Abstract
O2- and O4-methylthymidine (O2-MdT and O4-MdT) can be induced in tissues of laboratory animals exposed with N-methyl-N-nitrosourea, a known carcinogen. These two O-methylated DNA adducts have been shown to be poorly repaired and may contribute to the mutations arising from exposure to DNA methylating agents. Here, in vitro replication studies with duplex DNA substrates containing site-specifically incorporated O2-MdT and O4-MdT showed that both lesions blocked DNA synthesis mediated by three different DNA polymerases, including the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I (Kf−), human DNA polymerase κ (pol κ) and Saccharomyces cerevisiae DNA polymerase η (pol η). Results from steady-state kinetic measurements and LC-MS/MS analysis of primer extension products revealed that Kf− and pol η preferentially incorporated the correct nucleotide (dAMP) opposite O2-MdT, while O4-MdT primarily directed dGMP misincorporation. While steady-state kinetic experiments showed that pol κ-mediated nucleotide insertion opposite O2-MdT and O4-MdT is highly promiscuous, LC-MS/MS analysis of primer extension products demonstrated that pol κ incorporated favorably the incorrect dGMP opposite both lesions. Our results underscored the limitation of the steady-state kinetic assay in determining how DNA lesions compromise DNA replication in vitro. In addition, the results from our study revealed that, if left unrepaired, O-methylated thymidine lesions may constitute important sources of nucleobase substitutions emanating from exposure to alkylating agents.
Introduction
Low levels of N-nitroso compounds (NOCs) are present in a variety of sources including air, beer, food, water, chewing tobacco and tobacco smoke. Many of these NOCs are known carcinogens in both humans and laboratory animals and, after metabolic activation, they can result in DNA alkylation.1 Alkylation at nitrogen atoms on nucleobases has been shown to induce transversions, frameshift mutations, and small deletions.2 Alkylation at oxygen atoms, albeit occurring to a lesser extent than N-alkylation,3,4 primarily produces point mutations and is linked with the carcinogenic and mutagenic properties of many alkylating agents.5 For larger alkylating agents, the reactivity of oxygen atoms in nucleobases differs, with the O6 position of guanine being the most reactive, followed by the O2 and O4 positions of thymine, and then the O2 position of cytosine.3,6
Different O-alkylated DNA lesions can be repaired with varying efficiencies. For instance, O6-alkylguanine lesions are readily repaired, whereas the O-alkylpyrimidines are removed at much lower rates. In this vein, Escherichia coli O6-methylguanine-DNA methyltransferase (Ada) and mammalian O6-methylguanine-DNA methyltransferase (MGMT) repair O6-MdG 105 and ~103 times faster than O4-methylthymidine (O4-MdT), respectively.7–10 Various alkylated purine and pyrimidine bases, including O2- and O4-alkylthymines, have been found in tissues of rats treated with N,N-dimethylnitrosamine (DMN) or N-ethyl-N-nitrosourea (ENU).11 Interestingly the amount of O4-ethylthymine in tissues was lower than that of O2-ethylthymine, suggesting that the latter might be more resistant to repair.11 The poor repair of O-alkylthymidines may render these lesions highly persistent in the genome, thereby interfering with the cellular replication machinery.
To cope with unrepaired DNA lesions which stall DNA replication, cells are equipped with a number of translesion synthesis (TLS) DNA polymerases capable of replicating past damaged nucleobases.12,13 These specialized polymerases have lower fidelity and processivity than replicative DNA polymerases, largely owing to their more spacious active sites and lack of proofreading 3′→5′ exonuclease activities.14 However, in some instances DNA synthesis mediated by TLS polymerases is both accurate and efficient. For example, the DinB DNA polymerase (i.e., pol κ in mammalian cells and pol IV in Escherichia coli), a Y-family polymerase conserved in all three kingdoms of life, is capable of bypassing accurately and efficiently some minor-groove N2-substituted dG derivatives in vitro and in cells.15–18 Polymerase η, another Y-family DNA polymerase, incorporates, with high efficiency, the correct nucleotide opposite the cis-syn cyclobutane pyrimidine dimers induced by UV light,19–22 The importance of TLS DNA polymerases is manifested by the fact that mutational inactivation of pol η in humans leads to the variant form of xeroderma pigmentosum,23,24 a genetic disease characterized by an elevated predisposition for sunlight-induced skin cancers.25
In addition to their poor repair, several studies have shown that O4- and O2-alkylthymidine are both blocking and miscoding during DNA replication in vitro and in vivo. O4-alkylthymidines have been shown to primarily induce T→C mutations.2,6,10,26–30 The minor-groove lesion, O2-ethylthymidine was found to be highly blocking to the exonuclease-free Klenow fragment of E. coli DNA polymerase I (Kf−) and the lesion directs the polymerase to incorporate both dA and dT.31 In addition, O2-methylthymidine (O2-MdT) and O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dT) are strongly blocking to replication mediated by Kf− and Dpo4 in vitro and the replication machinery of E. coli cells.32–35 Moreover, both lesions exhibit strong miscoding potential in these experimental systems. 32,33
Herein we investigated how O2-MdT and O4-MdT perturb DNA replication by three purified polymerases in vitro. These included the Kf− and two Y-family polymerases, Saccharomyces cerevisiae DNA polymerase η (pol η) and human DNA polymerase κ. We chose these polymerases because Kf− is a widely used model DNA polymerase for examining how DNA lesions perturb DNA replication, pol η is capable of bypassing another major-groove O-alkyl product, O6-methylguanine,36 and pol κ can accurately and efficiently bypass a number of minor-groove N2-dG lesions.15,17,18
Materials and Methods
Materials
All enzymes and chemicals unless otherwise specified were purchased from New England Biolabs (Ipswich, MA) or Sigma-Aldrich (St. Louis, MO). Unmodified oligodeoxyribonucleotides (ODNs) used in this study were purchased from Integrated DNA Technologies (Coralville, IA). [γ-32P]ATP was obtained from Perkin Elmer (Piscataway, NJ). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP) was purchased from TCI America (Portland, OR). The phosphoramidite building block of O4-MdT, conventional phosphoramidites of unmodified nucleosides, and other reagents for solid-phase DNA synthesis were obtained from Glen Research (Sterling, VA). Kf− and human DNA polymerase κ were from New England Biolabs (Ipswich, MA) and Enzymax (Lexington, KY), respectively. Saccharomyces cerevisiae DNA polymerase η was expressed and purified following previously published procedures.37,38
Preparation of Lesion-containing Substrates
The O2-MdT phosphoramidite building block was synthesized following previously published procedures.39 ODNs containing a site-specifically incorporated O2-MdT or O4-MdT [17mer, d(CCATGGCAXGAGAATTC), X = O2-MdT or O4-MdT] were synthesized on a Beckman Oligo 1000S DNA Synthesizer (Fullerton, CA). The ODNs were cleaved from the controlled pore glass (CPG) support, deprotected by treatment with 10% 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) in methanol in the dark at 23°C for five days and purified by 20% denaturing polyacrylamide gel electrophoresis (PAGE). The structures of the ODNs were confirmed by electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) analyses (Figure S1). The 28mer substrates, d(CCATGGCAXGAGAATTCTATGGTCCTAG) (‘X’ = dT, O2-MdT or O4-MdT) were obtained by ligating the above-described 17-mer ODNs with a 5′-phosphorylated d(TATGGTCCTAG) in the presence of a template ODN following previously published procedures.40 The desired lesion-containing 28mer ODNs were purified using PAGE and desalted by ethanol precipitation. The purity of the product was further confirmed by PAGE analysis.
Steady-state Kinetic Measurements – Nucleotide Insertion
In-vitro replication experiments were performed following previously described procedures.41,42 Briefly, the 28mer template, d(CCATGGCAXGAGAATTCTATGGTCCTAG) (‘X’ = dT, O2-MdT or O4-MdT, 20 nM), was annealed with a 5′-32P-labeled 20-mer primer, d(GCTAGGACCATAGAATTCTC) (10 nM) for steady-state kinetic measurements of nucleotide insertion opposite the lesions and unmodified dT. For Kf−, the reactions were carried out in a buffer containing 50 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 2 mM EDTA, 1.6 mM β-mercaptoethanol, and 5 μg/mL BSA. For pol η and pol κ, the reactions were conducted in a buffer containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 7.5 mM DTT.
For steady-state kinetic measurements monitoring the nucleotide insertion opposite the lesion, the primer-template complex was incubated with Kf− (0.7–7 nM), human pol κ (1–5 nM) or yeast pol η (7.5 nM) in the presence of an individual dNTP at varying concentrations. The reaction was continued at 23°C (pol κ) or 37°C (Kf− and pol η) for 10 min in the same reaction buffers as described above, and terminated by adding a 2-volume excess of formamide gel-loading buffer. The buffer contained 80% formamide, 10 mM EDTA (pH 8.0), 1 mg/mL xylene cyanol and 1 mg/mL bromophenol blue. The products were resolved on 20% (29:1) cross-linked polyacrylamide gels containing 8 M urea. Gel band intensities for the substrates and products were quantified using a Typhoon 9410 variable-mode imager (Amersham Biosciences Co.) and ImageQuant version 5.2 (Amersham Biosciences Co.). The dNTP concentration was optimized for different insertion reactions to allow for approximately 20% primer extension.43 The observed rate of dNTP incorporation (Vobs) was plotted versus the dNTP concentration, and the apparent steady-state kinetic parameters (Km and Vmax) for the incorporation of the correct and incorrect nucleotides were determined by fitting the rate data with the Michaelis-Menten equation:
The kcat values were then calculated by dividing the Vmax values with the concentration of the polymerase used. The efficiency of nucleotide incorporation was determined by the ratio of kcat/Km, and the fidelity of nucleotide incorporation was gauged by the frequency of misincorporation (finc), which was calculated using the following equation:44
Steady-state Kinetic Measurements - Nucleotide Extension Mediated by Human pol κ
The dT-, O2-MdT-, or O4-MdT-containing templates (20 nM) were incubated with a 5′-32 P-21 mer primer d(GCTAGGACCATAGAATTCTCN) (10 nM), where N is an A or G. The primer/template complex was incubated in the above-described buffer with pol κ (1–5 nM) and varying concentrations of the correct dTTP at 23°C for 10 min. The reaction was again terminated with formamide gel-loading buffer. The efficiency of extension was calculated by taking the ratio of kcat/Km, while the frequencies of mis-extension were determined by dividing the kcat/Km values for the extension with the incorrect nucleotide over that with the correct nucleotide.
Primer extension assays monitored by gel electrophoresis
For primer extension assays, all four dNTPs and varying concentrations of a DNA polymerase, as indicated in the figures, were subsequently added to the duplex mixture and incubated for 60 min. The reaction was terminated by adding a 2-volume excess of formamide gel-loading buffer. The products were resolved on 20% (29:1) cross-linked polyacrylamide gels containing 8 M urea. Gel band intensities for the substrates and products were quantified by using a Typhoon 9410 variable-mode imager and ImageQuant version 5.2.
Primer extension assays monitored by LC-MS/MS
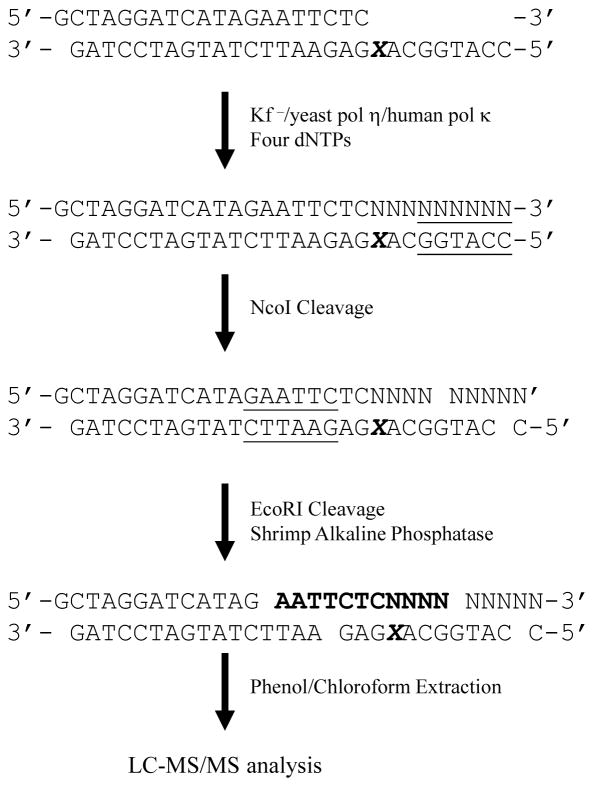
The primer and lesion-containing templates (0.50 μM each) were annealed and incubated overnight in a 50-μL solution with pol κ (0.16 μM) at 23°C, and with pol η (0.24 μM) and Kf− (55 nM) at 37°C. The reactions were conducted in the presence of all four dNTPs (1 mM each) in the same buffers as described above. Two additional primer extension reactions were carried out for the O4-MdT-containing substrate using pol η under the same conditions except that lower concentrations of dNTPs (50 μM each) or a shorter reaction time (6 hrs) was employed. Each replication reaction was subsequently terminated by chloroform extraction and the aqueous layer dried using a Speed-Vac. The resulting replication products were cleaved sequentially with two restriction enzymes as shown in Scheme 1. In this regard, the replication mixture was first incubated with NcoI (40 U) in a buffer containing 50 mM NaCl, 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, and 1 mM DTT for 4 hrs. The NcoI was subsequently removed by chloroform extraction and the aqueous layer dried using a Speed-Vac. EcoRI (50 U) and shrimp alkaline phosphatase (20 U) were then added to the reaction mixture and incubated at 23°C for overnight in a buffer containing 50 mM NaCl, 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, and 1 mM DTT. The proteins in the mixture were again removed by chloroform extraction, and the aqueous layer dried using a Speed-Vac. The dried residue was reconstituted in 50-μL H2O, and a 10-μL aliquot was injected for LC-MS/MS analysis on an Agilent Zorbax SB-C18 column (0.5 × 250 mm, 5 μm in particle size). The gradient for the HPLC elution was 5-min of 5–20% methanol followed by 35-min of 20–50% methanol in 400 mM HFIP (pH adjusted to 7.0). The temperature for the ion-transport tube was maintained at 350°C to minimize the formation of HFIP adducts of the ODNs.
Scheme 1.
To identify the replication products, samples were first analyzed in the data-dependent scan mode, where the most abundant ion found in MS was chosen for fragmentation in MS/MS. The fragment ions found in the MS/MS were manually assigned and the sequences of the ODNs determined. The mass spectrometer was subsequently set up for monitoring specifically the fragmentation of the precursor ions for the extended fragments of the primer strand.
To correct for the varied ionization efficiencies of different ODNs, we constructed calibration curves using mixtures with varying concentrations of the standard synthetic ODNs identified in the reaction mixtures and a constant amount of the d(AATTCTCATGC) (11A, which represents the fully extended, unmutated product). Areas were determined for the peaks found in the selected-ion chromatogram (SIC) for monitoring the formation of three abundant fragment ions for each ODN. The peak areas of individual ODNs were then normalized to that of the 11A and plotted against the molar ratios of these ODNs over 11A to give calibration curves (Figure S2). The corresponding normalized ratios for the replication samples were also determined, from which we measured the molar ratios for each extended product over 11A based on the calibration curves. The percentage of each product was then calculated based on the molar ratios of all products detected in the replication mixture.
Results
Primer extension assay monitored by PAGE
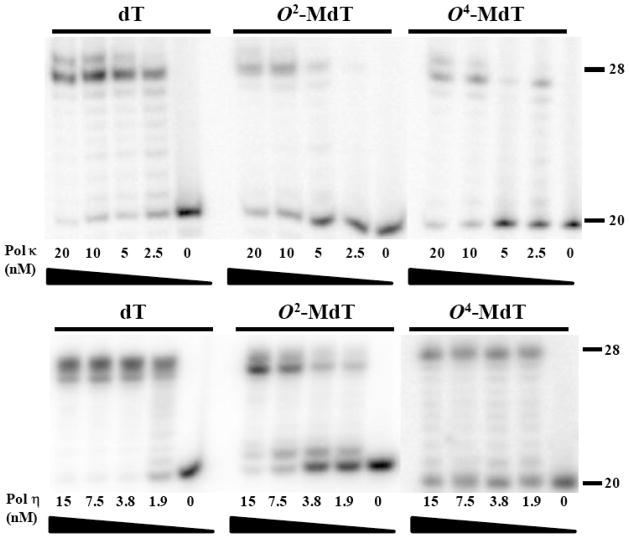
To examine the effects of O2-MdT and O4-MdT on DNA replication, we performed in vitro replication studies using three DNA polymerases, the exonuclease-free Klenow fragment of E. coli DNA polymerase I, human DNA polymerase κ and S. cerevisiae DNA polymerase η. The results showed that, while full-length products were observed with the use of high concentrations of the enzymes, O2-MdT and O4-MdT are moderately blocking to all three polymerases (Figure 1).
Figure 1.
Primer extension assays for O2-MdT- and O4-MdT-bearing substrates and the control undamaged substrate with Kf− (A), human polymerase κ (B), and yeast polymerase η (C). The sequences for the templates are d(CCATGGCAXGAGAATTCTATGATCCTAG) (‘X’ represents dT, O2-MdT or O4-MdT), and a 5′-[32P]-labeled d(GCTAGGATCATAGAATTCTC) was used as the primer.
Steady-state Kinetic Studies
Steady-state kinetic parameters for Kf−-, pol κ- and pol η-mediated nucleotide incorporation opposite O2-MdT, O4-MdT and corresponding unmodified dT in the above substrates were determined. Relative to the unmodified substrate, the efficiencies for Kf− to incorporate the correct dAMP opposite O2-MdT and O4-MdT were reduced, with the kcat/Km values being 140, 5.7, and 2.0 × 10−2 μM−1 min−1 for substrates containing dT, O2-MdT and O4-MdT, respectively (Table 1, Figure S3). Thus, methylation at the O2- and O4-positions of thymidine confers a reduction in efficiency for dAMP insertion by approximately 25 and 7000 folds, respectively. The magnitude of the reduction in efficiency for dAMP insertion opposite O2-MdT relative to unmodified dT is less than what was previously reported,34 which may be attributed to the different sequence contexts used for this and previous studies; sequence context is known to affect the efficiencies of nucleotide insertion.29 Kf− incorporates the other three nucleotides opposite O2-MdT at much lower efficiencies than dAMP, i.e., 200 times lower for dCMP and dTMP, and 3 orders of magnitude lower for dGMP (Table 1). On the other hand, we found that Kf− incorporates dAMP, dCMP and dTMP opposite O4-MdT at similar efficiencies, though the polymerase displays a much stronger preference (by ~100-fold) for incorporation of dGMP over dAMP (Table 1). Together, Kf− preferentially inserts the correct dAMP opposite O2-MdT, but the incorrect dGMP opposite O4-MdT.
Table 1.
Efficiency and fidelity of E. coli Kf−-mediated nucleotide incorporation opposite undamaged dT, O2-MdT and O4-MdT as determined by steady-state kinetic measurements.*
| dNTP | Kcat (min−1) | Km (μM) | Kcat/Km (μM−1 min−1) | fInc |
|---|---|---|---|---|
| Undamaged dT-containing Substrate | ||||
| dATP | 2.6 ± 0.5 | 0.018 ± 0.001 | 140 | 1 |
| dCTP | 3.9 ± 0.3 | 160 ± 30 | 2.4 × 10−2 | 1.7 × 10−4 |
| dGTP | 5.5 ± 0.8 | 150 ± 20 | 3.7 × 10−2 | 2.5 × 10−4 |
| dTTP | 5.5 ± 0.6 | 490 ± 30 | 1.1 × 10−2 | 7.8 × 10−5 |
| O2-MdT-containing Substrate | ||||
| dATP | 2.5 ± 0.3 | 0.44 ± 0.07 | 5.7 | 1 |
| dCTP | 2.9 ± 0.4 | 130 ± 50 | 2.2 × 10−2 | 3.9 × 10−3 |
| dGTP | 3.5 ± 0.8 | 24000 ± 2000 | 1.5 × 10−4 | 2.6 × 10−5 |
| dTTP | 5.0 ± 0.6 | 2300 ± 200 | 2.2 × 10−3 | 3.8 × 10−4 |
| O4-MdT-containing Substrate | ||||
| dATP | 5.1 ± 0.4 | 260 ± 40 | 2.0 × 10−2 | 1 |
| dCTP | 2.6 ± 0.2 | 230 ± 10 | 1.1 × 10−2 | 0.58 |
| dGTP | 5.2 ± 0.3 | 1.9 ± 0.1 | 2.7 | 140 |
| dTTP | 3.3 ± 0.1 | 1100 ± 100 | 3.0 × 10−3 | 0.15 |
The Km and kcat were average values based on three independent measurements
Relative to the unmodified substrate, the efficiencies for human pol κ to incorporate the correct nucleotide, dAMP, opposite O2-MdT and O4-MdT were diminished by ~270 and 460 fold, respectively, with the kcat/Km values being 210, 0.77, and 0.46 μM−1 min−1 for dT, O2-MdT, and O4-MdT, respectively (Table 2 and Figure S4). The efficiencies for human pol κ to incorporate three other nucleotides, i.e., dGMP, dCMP, dTMP opposite unmodified dT were much lower than that for dAMP insertion (Table 2). Incorporation efficiencies for the O2-MdT- and O4-MdT-containing templates displayed only slight decreases, i.e., by 2–5 fold, for the other three nucleotides (Table 2). On the other hand, the efficiencies for the insertion of dCMP and dTMP opposite O4-MdT were 2 fold lower than that for dAMP incorporation. These results demonstrate that human pol κ-mediated nucleotide incorporation opposite O2-MdT and O4-MdT is error-prone.
Table 2.
Efficiency and fidelity of human polymerase κ-mediated nucleotide incorporation opposite undamaged dT, O2-MdT and O4-MdT as determined by steady-state kinetic measurements.*
| dNTP | Kcat (min−1) | Km (μM) | Kcat/Km (μM−1 min−1) | fInc |
|---|---|---|---|---|
| Undamaged dT-containing Substrate | ||||
| dATP | 5.9 ± 0.2 | 0.028 ± 0.002 | 210 | 1 |
| dCTP | 4.4 ± 0.1 | 0.66 ± 0.07 | 6.7 | 0.032 |
| dGTP | 4.3 ± 0. 4 | 6.0 ± 0.3 | 0.72 | 0.0034 |
| dTTP | 1.3 ± 0.2 | 63 ± 4 | 2.2 × 10−2 | 9.5 × 10−5 |
| O2-MdT-containing Substrate | ||||
| dATP | 2.8 ± 0.1 | 3.7 ± 0.2 | 0.76 | 1 |
| dCTP | 4.1 ± 0.1 | 10 ± 1 | 0.41 | 0.54 |
| dGTP | 3.0 ± 0.2 | 19 ± 1 | 0.16 | 0.21 |
| dTTP | 3.8 ± 0.6 | 21 ± 3 | 0.18 | 0.24 |
| O4-MdT-containing Substrate | ||||
| dATP | 1.8 ± 0.7 | 4.0 ± 0.1 | 0.45 | 1 |
| dCTP | 2.9 ± 0.1 | 15 ± 1 | 0.19 | 0.43 |
| dGTP | 4.0 ± 0.3 | 34 ± 2 | 0.12 | 0.26 |
| dTTP | 2.9 ± 0.7 | 14 ± 1 | 0.21 | 0.46 |
The Km and kcat were average values based on three independent measurements
The efficiencies for yeast pol η to incorporate the correct nucleotide, dAMP, opposite dT, O2-MdT, and O4-MdT were 180, 0.34, and 7.6×10−3 μM−1 min−1, respectively (Table 3 and Figure S5). Interestingly, yeast pol η inserts dGMP (0.57 μM−1 min−1) opposite O4-MdT ~80 times more efficiently than dAMP (7.6×10−3 μM−1 min−1, Table 3); the incorporation efficiencies for dCMP and dTMP were, however, 2–3 times lower than that for dAMP insertion. By contrast, we observed lower efficiencies for the incorporation of incorrect nucleotides opposite O2-MdT; relative to dAMP insertion, the efficiency was approximately 10 times lower for dGMP and 20–30 times lower for dCMP and dTMP (Table 3). These results support that yeast pol η-mediated nucleotide incorporation opposite O2-MdT in the template strand is accurate, whereas the template O4-MdT primarily directs dGMP misincorporation.
Table 3.
Efficiency and fidelity of yeast polymerase η-mediated nucleotide incorporation opposite undamaged dT, O2-MdT and O4-MdT as determined by steady-state kinetic measurements.*
| dNTP | Kcat (min−1) | Km (μM) | Kcat/Km (μM−1 min−1) | fInc |
|---|---|---|---|---|
| Undamaged dT-containing Substrate | ||||
| dATP | 1.8 ± 0.2 | 0.01 ± 0.001 | 180 | 1 |
| dCTP | 1.2 ± 0.2 | 0.98 ± 0.03 | 1.2 | 6.7 × 10−3 |
| dGTP | 2.1 ± 0.2 | 5.3 ± 0.4 | 0.39 | 2.2 × 10−3 |
| dTTP | 3.0 ± 0.3 | 3.8 ± 0.3 | 0.80 | 4.4 × 10−3 |
| O2-MdT-containing Substrate | ||||
| dATP | 2.6 ± 0.1 | 7.6 ± 0.2 | 0.34 | 1 |
| dCTP | 4.6 ± 0.1 | 230 ± 30 | 2.0 × 10−2 | 5.8 × 10−2 |
| dGTP | 8.5 ± 0.1 | 510 ± 10 | 1.7 × 10−2 | 4.9 × 10−2 |
| dTTP | 3.6 ± 0.1 | 310 ± 50 | 1.2 × 10−2 | 3.4 × 10−2 |
| O4-MdT-containing Substrate | ||||
| dATP | 2.2 ± 0.1 | 290 ± 70 | 7.6 × 10−3 | 1 |
| dCTP | 2.5 ± 0.3 | 2100 ± 400 | 1.2 × 10−3 | 1.6 × 10−1 |
| dGTP | 0.85 ± 0.07 | 1.5 ± 0.03 | 0.57 | 75 |
| dTTP | 3.3 ± 0.4 | 840 ± 50 | 3.9 × 10−3 | 5.2 × 10−1 |
The Km and kcat were average values based on three independent measurements
Primer extension studies with the use of LC-MS/MS
The above steady-state kinetic analysis provides useful information about how O2- and O4-MdT direct DNA polymerases to insert nucleotides opposite these lesions. However, such analysis may not reflect faithfully the nucleotide incorporation conditions in cells where DNA synthesis occurs in the presence of all four canonical nucleotides. Additionally, the primers carrying the correct or wrong nucleotide opposite the lesions may be extended at different efficiencies.45 Thus, we employed LC-MS/MS to interrogate the extension products following previously described methods with some modifications.46,47 In this regard, primer extension reactions were conducted using the same primer/template complex as described above except that the primer was not radiolabeled. Instead of employing a uracil-containing primer, which can be subsequently cleaved using uracil DNA glycosylase followed with hot piperidine treatment,46,47 we digested the reaction mixtures with two restriction enzymes to give shorter extension products for LC-MS/MS analysis (Scheme 1).
Similar as what we reported previously,48 the LC-MS/MS results showed a detection limit of ~10 fmol for the 7mer, 8A, 8G, 10 Del, 11T, 11C, and 11G at a signal-to-noise ratio greater than 20. Generally, when operated in the data-dependent scan mode, the LC-MS/MS method allows for the detection of species present at 1% or above when a 10-pmol reaction mixture was injected for LC-MS/MS analysis.
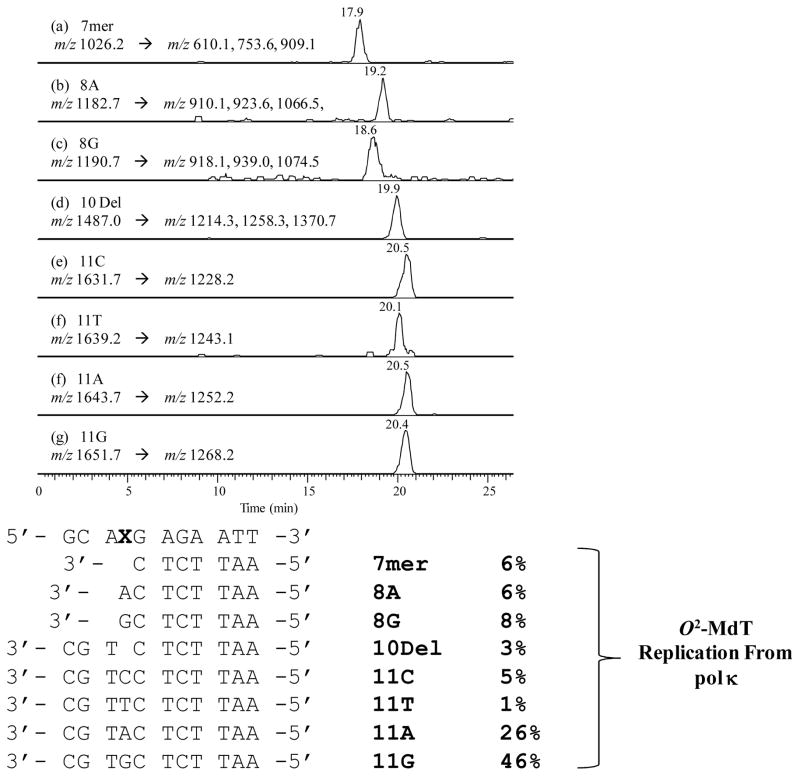
LC-MS/MS analysis of the extension products for both O2-MdT- and O4-MdT-containing substrates revealed the presence of the unextended or incompletely extended primer along with full-length replication products. Here we use the human pol κ-catalyzed primer extension of the O2-MdT-carrying substrate as an example to illustrate how we employ LC-MS/MS for the identification and quantification of replication products. The total-ion chromatogram (Figure S6) reveals the 7mer unextended primer d(AATTCTC), the +1 products (8A and 8G), the 10mer deletion product, and the full-length extension products (including 11C, 11T, 11A and 11G) eluting at 17.9, 19.2, 19.9 and 20.6 min, respectively (Figure 2). The digested damage-containing template d(CCATGGCAXGAG), where ‘X’ is O2-MdT, and the 5′ portion of the original primer d(GCTAGGATCATAG) also elute at 20.6 min (ESI-MS averaged from this retention time is shown in Figure S6, and the sequences for the identified products are listed in Table 4). The identities of the aforementioned ODNs were established from ESI-MS and MS/MS analyses (MS/MS shown in Figures S7 and S8). Using the same LC-MS/MS analysis, we were able to identify the replication products arising from the other eight in vitro replication reactions (Table 4, and selected-ion chromatograms are shown in Figures S9–S11). It is worth noting that the LC-MS/MS results revealed that only ~5% of the O2-MdT was degraded to the corresponding unmodified dT-containing substrate, supporting that the damage-containing substrate remains largely intact during the primer extension and restriction digestion conditions (Figures S12 and S13).
Figure 2.
Selected-ion chromatograms obtained from the LC-MS and MS/MS analysis of the human pol κ-induced replication products that have been treated with two restriction enzymes, NcoI and EcoRI together with shrimp alkaline phosphatase. A 10-pmol replication mixture was injected for analysis.
Table 4.
Summary of the percentages of replication products produced for dT-, O2-MdT- and O4-MdT-containing substrates as determined by LC-ESI-MS/MS experiments.
| Name | Sequence | dT | O2-MdT | O4-MdT |
|---|---|---|---|---|
| Klenow Fragment (exo−) | ||||
| 7mer | d(AATTCTC) | 2 | 2 | 7 |
| 8mer | d(AATTCTCA) | 9 | ||
| 8mer | d(AATTCTCG) | 4 | 5 | |
| 11A | d(AATTCTCATGC) | 98 | 77 | 7 |
| 11G | d(AATTCTCGTGC) | 7 | 80 | |
|
| ||||
| Yeast Polymerase η | ||||
| 7mer | d(AATTCTC) | 4 | 6 | 6 |
| 8mer | d(AATTCTCA) | 2 | ||
| 8mer | d(AATTCTCG) | 6 | 2 | |
| 10Del | d(AATTCTCTGC) | 1 | ||
| 11C | d(AATTCTCCTGC) | 5 | ||
| 11T | d(AATTCTCTTGC) | 6 | ||
| 11A | d(AATTCTCATGC) | 96 | 45 | 9 |
| 11G | d(AATTCTCGTGC) | 27 | 83 | |
|
| ||||
| Human Polymerase κ | ||||
| 7mer | d(AATTCTC) | 1 | 6 | 11 |
| 8mer | d(AATTCTCA) | 6 | ||
| 8mer | d(AATTCTCG) | 8 | 18 | |
| 10Del | d(AATTCTCTGC) | 3 | ||
| 11C | d(AATTCTCCTGC) | 5 | 2 | |
| 11T | d(AATTCTCTTGC) | 1 | 3 | |
| 11A | d(AATTCTCATGC) | 99 | 26 | 8 |
| 11G | d(AATTCTCGTGC) | 46 | 58 | |
The template was d(CCATGGCAXGAGAATTCTATGATCCTAG), where ‘X’ represents dT, O2-MdT or O4-MdT.
Extension products arising from replication by all three DNA polymerases opposite the undamaged template resulted in, as expected, a single extension product with the insertion of the correct nucleotide (i.e., dAMP) opposite the unmodified dT (Table 4). On the other hand, LC-MS/MS analysis of primer extension products for substrates containing O2- and O4-MdT revealed the presence of unextended primer, incompletely extended primer (+1 product), and full-length extension products.
O4-MdT was blocking to the DNA polymerases; the total amount of unextended primer and +1 product (8G) accounted for 12%, 8%, and 29% of all products identified in mixtures arising from replication by Kf−, yeast pol η, and human pol κ, respectively. Full-length DNA synthesis mediated by Kf−, yeast pol η and human pol κ resulted in the incorrect dGMP being predominately incorporated opposite O4-MdT, with 11G accounting for 80%, 83% and 58% of all the identified replication products, respectively (Table 4). Extension products with the correct dAMP opposite O4-MdT represent a relatively small proportion of full-length products from reactions with Kf − (7%), yeast pol η (9%) and human pol κ (8%). Consistent with our steady-state kinetic results, pol κ also induced full-length extension products with incorporation of dCMP (2%) and dTMP (3%) opposite O4-MdT.
O2-MdT was blocking to DNA synthesis mediated by all three DNA polymerases studied, which is reflected by the presence of more unextended primer and +1 extension products (8A and 8G, Table 4) than replication opposite O4-MdT; the total amounts of unextended primer and +1 extension products represent 15%, 15%, and 23% of all products arising from replication by Kf−, yeast pol η, and human pol κ, respectively. However, all three polymerases were capable of producing full-length extension products. Kf− predominately yielded full-length product with the correct dAMP (77%) being inserted opposite O2-MdT, while misincorporation of dGMP (7%) occurs at a moderate frequency. Yeast pol η also preferentially extended the primer with the correct dAMP (45%) opposite O2-MdT, though full-length products with dCMP (5%), dTMP (6%) and dGMP (27%) being incorporated opposite the lesion were also observed. Nevertheless, human pol κ predominately produced full-length products with the incorrect dGMP (46%) being inserted opposite O2-MdT, followed by dAMP (26%), dCMP (5%) and dTMP (1%). Interestingly, replication mediated by yeast pol η and human pol κ also resulted in a −1 deletion product at frequencies of 1% and 3%, respectively, where the polymerase skipped the O2-MdT site and continued replication past the damaged nucleoside (Table 4, Figures 2 and S10).
It is worth noting that, in order to obtain a relatively large amount of full-length replication products and enable their facile identification by LC-MS/MS, the above primer extension reactions were carried out with relatively high concentrations of dNTPs (1 mM each) and for a relatively long period of time (overnight), which may deviate from cellular DNA replication conditions. To assess whether the dNTP concentration and reaction time affect significantly the distribution of reaction products, we also conducted the corresponding experiments for the pol η-mediated replication of the O4-MdT-bearing substrate where the individual dNTP concentrations were lowered to a more biologically relevant level (50 μM each) or the reaction time was shortened to 6 hrs. Similar to what were observed for the reaction conditions described above, the use of 50 μM dNTPs or a 6-hr incubation time mainly yield full-length products with the misincorporation of dGMP opposite the lesion at 73% and 62%, respectively (Table S1). On the other hand, both reaction conditions resulted in higher proportions of unextended primer, which represent 17% and 35% (for 6-hr reaction) of all identified products for the reactions with lower dNTP concentration and shorter reaction time, respectively (Table S1 and Figure S14). Together, these results suggest that the primer extension conditions described above are suitable for revealing the miscoding potential of the DNA lesions under investigation.
Differences in Preferences for Nucleotide Insertion as Revealed by Steady-state Kinetic Assay and LC-MS/MS Analysis of Primer Extension Products
The above results obtained from the steady-state kinetic assay and LC-MS/MS analysis of primer extension reaction mixture revealed some notable differences for pol κ-mediated replication of O2-MdT- and O4-MdT-containing substrates, and for pol η-mediated replication of O2-MdT-bearing substrate (Tables 2–4). For instance, steady-state kinetic assay showed that human pol κ preferentially inserted dAMP over dGMP opposite O2-MdT (by ~4.8 fold) and O4-MdT (by ~3.8 fold, Table 2). LC-MS/MS data, however, showed that the full-length replication products carry mainly a dGMP opposite the two lesions; 46% 11G and 26% 11A were observed for the O2-MdT-containing substrate, while 58% 11G and 8% 11A were found for the substrate harboring an O4-MdT (Table 4). Choi et al. 49 made a similar finding for pol η-mediated replication of an O6-methylguanine-housing substrate; steady-state kinetic experiment shows a slightly more preferential incorporation of dCMP over dTMP, but LC-MS/MS analysis of extension products revealed that dTMP is inserted opposite the lesion 3 times more preferentially than dCMP.
We reason that several factors may contribute to these differences. As stated previously, the steady-state kinetic experiments were performed in the presence of a single nucleotide, whereas replication reactions for LC-MS/MS analyses were performed in the presence of all four natural nucleotides. The selectivity in nucleotide insertion under the latter reaction condition may not be faithfully recapitulated in steady-state kinetic assay where there is an absence of competition for the insertion of different nucleotides. Second, the primer with dAMP or dGMP being inserted opposite the lesion may be extended at different efficiencies. To assess the degree to which the second factor contributes to the differences in findings made from the two different assays, we conducted steady-state kinetic experiments for pol κ to insert the correct nucleotide, dTMP, opposite the adjoining 5′ nucleoside of the lesion (dA). It turned out that the primer with dG being placed opposite O2-MdT is extended 1.4 times more efficiently than the corresponding primer with a dA opposite the lesion, whereas the primer with dG being situated across O4-MdT is extended at an efficiency that is 5.2 fold more than the corresponding primer with a dA (Table S2 and Figure S15). Thus, the difference in extension efficiency alters the distribution of full-length products, which, however, does not fully account for the differences observed from the LC-MS/MS and steady-state kinetic assays. On the basis of this observation, we may also conclude that the relative efficiencies for nucleotide incorporation opposite a lesion, as revealed by steady-state kinetic assay, may not reflect faithfully the selectivity in nucleotide insertion opposite the lesion in cells where all four natural nucleotides are simultaneously present.
Discussion
Alkylating agents are known to react extensively with DNA, producing mutagenic and carcinogenic adducts in a variety of prokaryotic and eukaryotic organisms.26 O2-MdT and O4-MdT can be produced in cellular DNA upon treatment with the carcinogenic N-methyl-N-nitrosourea.6 Despite representing a small percentage of the observed products (~0.1% of total alkylation in vivo), O2-MdT and O4-MdT are not easily repaired in vivo6,10 and therefore might contribute significantly to the observed nucleobase substitutions and deletions induced from exposure to DNA methylating agents.
O4-MdT was found to be highly mutagenic in vitro and in vivo, producing mainly T→C transitions,2,6,10,27–30,50 and our results are in agreement with these previous observations. Both Kf− and yeast pol η exhibited a strong preference for incorporating the incorrect dGMP opposite O4-MdT. This is in line with our recent observation that O4-carboxymethylthymidine (O4-CMdT) directed yeast pol η to insert preferentially the incorrect dGMP.45 On the other hand, steady-state kinetic assay results showed that human pol κ predominately incorporated the correct dAMP, followed by dCMP and dTMP, opposite O4-MdT (Table 2). LC-MS/MS analysis of the primer extension results unveiled that the full-length products yielded by all three DNA polymerases carry mainly a dG opposite the O4-MdT (Table 4). This finding suggests that the placement of a dG opposite the lesion renders efficient DNA synthesis beyond the lesion site.. This is particularly true for human pol κ; although, among the four natural nucleotides, dGMP is inserted opposite O4-MdT at the lowest efficiency (Table 2), LC-MS/MS analysis revealed that incorporation of a dG opposite the lesion was the most favored (Table 4). The facile incorporation of dGMP opposite O4-MdT may be attributed to the fact that thymine with a 4-alkyl group has been proposed to adopt a wobble conformation with guanine, thus allowing O4-MdT to code as a cytosine.51 Taken together, these results suggest that O4-MdT mainly induces T→C transitions with all three DNA polymerases studied.
Jasti et al.33 showed recently that O2-MdT constitutes a strong block to DNA synthesis in E. coli cells even under SOS conditions, and the lesion is capable of inducing targeted base substitutions and, to a small degree, frameshift mutations. Our results revealed that incorporation of nucleotides opposite, and extension past, O2-MdT is relatively inefficient by Kf−, yeast pol η, or human pol κ. Additionally, base substitutions and, to a small degree, a −1 deletion were observed for DNA synthesis mediated by both yeast pol η and human pol κ opposite O2-MdT (Tables 2–4). This finding is in stark contrast with previous observations that several minor-groove N2-substituted dG derivatives can be efficiently and accurately bypassed by DinB family polymerases.15–18 X-ray crystal structure of the catalytic core of human pol κ in complex with the primer/template and incoming nucleotide revealed the lack of steric hindrance in the minor groove at the primer-template junction.52 This structure feature may permit pol κ to accommodate O2-MdT into the active site of the polymerase. However, the unique hydrogen bonding property of O2-MdT may not favor its base pairing with any of the four canonical nucleotides, thereby preventing efficient incorporation of any nucleotide opposite the lesion. The ability of O2-MdT in stalling DNA replication and in directing nucleotide misincorporation, in conjunction with the poor repair of O2-alkylthymidine lesions,10,53,54 suggests that this lesion may bear significant biological consequences.
Although O2-MdT and O4-MdT only constitute a small fraction of DNA lesions produced by methylating agents, these lesions, if left unrepaired, may lead to mutations in the genome. Further investigation is needed for addressing whether other O2-alkylthymidine lesions are capable of inhibiting DNA replication and inducing mutations in vitro, and how these lesions compromise DNA replication in cells.
Supplementary Material
Acknowledgments
Funding. This work was supported by the National Institutes of Health (Grant # R01 DK082779 to Y.W.) and Tobacco-Related Disease Research Program (Grant # 19D-0009 to N.A.).
Footnotes
Supporting Information Available: LC-MS/MS primer extension and steady-state kinetic assay results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tannenbaum SR. N-nitroso compounds: A perspective on human exposure. Lancet. 1983;1:629–632. doi: 10.1016/s0140-6736(83)91801-9. [DOI] [PubMed] [Google Scholar]
- 2.Preston BD, Singer B, Loeb LA. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986;83:8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 4.Singer B, Essigmann JM. Site-specific mutagenesis: Retrospective and prospective. Carcinogenesis. 1991;12:949–955. doi: 10.1093/carcin/12.6.949. [DOI] [PubMed] [Google Scholar]
- 5.Vogel EW, Nivard MJ. International commission for protection against environmental mutagens and carcinogens. The subtlety of alkylating agents in reactions with biological macromolecules. Mutat Res. 1994;305:13–32. doi: 10.1016/0027-5107(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 6.Singer B. O-alkyl pyrimidines in mutagenesis and carcinogenesis: Occurrence and significance. Cancer Res. 1986;46:4879–4885. [PubMed] [Google Scholar]
- 7.Zak P, Kleibl K, Laval F. Repair of O6-methylguanine and O4-methylthymine by the human and rat O6-methylguanine-DNA methyltransferases. J Biol Chem. 1994;269:730–733. [PubMed] [Google Scholar]
- 8.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Alkylation-induced apoptosis of embryonic stem cells in which the gene for DNA-repair, methyltransferase, had been disrupted by gene targeting. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 9.Altshuler KB, Hodes CS, Essigmann JM. Intrachromosomal probes for mutagenesis by alkylated DNA bases replicated in mammalian cells: A comparison of the mutagenicities of O4-methylthymine and O6-methylguanine in cells with different DNA repair backgrounds. Chem Res Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 10.Dosanjh MK, Singer B, Essigmann JM. Comparative mutagenesis of O6-methylguanine and O4-methylthymine in Escherichia coli. Biochemistry. 1991;30:7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- 11.Den Engelse L, Menkveld GJ, De Brij RJ, Tates AD. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis. 1986;7:393–403. doi: 10.1093/carcin/7.3.393. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 13.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 14.Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 15.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 16.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 17.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan B, You C, Andersen N, Jiang Y, Moriya M, O’Connor TR, Wang Y. The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J Biol Chem. 2011;286:17503–17511. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 21.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci U S A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 25.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 26.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffhill R. In vitro miscoding of alkylthymines with DNA and RNA polymerases. Chem Biol Interact. 1985;53:121–130. doi: 10.1016/s0009-2797(85)80090-9. [DOI] [PubMed] [Google Scholar]
- 28.Menichini P, Mroczkowska MM, Singer B. Enzyme-dependent pausing during in vitro replication of O4-methylthymine in a defined oligonucleotide sequence. Mutat Res. 1994;307:53–59. doi: 10.1016/0027-5107(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 29.Rao S, Chenna A, Slupska M, Singer B. Replication of O4-methylthymine-containing oligonucleotides: Effect of 3′ and 5′ flanking bases on formation and extension of O4-methylthymine-guanine basepairs. Mutat Res. 1996;356:179–185. doi: 10.1016/0027-5107(96)00052-8. [DOI] [PubMed] [Google Scholar]
- 30.Encell LP, Loeb LA. Enhanced in vivo repair of O4-methylthymine by a mutant human DNA alkyltransferase. Carcinogenesis. 2000;21:1397–1402. [PubMed] [Google Scholar]
- 31.Grevatt PC, Solomon JJ, Bhanot OS. In vitro mispairing specificity of O2-ethylthymidine. Biochemistry. 1992;31:4181–4188. doi: 10.1021/bi00132a005. [DOI] [PubMed] [Google Scholar]
- 32.Gowda AS, Krishnegowda G, Suo Z, Amin S, Spratt TE. Low fidelity bypass of O2-(3-pyridyl)-4-oxobutylthymine, the most persistent bulky adduct produced by the tobacco specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by model DNA polymerases. Chem Res Toxicol. 2012;25:1195–1202. doi: 10.1021/tx200483g. [DOI] [PubMed] [Google Scholar]
- 33.Jasti VP, Spratt TE, Basu AK. Tobacco-specific nitrosamine-derived O2-alkylthymidines are potent mutagenic lesions in SOS-induced Escherichia coli. Chem Res Toxicol. 2011;24:1833–1835. doi: 10.1021/tx200435d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakash GASKG, Suo Z, Amin S, Spratt T. Low fidelity bypass of O2-(3-pyridyl)-4-oxobutylthymine, the most persistent bulky adduct produced by the tobacco specific nitrosamine 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone by model DNA polymerases. Biochemistry. 2012;25:1195–1202. doi: 10.1021/tx200483g. [DOI] [PubMed] [Google Scholar]
- 35.Grevatt PC, Solomon JJ, Bhanot OS. In vitro mispairing specificity of O2-ethylthymidine. Biochemistry. 1992;31:4181–4188. doi: 10.1021/bi00132a005. [DOI] [PubMed] [Google Scholar]
- 36.Haracska L, Prakash S, Prakash L. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannistraro VJ, Taylor JS. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase η. J Biol Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu YZ, Swann PF. Oligodeoxynucleotides containing O2-alkylthymine: Synthesis and characterization. Tetrahedron Lett. 1994;35:303–306. [Google Scholar]
- 40.Gu C, Wang Y. LC-MS/MS identification and yeast polymerase η by pass of a novel γ-irradiation-induced intrastrand cross-link lesion G[8–5]C. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 41.Lao YB, Yu NX, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with R- and S-N′-nitrosonornicotine. Chem Res Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Den Engelse L, De Graaf A, De Brij RJ, Menkveld GJ. O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis. 1987;8:751–757. doi: 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- 43.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 44.Boosalis MS, Petruska J, Goodman MF. DNA-polymerase insertion fidelity: Gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 45.Swanson AL, Wang J, Wang Y. In vitro replication studies of carboxymethylated DNA lesions with Saccharomyces cerevisiae polymerase η. Biochemistry. 2011;50:7666–7673. doi: 10.1021/bi2007417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu C, Wang Y. In vitro replication and thermodynamic studies of methylation and oxidation modifications of 6-thioguanine. Nucleic Acids Res. 2007;35:3693–3704. doi: 10.1093/nar/gkm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J Biol Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 48.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J Biol Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 50.Dosanjh MK, Essigmann JM, Goodman MF, Singer B. Comparative efficiency of forming m4T-G versus m4T-A base pairs at a unique site by use of Escherichia coli DNA polymerase I (Klenow fragment) and Drosophila melanogaster polymerase α-primase complex. Biochemistry. 1990;29:4698–4703. doi: 10.1021/bi00471a026. [DOI] [PubMed] [Google Scholar]
- 51.Swann PF. Why do O6-alkylguanine and O4-alkylthymine miscode? The relationship between the structure of DNA containing O6-alkylguanine and O4-alkylthymine and the mutagenic properties of these bases. Mutat Res. 1990;233:81–94. doi: 10.1016/0027-5107(90)90153-u. [DOI] [PubMed] [Google Scholar]
- 52.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 53.McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brent TP, Dolan ME, Fraenkelconrat H, Hall J, Karran P, Laval F, Margison GP, Montesano R, Pegg AE, Potter PM, Singer B, Swenberg JA, Yarosh DB. Repair of O-alkylpyrimidines in mammalian cells: A present consensus. Proc Natl Acad Sci U S A. 1988;85:1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.