentration tested in the present experiments.
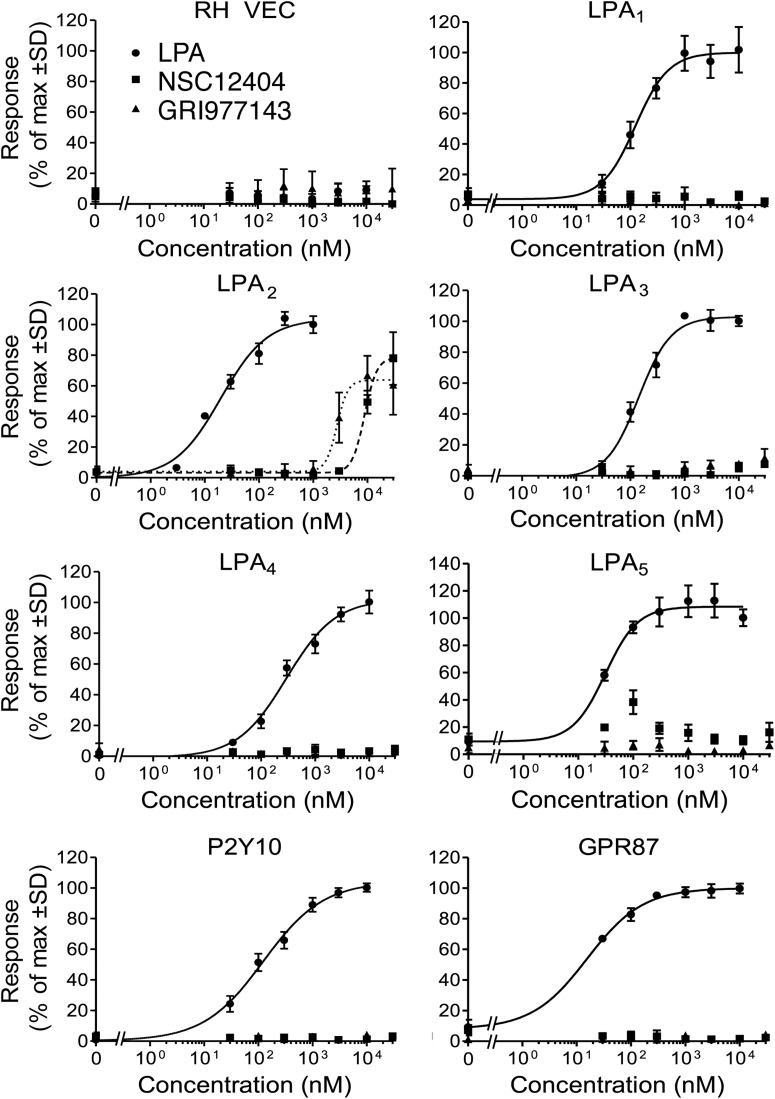
Fig. 1.
Receptor specificity of the prototype hit compound NSC12404 and in silico hit compound GRI977143 indicated by LPA GPCR-activated Ca2+-transients in cell lines expressing the individual LPA GPCR subtypes. The curves shown in this figure are representative of at least two experiments. RH VEC, vector-transfected RH7777 cells.
Similarity searches were performed separately using each similarity fingerprint to quantitate similarity. Hits meeting the 80% similarity threshold from each search were ranked based on the Tanimoto coefficient measure of similarity to the target molecule NSC12404, and the top 75 unique hits from each fingerprint search were selected for further analysis. The 225 compounds selected for further analysis were clustered on the basis of Tanimoto coefficients calculated using Molecular ACCess System-key fingerprints (MACCS keys) and evaluated using the diversity subset function implemented in MOE. This process selected a diverse subset of 27 compounds for biological evaluation by choosing the middle compounds in each cluster. These 27 compounds were tested in Ca2+ mobilization assays at a concentration of 10 μM using stable cell lines individually expressing LPA2 and also in vector-transfected control cells (Fig. 1; Table 1). Hits activating LPA2 were further tested using cells expressing the other established and putative LPA GPCRs. Experimental testing of the selected compounds identified three new selective LPA2 agonists: GRI977143, H2L5547924, and H2L5828102 (Table 1). NSC12404, H2L5547924, H2L5828102, and GRI977143 only activated LPA2 and failed to activate any of the other established and putative LPA GPCRs when applied up to 10 μM. A 10 μM concentration of these compounds has also been tested for the inhibition of the Ca2+ response elicited by the ∼EC75 concentration of LPA 18:1 at those receptors that the compound failed to activate when applied at 10 μM. We found that at this high concentration NSC12404 and GRI977143 inhibited LPA3, but none of the other receptors we tested was either activated or inhibited by these two compounds. H2L5547924 activated LPA2 but partially inhibited LPA1, LPA3, LPA4, GPR87, and P2Y10. Although H2L5828102 was a specific agonist of LPA2, it fully inhibited LPA3 and partially inhibited LPA1, GPR87, and P2Y10 (Table 1). On the basis of its lower EC50 concentration to activate the LPA2 receptor compared with NSC12404 and because it only inhibited the LPA3 receptor compared with the H2L compounds, we selected GRI977143 for further characterization in cell-based assays.
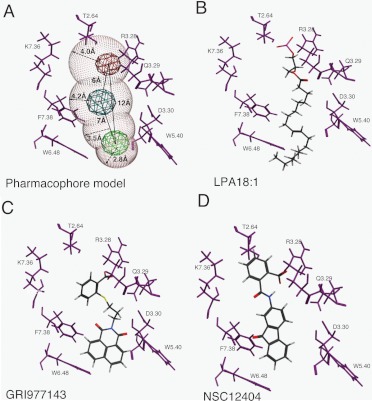
The LPA2 computational model docked with LPA 18:1 suggests 13 residues that comprise the ligand binding pocket (Fig. 2, B–D; Table 2). Computational docking of the four compounds listed in Table 1 indicates that these LPA2 ligands interact with some additional residues unique to a specific agonist in addition to the 13 common residues (Table 2; Fig. 2, B–D). The model of GRI977143 docked to the LPA2 structure is shown in Fig. 2. The docked structure shows that GRI977143 docks in the vicinity of the key residues Arg3.28, Gln3.29, Lys7.36, and W4.64, that we have previously shown are required for ligand activation of LPA2 (Valentine et al., 2008). In addition, the model predicted an interaction with Trp5.40 that was unique to this ligand.
Fig. 2.
Pharmacophore development for the LPA2 GPCR. The three-dimensional pharmacophore generated (A) was based on the common structural features of docked LPA (B), GRI977143 (C), and NSC12404 (D). A, pharmacophore properties are shown in red (anionic), blue (hydrogen bond acceptor), and green (hydrophobic interaction). The volume of the binding site is shown in white spheres. B–D, the three agonists (ball and stick) used for pharmacophore development are shown with interactions with key amino acid residues (purple) within 4.5 Å of the previously validated ligand binding pocket.
TABLE 2.
Title to come
| EC50 | Residues Common to All Agonistsa | Residues Unique to a Specific Liganda | |
|---|---|---|---|
| μM | |||
| LPA 18:1 | 0.014 | Arg3.28, Gln3.29, Gly3.30, Leu3.32, Asp3.33, Leu3.34, Trp4.64, Leu5.37, Arg5.38, Trp6.48, Lys7.36, Phe7.38, Leu7.39 | |
| OTP | 0.09 | Trp5.40, Leu6.55 | |
| NSC12404 | 9.5 | Thr2.64 | |
| GRI977143 | 3.3 | Trp5.40 | |
| H2L5547924 | 2.9 | Interacts with all common residues but some interactions are less favorable than with LPA or OTP | |
| H2L5828102 | 16.7 | Leu7.32, Thr2.64, Ser270 |
Residues predicted to be within 4.5 Å of the docked ligand.
A structure-based pharmacophore was developed using the docking function of the MOE software (Chemical Computing Group, 2002). Compound NSC12404 and LPA were docked into a homology model of LPA2 (Sardar et al., 2002; Valentine et al., 2008). In the pharmacophore model, we identified three feature sites based on the interactions between the agonists and the protein. We defined the key residues as those within 4.5 Å of our LPA2 agonists. The pharmacophore features and the corresponding amino acid residues involved in ligand interactions are shown in Fig. 2A. This pharmacophore model has three features: a hydrophobic feature (green), a hydrogen bond acceptor (blue), and an anionic (red) feature. The four volume spheres in the pharmacophore with radii in the 2.8 to 4.2 Å range delineate the regions ideal for different types of chemical interactions with the ligand in the binding pocket. The distances between chemical features along with the radii of the four volume spheres are shown in Fig. 2A.
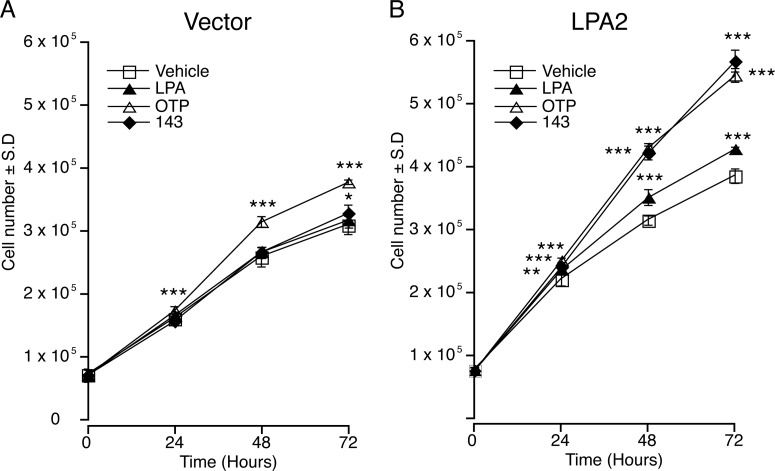
Effect of GRI977143 on Cell Growth.
LPA can function as a mitogen or an antimitogen, depending on the cell type and the receptors it expresses (Tigyi et al., 1994). We tested GRI977143 for its effect on cell proliferation of vector-transduced (Fig. 3A) and LPA2-transduced MEF cells (Fig. 3B). LPA had no significant effect on the proliferation of empty vector-transduced MEF cells. Likewise, GRI977143 did not cause a significant increase in vector cell proliferation except at 72 h (p < 0.05). In contrast, OTP significantly (p < 0.001) increased the growth of empty vector-transduced MEF cells from 24 h onward. The effects of LPA, OTP, and GRI977143 on the growth of LPA2-transduced MEF were all significant from 24 h onward.
Fig. 3.
Effects of LPA (1 μM), OTP (1 μM), and GRI977143 (10 μM) on fibroblast growth. Growth curves of the vector-tranfected (A) and the LPA2-transduced MEF cells (B). Values are means ± S.D and are representative of two independent experiments 143, GRI977143. (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
Effect of GRI977143 on MM1 Hepatoma Cell Invasion.
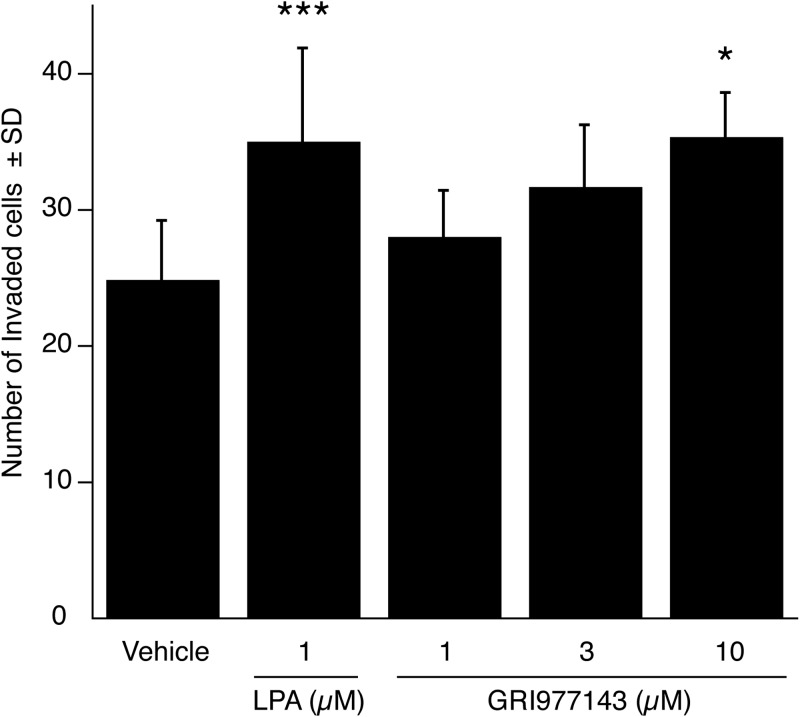
The highly invasive rat hepatoma MM1 cells invade mesothelial cell monolayers in an LPA-dependent manner (Mukai and Akedo, 1999; Mukai et al., 2003; Uchiyama et al., 2007). LPA2 receptor is abundantly expressed in MM1 cells (Gupte et al., 2011). Thus, we posed the question whether GRI977143-mediated activation of LPA2 could stimulate the invasion of HUVEC monolayers by MM1 cells. Our results showed that whereas 1 μM LPA caused a significant increase in MM1 cell invasion, a higher 10 μM concentration of GRI977143 was required to elicit the same significant increase in invasion (Fig. 4).
Fig. 4.
Effect of GRI977143 on the invasion of HUVEC monolayers by MM1 hepatocarcinoma cells. Data are the means of five nonoverlapping fields and are representative of two independent experiments (*, p ≤ 0.05; ***, p ≤ 0.001).
Effect of GRI977143 on LPA2-Mediated Protection against Doxorubicin-Induced Apoptosis.
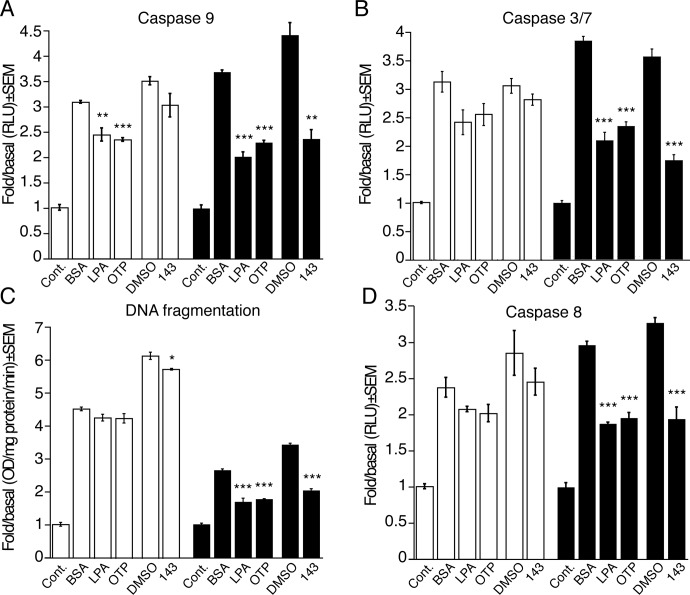
We examined the antiapoptotic properties of GRI977143 using doxorubicin to induce apoptosis. GRI977143 (10 μM) decreased caspase 9 activation in LPA2-transduced MEF cells by 46 ± 4%; this decrease was similar in its magnitude to that of 1 μM LPA, whereas 1 μM OTP resulted in a slightly smaller 38 ± 1% decrease (Fig. 5A). GRI977143 did not affect caspase 9 activation in the vector-transduced cells, whereas LPA and OTP even at a 1 μM concentration reduced caspase 9 activation by 20 to 24% (Fig. 5A). To guide our dosing considerations in the apoptosis assays, we also tested the dose-response relationship of our test compounds on doxorubicin-induced caspase 3 and 7 activation in vector- and LPA2-transduced MEF cells. In the LPA2-transduced MEF cells, GRI977143 elicited a dose-dependent and significant protection at ≥3 μM (p < 0.01; Supplemental Fig. 1). LPA and OTP dose dependently protected LPA2-transduced MEF cells starting from a concentration as low as 30 nM; however, at the highest 10 μM concentration tested, LPA also had an inhibitory effect in the vector-transduced cells (Supplemental Fig. 1, A and B). In contrast, when applied at 10 μM, GRI977143 and OTP did not attenuate caspase 3 or caspase 7 in the vector-transduced cells (Supplemental Fig. 1A). When applied at a 10 μM concentration, GRI977143 reduced caspase 3 and 7 activation on LPA2-transduced MEF cells by 51 ± 3% and was approximately as potent as 3 μM LPA or OTP (Fig. 5B; Supplemental Fig. 1).
Fig. 5.
Effects of LPA and OTP (A and D: 1 μM; B and C: 3 μM) and GRI977143 (10 μM) on doxorubicin-induced apoptotic signaling in vector-transduced (□) or LPA2-transduced (■) MEF cells. Bars represent the mean of triplicate wells, and the data are representative of three independent experiments. (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). Cont., control; BSA, bovine serum albumin; DMSO, dimethyl sulfoxide; 143, GRI977143; RLU, relative light units.
To further characterize the effect of GRI977143 on apoptosis, we measured doxorubicin-induced DNA fragmentation in vector- and LPA2-transduced MEF cells. In LPA2-transduced MEF cells, GRI977143 reduced DNA fragmentation by 41 ± 2% (p < 0.001) compared with a modest 7 ± 1% protection in the vector-transduced cells (p < 0.05); 3 μM LPA and 3 μM OTP also protected LPA2-transduced MEF cells by decreasing DNA fragmentation by 35 ± 4 and 32 ± 1%, respectively (Fig. 5C).
We also examined the effect of GRI977143 on caspase 8 activation in the doxorubicin-induced apoptosis model. Administration of 10 μM GRI977143 resulted in a 41 ± 5% decrease in caspase 8 activation in LPA2-transduced MEF cells. Treatments with 1 μM LPA or 1 μM OTP decreased caspase 8 activation by 36 ± 1 and 33 ± 2%, respectively. A similar but lesser effect of LPA and OTP was noted in the vector-transduced cells, amounting to 12 ± 2 and 15 ± 5% decreases, respectively (Fig. 5D). These findings together establish that selective activation of LPA2 receptor signaling by GRI977143 protects against doxorubicin-induced apoptosis by inhibiting caspase 3, 7, 8, and 9 and reducing DNA fragmentation.
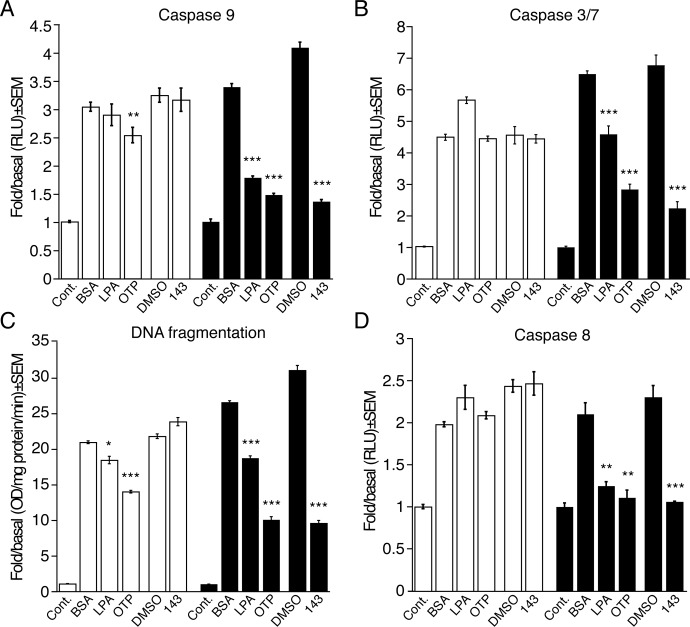
GRI977143 Reduces Apoptosis Induced by Serum Withdrawal in MEF Cells.
We also examined whether GRI977143 could provide the necessary trophic support to serum-starved MEF cells expressing or lacking LPA2 receptors. Experiments with this paradigm showed that 10 μM GRI977143 was highly effective in reducing caspase 3, 7, 8, and 9 activation and also attenuated DNA fragmentation (Fig. 6). GRI977143 failed to cause any reduction in these apoptotic indicators in vector-transduced MEF cells. In contrast, LPA and OTP protected the vector-transduced MEF cells too. These results mirrored our findings in the doxorubicin-induced apoptosis paradigm, extending the role of LPA2 activation to the prevention of serum withdrawal-induced apoptosis.
Fig. 6.
Effects of LPA (A and B: 3 μM; C and D: 10 μM), OTP (3 μM), and GRI977143 (10 μM) on serum withdrawal-induced apoptotic signaling in vector-transduced (□) or LPA2-transduced (■) MEF cells. Bars represent the mean of triplicate wells, and the data are representative of three independent experiments. (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). Cont., control; BSA, bovine serum albumin; DMSO, dimethyl sulfoxide; 143, GRI977143;RLU, relative light units.
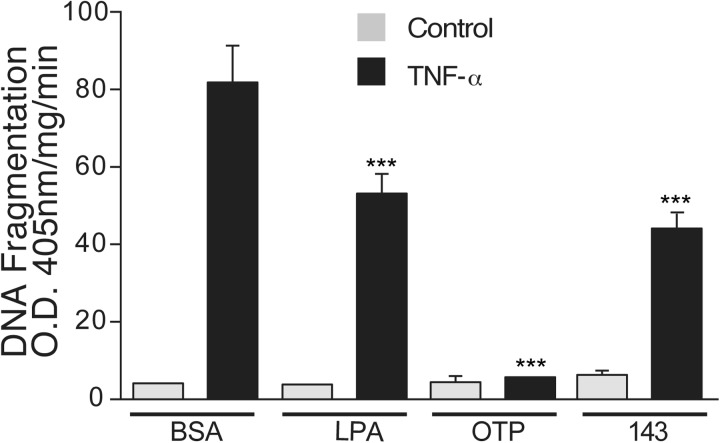
GRI977143 Inhibits TNF-α-Induced Apoptosis in IEC-6 Cells.
We showed earlier that LPA and OTP protects and rescues nontransformed crypt-like IEC-6 cells from TNF-α-induced apoptosis (Deng et al., 2002, 2003, 2004, 2007). IEC-6 cells endogenously express LPA1/2/3/4 GPCRs, GPR87, and P2Y5 (Valentine et al., 2010). Thus, we tested the effect of GRI977143 in this model of extrinsic apoptosis. Treatment with TNF-α/CHX increased DNA fragmentation more than 20-fold; the fragmentation was completely blocked by 10 μM OTP and significantly reduced by 1 μM LPA or 10 μM GRI977143 treatment (Fig. 7). Neither LPAR agonist caused any detectable change in DNA fragmentation when added to the cultures in the absence of TNF-α/CHX. These results extend our previous observations obtained in the doxorubicin-induced intrinsic apoptosis model to the TNF-α-induced model mediated via the extrinsic apoptosis pathway.
Fig. 7.
Effects of LPA (1 μM), OTP (10 μM), and GRI977143 (10 μM) on DNA fragmentation elicited via extrinsic apoptosis induced by TNF-α and CHX treatment in IEC-6 cells. Bars represent the mean of triplicate wells, and the data are representative of two experiments. BSA, bovine serum albumin; 143, GRI977143. (***, p ≤ 0.01).
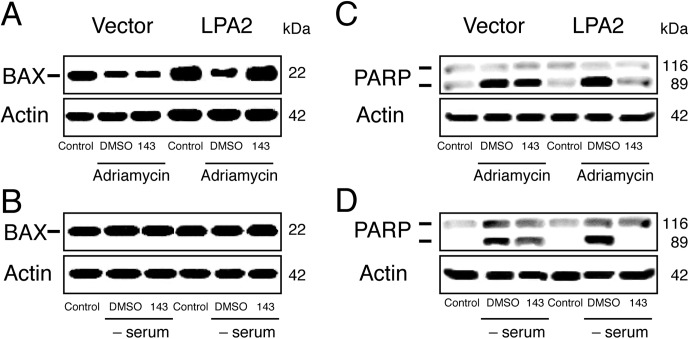
Effect of GRI977143 on Bax Translocation and PARP-1 Cleavage Induced by Doxorubicin or Serum Withdrawal.
Because GRI977143 reduced activation of caspases 3, 7, 8, and 9, we tested the effect of 10 μM GRI977143 on Bax translocation to the mitochondria induced by doxorubicin or serum withdrawal. As shown in Fig. 8A, 10 μM GRI977143 treatment maintained a high level of Bax in the cytoplasm of LPA2-transduced MEF cells after doxorubicin treatment, consequently reducing its translocation to the mitochondria. GRI977143 failed to reduce Bax translocation in the vector-transduced MEFs. In the serum withdrawal model of apoptosis we did not detect any change in the cytosolic Bax level (Fig. 8B).
Fig. 8.
Effects of GRI977143 on cytoplasmic Bax levels and PARP-1 cleavage in vector- or LPA2-transduced MEF cells after doxorubicin- or serum withdrawal-induced apoptosis. The Western blots shown are representative of three experiments. Adriamycin, doxorubicin; DMSO, dimethyl sulfoxide; 143, GRI977143.
GRI977143 treatment (10 μM) also reduced PARP-1 cleavage after both apoptosis-inducing treatments (Fig. 8, C and D). This effect was not observed in the vector-transduced cells. These experiments are consistent with the hypothesis that GRI977143 attenuates the activation of the mitochondrial apoptosis pathway through a mechanism that requires the LPA2 receptor.
Effect of GRI977143 on ERK1/2 Activation.
To elucidate some of the molecular mechanisms responsible for the antiapoptotic effect of GRI977143, we investigated its effect on the activation of ERK1/2 kinases, which is a required step in LPA2 receptor-mediated antiapoptotic signaling (Deng et al., 2002, 2003; E et al., 2009). Treatment with 10 μM GRI977143 for 10 min increased ERK1/2 activation 9.6-fold in LPA2-transduced MEF cells but did not alter the basal activity of these kinases in the vector-transduced cells (Fig. 9, A and B). This result supports the hypothesis that the prosurvival effect of GRI977143 detected in different intrinsic and extrinsic apoptosis models is mediated by the LPA2 receptor and involves ERK1/2 activation.
Fig. 9.
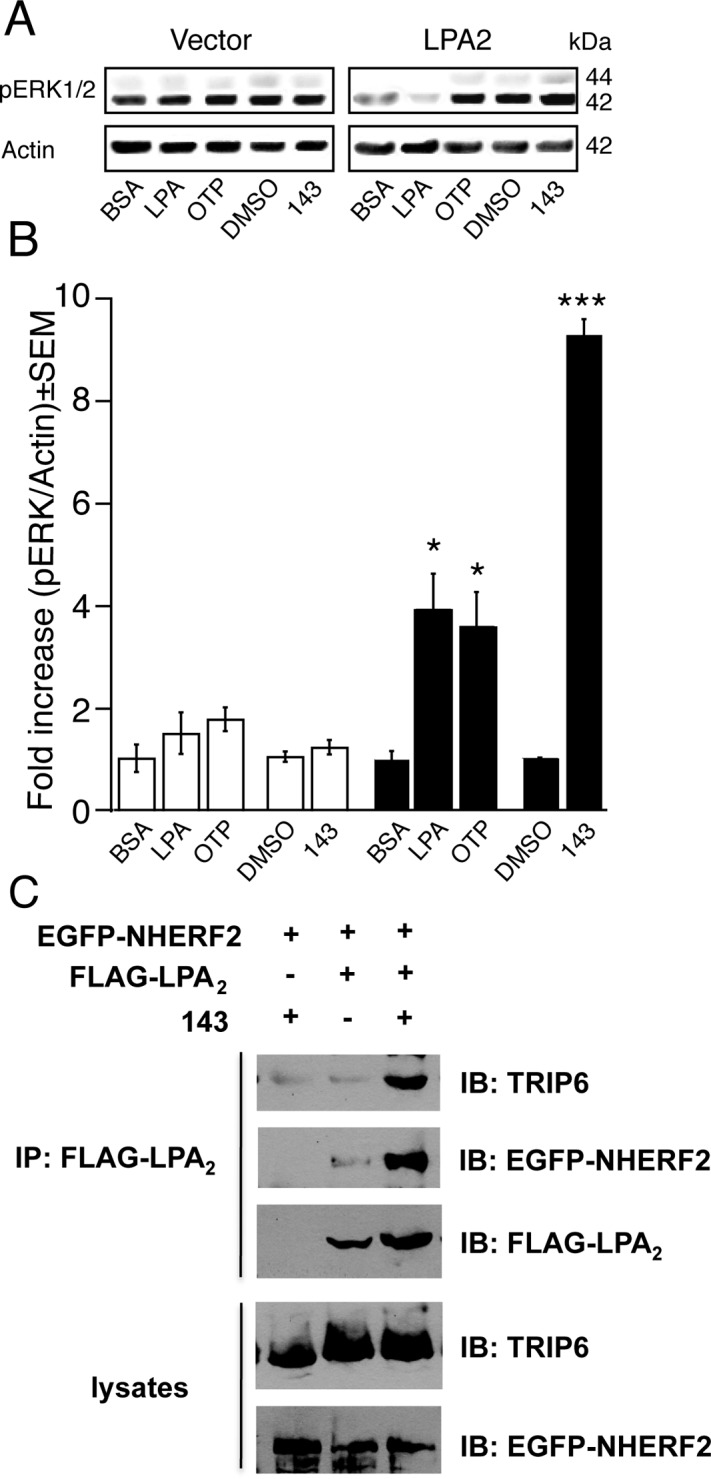
Signaling pathways activated by LPA (1 μM), OTP (1 μM), or GRI977143 (10 μM). Representative Western blots (A) and densitometry (B) of the mean ERK1/2 activation in vector-transduced (□) and LPA2-transduced (■) MEF cells after GRI977143 treatment. Data were normalized for equal loading based on actin and are representative of three independent experiments (*, p ≤ 0.05; ***, p ≤ 0.001). C, GRI977143 elicits macromolecular complex assembly between FLAG-LPA2, EGFP-NHERF2, and endogenous TRIP6. The blot shown is representative of two cotransfection experiments. BSA, bovine serum albumin; DMSO, dimethyl sulfoxide; 143, GRI977143; IP, immunoprecipitation; IB, immunoblot.
Effect of GRI977143 on the Assembly of a Macromolecular Complex between LPA2, TRIP6, and NHERF2.
LPA2 receptor-mediated supramolecular complex formation is required for the protection against doxorubicin-induced apoptosis (E et al., 2009). To further elucidate molecular mechanisms activated by GRI977143, we investigated its effect on agonist-induced signalosome assembly between TRIP6, NHERF2, and the C terminus of LPA2. This macromolecular complex plays an important role in the antiapoptotic effect via stimulation of the ERK1/2 and protein kinase B (Akt)-nuclear factor-κB survival pathways. GRI977143 elicited the assembly of the macromolecular complex indicated by the recruitment of TRIP6 and EGFP-NHERF2 to the LPA2 receptor (Fig. 9C). Only trace amounts of the ternary complex were detected in the vehicle-treated cell lysates, indicating that activation of LPA2 by GRI977143 elicited the assembly of the signaling complex.
Discussion
The growth factor-like actions and its simple chemical structure make LPA an ideal candidate for drug discovery. A major obstacle in development of LPA analogs is their high degree of hydrophobicity that makes these agents nonideal drug candidates. Another complicating factor is the multiplicity of LPA GPCRs, which represents a significant challenge to the development of compounds specific to a single target such as LPA2. Our group has been developing and validating computational models of the putative ligand binding pockets of LPA GPCRs (Parrill et al., 2000; Wang et al., 2001; Fujiwara et al., 2005, 2007; Valentine et al., 2008). Our previous work, aimed at the virtual discovery of LPA1-specific compounds, has serendipitously identified NSC12404, which is a weak but specific agonist of LPA2 (Perygin, 2010). In the present study, we used this hit for virtual screening of the Genome Research Institute and H2L chemical libraries. This approach identified three new selective nonlipid LPA2 agonists: GRI977143, H2L5547924, and H2L5828102 (Table 1).
We selected GRI977143 for initial characterization in cell-based assays and compared its pharmacological and signaling properties with those of LPA and the previously identified LPA mimic OTP. The other compounds, NSC12404, H2L5547924, and H2L5828102, might be worthy of detailed characterization and synthetic improvements in the future. GRI977143 was a specific agonist of only LPA2 when tested for agonist or antagonist activity at up to a 10 μM concentration at five established and two putative LPA GPCRs. It is noteworthy that this compound at >10 μM also showed modest partial inhibition of LPA3. One of our strategies used MEF cells derived from LPA1&2 double-knockout mice (Lin et al., 2007). The parental MEF cells do not express functional LPA1/2/3 receptors but express LPA4/5/6 transcripts. Thus, these MEF cells can be considered an LPA receptor-null host cell line for LPA1/2/3, which belong to the endothelial differentiation gene family of LPA receptors. Knock in of LPA2 rendered these MEF cells responsive to LPA with pharmacological properties similar to those of LPA2 established in other cell types endogenously expressing this receptor subtype (Lin et al., 2007; E et al., 2009). The modest but sometimes significant antiapoptotic responses that were elicited in the vector-transduced MEF cells in response to LPA and OTP are probably due to activation by LPA4/5/6 receptors. GRI977143 had no effect in the vector-transduced MEF cells with the exception of a minimal reduction in DNA fragmentation in the doxorubicin model of apoptosis (Fig. 5C). There was no such detectable effect of GRI977143 in the serum withdrawal-induced apoptosis model (Fig. 6C). We do not know the reason for the effect of GRI977143 on DNA fragmentation in the control MEF cells in the doxorubicin model only, but it might be due to some yet unknown off-target effect of the compound. Certainly, the lack of effect of GRI977143 in vector-transduced MEF cells on Ca2+ mobilization (data not shown), on caspases 3, 7, 8, and 9, on DNA fragmentation, and on ERK1/2 activation is consistent with the hypothesis that specific activation of LPA2 is responsible for these same responses that we consistently detected in the MEF cells expressing the LPA2 receptor. It is also important to recognize that GRI977143 protected IEC-6 cells, which endogenously express multiple LPA GPCR subtypes, from apoptosis. This result is the first evidence that we know of in the literature indicating that specific activation of LPA2 is sufficient to evoke an antiapoptotic effect, and this effect is not limited to the LPA2 knock-in MEF cells.
We also showed that specific stimulation of the LPA2 receptor subtype promotes cell growth (Fig. 3). This is the first pharmacological evidence that this receptor subtype mediates mitogenesis. Surprisingly, the LPA receptor panagonist OTP and GRI977143 had equally robust activity on cell proliferation. We note that OTP and GRI977143 after 3 days also promoted the growth of vector-transduced MEF cells, which might be due to off-target or indirect effects. We cannot exclude the possibility that OTP and GRI977143 somehow potentiated the effect of the 1.5% serum present in the medium. There might be differences in the pharmacokinetic properties of these ligands, which could explain the differences we noted. Future experiments will have to address the difference effects on cell growth observed between these ligands.
LPA has been shown to promote cancer cell invasion and metastasis. We tested the effect of GRI977143 in an in vitro invasion model that has been considered a realistic model of metastasis (Mukai et al., 2000, 2005; Uchiyama et al., 2007). Stimulation of MM1 hepatocarcinoma cells with GRI977143 elicited a dose-dependent increase in the number of cells that penetrated the HUVEC monolayer (Fig. 4). However, this effect, although significant at a 10 μM concentration of GRI977143, was modest compared with that of LPA. The MM1 cells express LPA2 ≫ LPA1 > LPA6 > LPA5 > LPA4 transcripts, whereas HUVECs express LPA5 ≫ LPA4 > GPR87 ∼ LPA1 > LPA2 transcripts determined by quantitative RT-PCR (S.-C. Lee and G. Tigyi, unpublished data). The increase in GRI977143-induced invasion of MM1 cells is likely to represent the effect of selective stimulation of LPA2 in the invading MM1 cells rather than in HUVECs because of the very low expression of this receptor subtype in the cells of the monolayer (Gupte et al., 2011). Thus, our present results provide new pharmacological evidence that activation of LPA2 can promote invasion and metastasis.
Studies have already established the role of the LPA2 receptor in protecting cells from programmed cell death (Deng et al., 2002; Lin et al., 2007; Taghavi et al., 2008; Yu et al., 2008; E et al., 2009; Sun et al., 2010). The LPA2-specific agonist properties of GRI977143 allowed us to test this hypothesis in the LPA2 knock-in MEF cells and in IEC-6 cells, the latter of which endogenously express multiple LPA GPCRs (Deng et al., 2002, 2003, 2004, 2007). GRI977143 reduced the activation of the executioner caspases 3 and 7 and upstream regulator caspases 8 and 9. Attenuation of these caspases explains the reduction of PARP-1 cleavage and DNA fragmentation we observed. These mechanisms were common to GRI977143 regardless of whether apoptosis was elicited via the extrinsic pathway or the intrinsic pathway (Figs. 5, 6, and 7). Both LPA and OTP activate these same mechanisms but also activate multiple LPA GPCRs (Deng et al., 2002, 2003, 2004, 2007). Thus, we propose that specific activation of LPA2 is sufficient to protect cells from apoptosis. The specific agonist properties of GRI977143 might represent an advantage over LPA and other receptor nonselective LPA mimics that also stimulate LPA1 receptor subtype activation, which has been shown to promote cell death via anoikis in tumor cells (Furui et al., 1999), in cardiac myocytes (Chen et al., 2006), and in pulmonary epithelial cells (Funke et al., 2012).
LPA2-mediated activation of the ERK1/2 prosurvival kinases is a required event in antiapoptotic signaling (Deng et al., 2004; Lin et al., 2007; E et al., 2009). Consistent with our previous results obtained with LPA and OTP (Deng et al., 2007; Lin et al., 2007; E et al., 2009), GRI977143 treatment resulted in a robust ERK1/2 activation. We have previously shown that in addition to the Gi protein-mediated signals demonstrated by the partial pertussis toxin sensitivity of the effect (Deng et al., 2002, 2004), the LPA2-mediated antiapoptotic effect requires additional ligand-induced assembly of a C-terminal macromolecular complex consisting of LPA2, TRIP6, and a homodimer of NHERF2 (Lai et al., 2005; Lin et al., 2007; E et al., 2009). Activation of LPA2 regulates the c-Src-mediated phosphorylation of TRIP6 at the Tyr55 and Pro58 residue, which in turn promotes LPA-induced ERK1/2 activation (Lai et al., 2005). We found that GRI977143 elicited the assembly of this signalosome (Fig. 9C), which can explain the concomitant robust ERK1/2 activation (Fig. 9, A and B).
Taken altogether, the present findings indicate that nonlipid LPA2-specific agonists, such as those described here, represent an excellent starting point for the development of lead compounds with potential therapeutic utility for the prevention of programmed cell death involved in many types of degenerative and inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Drs. Jerold Chun (The Scripps Research Institute, La Jolla, CA) for providing the LPA1 and LPA2 mice, Fumikazu Okajima, Takao Shimizu, Norihisha Fujita, and Michiko Mukai for providing the different cell types used in the study, A. P. Naren for providing the anti-EGFP antibody, and William Seibel (University of Cincinnati, Cincinnati, OH) for his assistance with the similarity search of the UC-DCC library. The donation of the MOE program by the Chemical Computing Group and is greatly appreciated.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This research was supported by the National Institutes of Health National Institute of Allergy [Medical Countermeasures Radiological and Nuclear Threats Program AI80405].
G.T. is a founder of RxBio, Inc.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- LPA
- lysophosphatidic acid
- S1P
- sphingosine-1-phosphate
- OTP
- octadecenyl thiophosphate
- GPCR
- G protein-coupled receptor
- Ki16425
- 3-(4-[4-([1-(2-chlorophenyl)ethoxy]carbonyl amino)-3-methyl-5-isoxazolyl] benzylsulfanyl) propanoic acid
- AM152
- (R)-1-phenylethyl-5-(4-biphenyl-4-cyclopropanecarboxylic acid)-3-methylisoxazole-4-yl carbamate sodium salt
- AM095
- sodium, (4′-(3-methyl-4-((R)-1-phenylethoxycarbonylamino)-isoxazol-5-yl)-biphenyl-4-yl)-acetate
- GRI977143
- 2-((3-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)propyl)thio)benzoic acid
- GRI
- Genome Research Institute
- Bax
- Bcl-2-associated X protein
- PARP-1
- poly(ADP-ribose)polymerase 1
- ERK1/2
- extracellular signal regulated kinases 1/2
- TRIP6
- thyroid receptor interacting protein 6
- NHERF2
- Na+-H+ exchange regulatory factor 2
- NSC12404
- 2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid
- H2L
- Hit2Lead
- H2L5547924
- 4,5-dichloro-2-((9-oxo-9H-fluoren-2-yl)carbamoyl)benzoic acid
- H2L5828102
- 2-((9,10-dioxo-9,10-dihydroanthracen-2-yl)carbamoyl)benzoic acid
- PBS
- phosphate-buffered saline
- MOE
- Molecular Operating Environment
- UC-DDC
- University of Cincinnati Drug Discovery Center
- TM
- transmembrane
- MEF
- mouse embryonic fibroblast
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- IEC-6
- intestinal epithelial cell line 6
- RH7777
- McArdle rat hepatoma cell line
- LPAR
- LPA receptor
- HUVEC
- human umbilical vein endothelial cell
- TNF-α
- tumor necrosis factor α
- CHX
- cycloheximide
- EGFP
- enhanced green fluorescent protein.
Authorship Contributions
Participated in research design: Kiss, Fells, Gupte, Lee, Liu, Nusser, Ray, Lin, Parrill, Sümegi, Miller, and Tigyi.
Conducted experiments: Kiss, Fells, Gupte, Lee, Liu, Nusser, Lim, Ray, Lin, Parrill, and Tigyi.
Performed data analysis: Kiss, Fells, Gupte, Lee, Liu, Nusser, Lim, Ray, Lin, Parrill, Miller, and Tigyi.
Wrote or contributed to the writing of the manuscript: Kiss, Fells, Lee, Nusser, Lim, Ray, Lin, Parrill, and Tigyi.
References
- Ballesteros JA, Weinstein H. (1995) Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci 25:366–425 [Google Scholar]
- Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, Aradhye S, Burtin P. (2010) Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat Rev Drug Discov 29:883–897 [DOI] [PubMed] [Google Scholar]
- Chen J, Han Y, Zhu W, Ma R, Han B, Cong X, Hu S, Chen X. (2006) Specific receptor subtype mediation of LPA-induced dual effects in cardiac fibroblasts. FEBS Lett 580:4737–4745 [DOI] [PubMed] [Google Scholar]
- Deng W, Balazs L, Wang DA, Van Middlesworth L, Tigyi G, Johnson LR. (2002) Lysophosphatidic acid protects and rescues intestinal epithelial cells from radiation- and chemotherapy-induced apoptosis. Gastroenterology 123:206–216 [DOI] [PubMed] [Google Scholar]
- Deng W, Poppleton H, Yasuda S, Makarova N, Shinozuka Y, Wang DA, Johnson LR, Patel TB, Tigyi G. (2004) Optimal lysophosphatidic acid-induced DNA synthesis and cell migration but not survival require intact autophosphorylation sites of the epidermal growth factor receptor. J Biol Chem 279:47871–47880 [DOI] [PubMed] [Google Scholar]
- Deng W, Shuyu E, Tsukahara R, Valentine WJ, Durgam G, Gududuru V, Balazs L, Manickam V, Arsura M, VanMiddlesworth L, et al. (2007) The lysophosphatidic acid type 2 receptor is required for protection against radiation-induced intestinal injury. Gastroenterology 132:1834–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Wang DA, Gosmanova E, Johnson LR, Tigyi G. (2003) LPA protects intestinal epithelial cells from apoptosis by inhibiting the mitochondrial pathway. Am J Physiol Gastrointest Liver Physiol 284:G821–G829 [DOI] [PubMed] [Google Scholar]
- Durgam GG, Tsukahara R, Makarova N, Walker MD, Fujiwara Y, Pigg KR, Baker DL, Sardar VM, Parrill AL, Tigyi G, et al. (2006) Synthesis and pharmacological evaluation of second-generation phosphatidic acid derivatives as lysophosphatidic acid receptor ligands. Bioorg Med Chem Lett 16:633–640 [DOI] [PubMed] [Google Scholar]
- E S, Lai YJ, Tsukahara R, Chen CS, Fujiwara Y, Yue J, Yu JH, Guo H, Kihara A, Tigyi G, et al. (2009) Lysophosphatidic acid 2 receptor-mediated supramolecular complex formation regulates its antiapoptotic effect. J Biol Chem 284:14558–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer DJ, Nusser N, Virag T, Yokoyama K, Wang Da, Baker DL, Bautista D, Parrill AL, Tigyi G. (2001) Short-chain phosphatidates are subtype-selective antagonists of lysophosphatidic acid receptors. Mol Pharmacol 60:776–784 [PubMed] [Google Scholar]
- Fujiwara Y, Osborne DA, Walker MD, Wang DA, Bautista DA, Liliom K, Van Brocklyn JR, Parrill AL, Tigyi G. (2007) Identification of the hydrophobic ligand binding pocket of the S1P1 receptor. J Biol Chem 282:2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Sardar V, Tokumura A, Baker D, Murakami-Murofushi K, Parrill A, Tigyi G. (2005) Identification of residues responsible for ligand recognition and regioisomeric selectivity of lysophosphatidic acid receptors expressed in mammalian cells. J Biol Chem 280:35038–35050 [DOI] [PubMed] [Google Scholar]
- Funke M, Zhao Z, Xu Y, Chun J, Tager AM. (2012) The lysophosphatidic acid receptor LPA1 promotes epithelial cell apoptosis after lung injury. Am J Respir Cell Mol Biol 46:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furui T, LaPushin R, Mao M, Khan H, Watt SR, Watt MA, Lu Y, Fang X, Tsutsui S, Siddik ZH, et al. (1999) Overexpression of edg-2/vzg-1 induces apoptosis and anoikis in ovarian cancer cells in a lysophosphatidic acid-independent manner. Clin Cancer Res 5:4308–4318 [PubMed] [Google Scholar]
- Gupte R, Patil R, Liu J, Wang Y, Lee SC, Fujiwara Y, Fells J, Bolen AL, Emmons-Thompson K, Yates CR, et al. (2011) Benzyl and naphthalene methylphosphonic acid inhibitors of autotaxin with anti-invasive and anti-metastatic activity. ChemMedChem 6:922–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, et al. (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YJ, Chen CS, Lin WC, Lin FT. (2005) c-Src-mediated phosphorylation of TRIP6 regulates its function in lysophosphatidic acid-induced cell migration. Mol Cell Biol 25:5859–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YJ, Lin WC, Lin FT. (2007) PTPL1/FAP-1 negatively regulates TRIP6 function in lysophosphatidic acid-induced cell migration. J Biol Chem 282:24381–24387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Dandridge KS, Di A, Marrs KL, Harris EL, Roy K, Jackson JS, Makarova NV, Fujiwara Y, Farrar PL, et al. (2005) Lysophosphatidic acid inhibits cholera toxin-induced secretory diarrhea through CFTR-dependent protein interactions. J Exp Med 202:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FT, Lai YJ, Makarova N, Tigyi G, Lin WC. (2007) The lysophosphatidic acid 2 receptor mediates down-regulation of Siva-1 to promote cell survival. J Biol Chem 282:37759–37769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA. (2003) Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Discov Today 8:12–16 [DOI] [PubMed] [Google Scholar]
- Chemical Computing Group (2002) Molecular Operating Environment, Chemical Computing Group, Montreal, QC, Canada [Google Scholar]
- Mukai M, Akedo H. (1999) Induction of tumor invasion by lysophosphatidic acid and inhibition of tumor invasion by cyclic phosphatidic acid. Tanpakushitsu Kakusan Koso 44 (8 suppl):1126–1131 [PubMed] [Google Scholar]
- Mukai M, Iwasaki T, Tatsuta M, Togawa A, Nakamura H, Murakami-Murofushi K, Kobayashi S, Imamura F, Inoue M. (2003) Cyclic phosphatidic acid inhibits RhoA-mediated autophosphorylation of FAK at Tyr-397 and subsequent tumor-cell invasion. Int J Oncol 22:1247–1256 [PubMed] [Google Scholar]
- Mukai M, Kusama T, Hamanaka Y, Koga T, Endo H, Tatsuta M, Inoue M. (2005) Cross talk between apoptosis and invasion signaling in cancer cells through caspase-3 activation. Cancer Res 65:9121–9125 [DOI] [PubMed] [Google Scholar]
- Mukai M, Nakamura H, Tatsuta M, Iwasaki T, Togawa A, Imamura F, Akedo H. (2000) Hepatoma cell migration through a mesothelial cell monolayer is inhibited by cyclic AMP-elevating agents via a Rho-dependent pathway. FEBS Lett 484:69–73 [DOI] [PubMed] [Google Scholar]
- Murakami M, Shiraishi A, Tabata K, Fujita N. (2008) Identification of the orphan GPCR, P2Y10 receptor as the sphingosine-1-phosphate and lysophosphatidic acid receptor. Biochem Biophys Res Commun 371:707–712 [DOI] [PubMed] [Google Scholar]
- Ohta H, Sato K, Murata N, Damirin A, Malchinkhuu E, Kon J, Kimura T, Tobo M, Yamazaki Y, Watanabe T, et al. (2003) Ki16425, a subtype-selective antagonist for EDG-family lysophosphatidic acid receptors. Mol Pharmacol 64:994–1005 [DOI] [PubMed] [Google Scholar]
- Parrill AL, Wang D, Bautista DL, Van Brocklyn JR, Lorincz Z, Fischer DJ, Baker DL, Liliom K, Spiegel S, Tigyi G. (2000) Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. J Biol Chem 275:39379–39384 [DOI] [PubMed] [Google Scholar]
- Perygin DH. (2010) Identification of non-lipid LPA receptor agonists and antagonists through in silico screening, in Chemistry, p 130, University of Memphis, Memphis, TN [Google Scholar]
- Ray RM, Bhattacharya S, Johnson LR. (2011) Mdm2 inhibition induces apoptosis in p53 deficient human colon cancer cells by activating p73- and E2F1-mediated expression of PUMA and Siva-1. Apoptosis 16:35–44 [DOI] [PubMed] [Google Scholar]
- Sardar VM, Bautista DL, Fischer DJ, Yokoyama K, Nusser N, Virag T, Wang DA, Baker DL, Tigyi G, Parrill AL. (2002) Molecular basis for lysophosphatidic acid receptor antagonist selectivity. Biochim Biophys Acta 1582:309–317 [DOI] [PubMed] [Google Scholar]
- Sun Y, Nam JS, Han DH, Kim NH, Choi HK, Lee JK, Rhee HJ, Huh SO. (2010) Lysophosphatidic acid induces upregulation of Mcl-1 and protects apoptosis in a PTX-dependent manner in H19–7 cells. Cell Signal 22:484–494 [DOI] [PubMed] [Google Scholar]
- Swaney JS, Chapman C, Correa LD, Stebbins KJ, Broadhead AR, Bain G, Santini AM, Darlington J, King CD, Baccei CS, et al. (2011) Pharmacokinetic and pharmacodynamic characterization of an oral lysophosphatidic acid type 1 receptor-selective antagonist. J Pharmacol Exp Ther 336:693–700 [DOI] [PubMed] [Google Scholar]
- Tabata K, Baba K, Shiraishi A, Ito M, Fujita N. (2007) The orphan GPCR GPR87 was deorphanized and shown to be a lysophosphatidic acid receptor. Biochem Biophys Res Commun 363:861–866 [DOI] [PubMed] [Google Scholar]
- Taghavi P, Verhoeven E, Jacobs JJ, Lambooij JP, Stortelers C, Tanger E, Moolenaar WH, van Lohuizen M. (2008) In vitro genetic screen identifies a cooperative role for LPA signaling and c-Myc in cell transformation. Oncogene 27:6806–6816 [DOI] [PubMed] [Google Scholar]
- Tigyi G. (2010) Aiming drug discovery at lysophosphatidic acid targets. Br J Pharmacol 161:241–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigyi G, Dyer DL, Miledi R. (1994) Lysophosphatidic acid possesses dual action in cell proliferation. Proc Natl Acad Sci USA 91:1908–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A, Mukai M, Fujiwara Y, Kobayashi S, Kawai N, Murofushi H, Inoue M, Enoki S, Tanaka Y, Niki T, et al. (2007) Inhibition of transcellular tumor cell migration and metastasis by novel carba-derivatives of cyclic phosphatidic acid. Biochim Biophys Acta 1771:103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine WJ, Fells JI, Perygin DH, Mujahid S, Yokoyama K, Fujiwara Y, Tsukahara R, Van Brocklyn JR, Parrill AL, Tigyi G. (2008) Subtype-specific residues involved in ligand activation of the endothelial differentiation gene family lysophosphatidic acid receptors. J Biol Chem 283:12175–12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine WJ, Kiss GN, Liu J, E S, Gotoh M, Murakami-Murofushi K, Pham TC, Baker DL, Parrill AL, Lu X, et al. (2010) (S)-FTY720-vinylphosphonate, an analogue of the immunosuppressive agent FTY720, is a pan-antagonist of sphingosine 1-phosphate GPCR signaling and inhibits autotaxin activity. Cell Signal 22:1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DA, Lorincz Z, Bautista DL, Liliom K, Tigyi G, Parrill AL. (2001) A single amino acid determines lysophospholipid specificity of the S1P1 (EDG1) and LPA1 (EDG2) phospholipid growth factor receptors. J Biol Chem 276:49213–49220 [DOI] [PubMed] [Google Scholar]
- Williams JR, Khandoga AL, Goyal P, Fells JI, Perygin DH, Siess W, Parrill AL, Tigyi G, Fujiwara Y. (2009) Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J Biol Chem 284:17304–17319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lai YJ, Lin WC, Lin FT. (2004) TRIP6 enhances lysophosphatidic acid-induced cell migration by interacting with the lysophosphatidic acid 2 receptor. J Biol Chem 279:10459–10468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Murph MM, Lu Y, Liu S, Hall HS, Liu J, Stephens C, Fang X, Mills GB. (2008) Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J Natl Cancer Inst 100:1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.