Abstract
Recent research using model organisms such as the nematode Caenorhabditis elegans has highlighted a critical role for several conserved signaling pathways in longevity determination. Here, we review three major endocrine- and nutrient-sensing signaling pathways with influence on lifespan, the insulin/insulin-like growth factor (IGF), target of rapamycin (TOR), and germline signaling pathways. Although these pathways engage distinct sets of transcription factors, the three pathways appear to modulate aging in C. elegans through partially overlapping effector mechanisms, including lipid metabolism and autophagy. This review highlights the latest advances in our understanding of how the insulin/IGF-1, TOR, and germline signaling pathways utilize different transcription factors to modulate aging in C. elegans with special emphasis on the role of lipid metabolism and autophagy.
Keywords: insulin/IGF-1, TOR, germline, autophagy, lipid metabolism, aging
Aging research in C. elegans
Studies in C. elegans have provided a wealth of information about the molecular mechanisms that modulate aging. C. elegans possesses a number of traits that make it an ideal model organism for aging research; 2–3 week lifespan, conserved developmental programs, transparency, small size, genetic tractability, and a fully sequenced genome [1]. Moreover, gene inactivation by RNAi has been extremely useful in identifying many new longevity genes [2], and has promoted a rapidly growing body of work that highlight multiple evolutionary conserved longevity paradigms. These include the insulin/IGF-1, TOR, and germline signaling pathways, which control many critical biological processes, including development, reproduction, metabolism, somatic maintenance, and stress resistance. Here, we review the most recent literature describing the molecular mechanisms by which these three pathways modulate C. elegans aging.
Insulin/IGF-1 signaling
The basics
Research on the genetics of aging in C. elegans began with the seminal finding that the organism’s lifespan was doubled by mutations in the age-1 or daf-2 genes, orthologs of the mammalian phosphoinositide 3-kinase (PI3K) and insulin/IGF-1 receptor (InR), respectively [3,4]. Both genes play key roles in insulin/IGF-1 signaling, and several additional pathway components have since been shown to modulate aging in flies, mice, and possibly humans, implying that the effects of the pathway on aging are conserved (reviewed in [5]).
Another important implication of these pioneering studies was that aging can be modulated by peptide hormones [6]. C. elegans expresses 40 insulin-like peptides [7], of which at least two are agonists (DAF-28 and INS-7) [8,9,10] and at least one functions as an antagonist (INS-1) of DAF-2/InR [7]. Ligand binding to DAF-2/InR activates its tyrosine kinase activity and initiates a cascade of sequential phosphorylation events that activate several kinases: AGE-1/PI3K, pyruvate dehydrogenase lipoamide kinase isozyme (PDK)-1, AKT-1/2, and serine/threonine-protein kinase (SGK)-1 (Figure 1A). Ultimately, AKT and SGK-1 phosphorylate and inactivate the FOXO transcription factor DAF-16 by preventing its translocation to the nucleus, thereby blocking transcription of target genes [11].
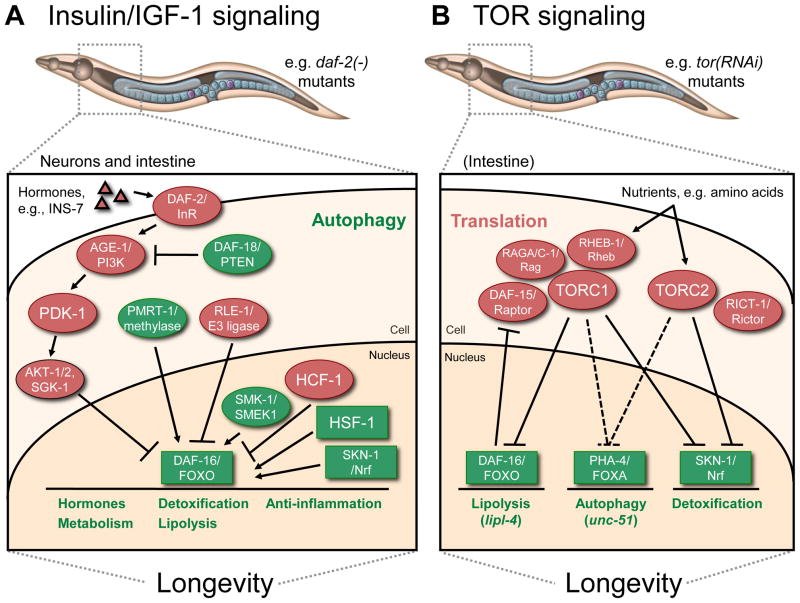
Figure 1. Overview of insulin/IGF-1 (A) and TOR (B) signaling in C. elegans aging.
A. In daf-2/InR insulin-receptor mutants, insulin-like peptides (e.g., INS-7) secreted from neurons reach intestinal cells and trigger the canonical insulin-signaling pathway, which prevents DAF-16/FOXO from entering the nucleus. Other mechanisms of DAF-16/FOXO regulation include ubiquitination (RLE-1/E3 ligase) and arginine methylation (PMRT-1/methylase). Nuclear-localized DAF-16/FOXO activity is enhanced by the action of SMK-1/SMEK and HSF-1, and is inhibited by HCF-1. The transcription factor SKN-1/Nrf is also required for longevity in daf-2/InR mutants. Collectively, these factors transcriptionally regulate multiple output processes as noted. Autophagy is another cellular process required for daf-2/InR mutants to live long. It is not yet known whether autophagy is a transcriptionally regulated process in daf-2/InR mutants.
B. TOR responds to nutrients and functions in two different complexes, TORC1 and TORC2. In analogy with mammalian studies, TORC1 is thought to interact with DAF-15/Raptor as well as Rag GTPases like RAGA-1, RAGC-1, and RHEB-1/Rheb. TORC1 and TORC1 specifically impair the activity of DAF-16/FOXO and SKN-1/Nrf, whereas it is not yet clear which of the two TOR complexes regulates PHA-4/FOXA (indicated by dashes lines). By using these transcription factors, TOR inhibits the expression of at least certain lipolysis-, autophagy-, and detoxification-associated genes. Listed as a cytoplasmic process, TOR signaling also likely modulates aging through a general suppression of translation. While the intestine has been linked to the longevity mediated by TORC1, the specific tissue requirements for TOR-dependent effects on aging have not yet been systematically investigated.
Factors with longevity-promoting effects are in green and those with lifespan-limiting effects are in red. Transcription factors are in boxes. See text for details.
As expected, reduced insulin/IGF-1 signaling (e.g., mutations in daf-2/InR) relieve this block and allow entry of DAF-16/FOXO into the nucleus, where it induces the expression of genes that increase lifespan and promote resistance to various stresses ([12], see below). Several additional mechanisms promote DAF-16/FOXO nuclear localization (Figure 1A). DAF-18/PTEN dephosphorylates and inhibits AGE-1/PI3K, thereby preventing DAF-16/FOXO phosphorylation by downstream kinases [13]. Nuclear translocation of DAF-16/FOXO can also be stimulated by alternate phosphorylation, for example via the kinase JNK-1 [14]. Nuclear maintenance of DAF-16/FOXO is promoted by PRMT-1, an arginine methyltransferease [15]. The stability of DAF-16/FOXO is also regulated by RLE-1, an E3 ubiquitin ligase that catalyzes ubiquitination of DAF-16/FOXO and targets it for degradation [16]. Collectively, these factors modulate DAF-16/FOXO activity to ensure the appropriate transcriptional response to specific environmental and hormonal cues (Figure 1A).
DAF-16 transcriptional coregulators
Once in the nucleus, several proteins function together with DAF-16/FOXO to modulate its activity in response to reduced insulin/IGF-1 signaling. Coregulators such as suppressor of MEK (SMK-1/SMEK) and host cell factor-1 (HCF-1) are critical for DAF-16/FOXO–mediated protection against microbial infection, DNA damage, and oxidative stress [17], whereas other transcription factors, such as heat-shock factor-1 (HSF-1), guide DAF-16/FOXO activity and cooperatively induce transcription of subsets of target genes, including heat-shock proteins involved in proteostasis [18]. The transcription factor SKiNhead (SKN)-1 (Nrf) regulates resistance to oxidative stress and expression of detoxification genes in response to reduced insulin/IGF-1 signaling, but extends lifespan independently of DAF-16/FOXO [19]. In daf-2/InR animals, HSF-1, SMK-1/SMEK and SKN-1/Nrf become nuclear localized independently of DAF-16/FOXO. Instead, HSF-1 activity is regulated via a repression complex [20], SMK-1/SMEK-1 is constitutively nuclear [17] and SKN-1/Nrf subcellular localization is controlled by phosphorylation via the p38 MAPK pathway [21,22]. In sum, multiple nuclear factors and parallel signaling pathways converge on DAF-16/FOXO to modulate longevity in long-lived insulin/IGF-1 pathway mutants (Figure 1A).
DAF-16 target genes
Gene expression profiling of daf-2/InR mutants has identified hundreds of DAF-16/FOXO–regulated genes involved in various cellular processes including metabolism, proteostasis, and stress responses such as oxidative stress, detoxification, and immunity [23,24,25,26]. Examples of genes with elevated expression in daf-2/InR mutants are members of the heat shock hsp-16 protein family, cytochrome P450 enzymes, glutathione-S-transferases, catalases, and the superoxide dismutase sod-3 (reviewed in [12]). In contrast, a number of genes show reduced expression in daf-2/InR mutants, and these include the insulin-like peptide ins-7 and the vitellogenin yolk proteins [23]. However, although DAF-16/FOXO directly regulates the expression of many of these genes [27], RNAi-mediated inhibition of any single target only modestly reduce the long lifespan of daf-2/InR mutants, suggesting that DAF-16/FOXO–dependent longevity likely requires the expression of multiple target genes [23].
Tissue specificity and endocrine signaling
Insulin/IGF-1 signaling in specific tissues in the worm, primarily neuronal and intestinal cells, is critical for the pathway to affect longevity systemically (reviewed in [11]) (Figure 1A). In brief, extensive genetic analysis has suggested that reduced DAF-2/InR activity in neurons leads to cell-autonomous DAF-16/FOXO activation and decreased production of hormones, including insulin-like peptide INS-7 [10]. In turn, reduced systemic levels of INS-7 signaling upregulates DAF-16/FOXO activity in the intestine, which increases the production of additional hormones while inhibiting the production of insulin-like peptides. Consequently, somatic cells acquire germline stem cell-like, stress-resistant properties conducive to longevity [28].
In sum, hormonal signaling is likely to regulate a number of cellular processes in long-lived daf-2/InR mutants. Below, we will focus on two recently identified processes with emerging roles in C. elegans longevity, namely energy/lipid metabolism and autophagy.
Cellular effector mechanisms
Energy and lipid metabolism
Insulin/IGF-1 signaling is a master regulator of metabolism that coordinates food intake and cellular energy homeostasis by stimulating glucose uptake and by driving anabolic processes. In C. elegans, glucose supplementation reduces lifespan by stimulating insulin signaling, and inhibiting DAF-16/FOXO–mediated transcription of longevity genes [29], while glucose restriction relieves this block and leads to lifespan extension [30]. Accordingly, impairing insulin signaling induces a survival response that alters feeding behavior [31] and promotes recycling of endogenous macromolecules. In support of this notion, daf-2/InR mutants show a DAF-16/FOXO-mediated increase in expression of the lipase LIPL-4 [32]. This lipase is not only partially required for daf-2/InR mutants to live long but also, when overexpressed, is sufficient to extend the lifespan of wild-type animals [32]. Taken together, these initial observations suggest that lipid breakdown is beneficial to daf-2/InR mutants. However, daf-2/InR mutants have increased lipid stores [33,34], indicating a complex role for lipid metabolism in these animals that remains to be investigated in more detail.
Energy metabolism also plays a central role in lifespan determination, as illustrated by the regulatory function of the energy-sensing enzyme AMP-activated kinase (AMPK). AMPK is activated by high intracellular AMP to ATP ratios, a condition that is observed in daf-2/InR mutants and in wild-type animals exposed to starvation or heat stress [35]. Consistent with this, a catalytic subunit of AMPK, AAK-2, is necessary for the long lifespan of daf-2/InR mutants [35], and AAK-2 overexpression is sufficient for lifespan extension of wild-type animals [35,36], an effect that is mediated at least partially through inhibition of the CREB-regulated transcriptional coactivatorCRTC- 1 [36].
Autophagy
Macroautophagy (hereafter referred to as autophagy) is a multistep process resulting in large-scale lysosomal degradation and recycling of vacuole-sequestered cytosolic cargos [37], and is emerging as a key player in the modulation of longevity in C. elegans. Autophagy is increased in daf-2/InR mutants and is essential for their lifespan extension [38,39], implying that these mutants recycle cargos that otherwise would be limiting to long-term survival. Consistent with this idea, increased autophagy and lysosomal degradation protects against protein aggregation [40]. This likely delays the eventual collapse of proteostasis observed during aging [41,42], because toxic protein aggregates are more efficiently removed.
Autophagy is at least partially regulated by AMPK activity in daf-2/InR mutants [43]. In mammals, AMPK stimulates autophagy by directly phosphorylating the autophagy initiating kinase, UNC-51 (ATG-1/ULK-1) [43]. Consistent with this, unc-51/ULK1, like aak-2/AMPK, is required for autophagy induction in daf-2/InR mutants [43]. Although recent work has suggested that autophagy can be transcriptionally regulated [44] (see section on Germline Signaling), DAF-16/FOXO appears to be unimportant for autophagy induction in daf-2/InR mutants [45]. Instead, DAF-16/FOXO could contribute to later stages of the autophagy process, as DAF-16/FOXO induces the expression of a lysosomal protein necessary for thermotolerance [46]. Future studies should investigate how autophagy is regulated to mediate the lifespan extension observed in daf-2/InR mutants.
Summary
Research over the last two decades has made the insulin/IGF-1 signaling pathway the most-studied longevity paradigm and has identified DAF-16/FOXO as a prominent transcription factor that, along with additional transcription factors and coregulators, controls the expression of multiple longevity genes. It has also become clear that endocrine signaling between tissues, such as the nervous system and the intestine, have systemic effects on lipid metabolism, autophagy and consequently longevity. Key issues for future research include understanding the insulin/IGF-1–dependent network of transcription factors and effector mechanisms, as well as the intracellular signaling events and feedback loops that coordinate insulin secretion and modulate aging.
TOR signaling
The basics
The TOR (Target-of-rapamycin) pathway controls growth and reproduction in response to the availability of amino acids and growth factors. As a nutrient sensor, TOR is a mediator of the metabolic response to dietary restriction, another conserved lifespan-extending paradigm with multiple links to the signaling pathways discussed here (reviewed in [47]). Consistent with this, inhibition of TOR activity extends lifespan in a variety of species [48]. TOR exists in two complexes, TORC1 and TORC2, which have different functions in mammalian cells; TORC1 integrates mitogen and nutrient signals to control cell proliferation and size, whereas TORC2 regulates cell shape [49]. TORC1 and TORC2 contain different co-activators, DAF-15/Raptor and RICT-1/Rictor, respectively [50] (Figure 1B), and regulate development, lipid storage, mRNA translation and autophagy in C. elegans [45,51,52,53,54,55]. TORC1 affects longevity in C. elegans at least in part via the GTPAses RAGA-1/RAGC-1 [53,56], RHEB-1/Rheb [57], and DAF-15/Raptor [54]. RNAi inhibition of raga-1 in the intestine is sufficient for lifespan extension [53], suggesting a role for this tissue in TOR-mediated longevity. Moreover, adult inhibition of rict-1/Rictor extends C. elegans lifespan [53]. The two transcription factors DAF-16/FOXO and SKN-1/Nrf are required for lifespan extension mediated by TORC1 inhibition, whereas SKN-1/Nrf is required for TORC1/rict-1–mediated lifespan extension [53]. TOR inhibition also requires the FOXA transcription factor PHA-4 to extend lifespan [58], but it has so far not been addressed whether this is primarily a TORC1 or TORC2 effect (Figure 1B).
Target genes
TOR regulates expression of the insulin-like peptide ins-7 [57], raising the possibility that TOR may, like DAF-2/InR, modulate aging through a systemic effect on hormones. Moreover, TORC1 regulates the expression of many daf-16/FOXO- and skn-1/Nrf target genes including detoxification genes [53]. Below, we discuss additional transcriptionally regulated targets that may contribute to TOR-mediated longevity in C. elegans (Figure 1B). TOR may also affect longevity by non-transcriptional mechanisms, including TORC1-mediated phosphorylation of S6 kinase (S6K), a key regulator of mRNA translation. Accordingly, inhibition of S6K as well as other regulators of mRNA translation, like tor inhibition, extends lifespan in multiple organisms [48]. While it is unclear how reduced protein synthesis extends lifespan, S6K may modulate longevity in C. elegans via both transcriptional (e.g., via PHA-4/FOXA) as well as post-translational mechanisms (e.g., involving AMPK) [48].
Cellular effector mechanisms
Lipid metabolism
Similar to daf-2/InR mutants, inhibition of TOR upregulates expression of the lipase LIPL-4 and increases lipolysis [44]. Moreover, lipl-4 expression in animals with reduced TOR levels is at least partly mediated by DAF-16/FOXO [44]. Whereas the contribution of LIPL-4 and lipid metabolism to TOR-mediated longevity remains unclear, TOR, daf-15/Raptor and rict-1/Rictor mutants all paradoxically accumulate more lipid droplets [51,54,55,59], as do daf-2/InR mutants.
Autophagy
As observed in other organisms, autophagy is induced by inhibition of TORC1 in C. elegans [45, 53]. This is partly due to increased expression of unc-51/ULK1, an upstream activator of the multistep autophagy process, via PHA-4/FOXA [44]. Notably, pha-4/FOXA itself is subject to transcriptional regulation by TOR [44,57]. Along these lines, autophagy genes [45,60] and pha-4/FOXA [58] are required for TOR/TORC1 mutants to live long, suggesting that TOR/TORC1 inhibition extends lifespan through autophagy. Because lifespan extension induced by rapamycin, a chemical that inhibits TOR, in flies also depends on autophagy genes [61], this longevity mechanism may well be conserved.
Overlap with insulin/IGF-1 signaling
TOR inhibition and reduced insulin/IGF-1 signaling engage overlapping transcriptional targets and effector mechanisms, and consistent with this, TOR inhibition does not further extend the long lifespan of daf-2/InR mutants [59,62]. On the other hand, DAF-16/FOXO negatively regulates the expression of the TORC1 coactivator daf-15/Raptor [51]. Moreover, as mentioned above, DAF-16/FOXO and SKN-1/Nrf are required for lifespan extension mediated by TORC1 inhibition [53]. It will be of interest to elucidate how signals from the insulin-signaling pathway (possibly via AKT as observed in mammals [50]) may modulate TOR in C. elegans, as well as to elucidate how DAF-16/FOXO and SKN-1/Nrf is regulated upon TOR inhibition.
Summary
Studies in several species demonstrate that TOR signaling affects aging in a conserved manner. Although the molecular mechanisms by which TOR affects C. elegans lifespan are only starting to be elucidated, it is clear that insulin/IGF-1 and TOR signaling converge at the level of transcription factors (DAF-16/FOXO and SKN-1/Nrf), target genes (the lipase lipl-4 and the insulin-like peptide ins-7), as well as potential effector mechanisms (autophagy and possibly lipid metabolism). Future investigations should systematically address the tissue-specific functions of TOR in longevity.
Germline signaling
The basics
In C. elegans, the germline integrates nutrient signaling and communicates with other tissues that modulate aging. Removal of germline cells by laser ablation of precursor cells extends the lifespan by 60%; however, this effect is nullified by concomitant removal of the somatic gonad, suggesting the existence of opposing signaling pathways originating from the germline and somatic gonad [63]. Germline-mediated regulation of lifespan has also been observed in flies, and signals from the reproductive system may affect lifespan in mice as well [64].
As in daf-2/InR mutants, daf-16/FOXO is required for the long lifespan of germline-less animals, yet in contrast to the insulin/IGF-1 pathway that affect DAF-16/FOXO in both neuronal and intestinal cells, germline ablation induces DAF-16/FOXO activation and translocation to the nucleus primarily in intestinal cells, a key site for modulation of longevity through germline signaling [64]. Several factors with functions in insulin/IGF-1 signaling are required for germline-less animals to live long, including DAF-18/PTEN [65], the co-factor SMK-1/SMEK-1 [17] and the transcription factor HSF-1 [66]. Other factors are specific to germline signaling, including the transcription elongation factor TCER-1/TCERG1 [67], and the ankyrin repeat-containing protein KRI-1/KRIT-1 [65]. KRI-1/KRIT-1 is required for nuclear localization of DAF-16/FOXO as well as upregulation of TCER-1/TCERG1 in the intestines of germline-less animals, but not in daf-2/InR animals. TCER-1/TCERG1 may be involved in lifespan modulation in germline-less animals through its interactions with PHI-62, a predicted RNA-binding protein that binds FTT-2/14-3-3, a known FOXO-interacting protein [68]. Moreover, the microRNA mir-71, which is produced in neurons, was recently found to regulate DAF-16/FOXO localization in the intestine [69], supporting a role for cell-nonautonomous regulation of lifespan in germline-less animals. These DAF-16/FOXO activators are all required for germline removal to extend lifespan (Figure 2).
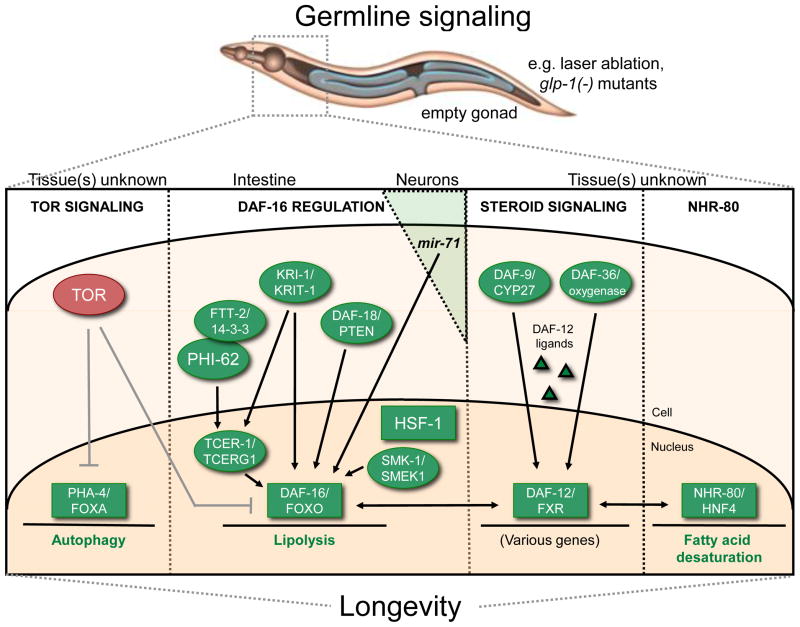
Figure 2. Overview of germline signaling in C. elegans aging.
Lifespan extension via germline removal, for example by mutation of glp-1/Notch, depends on at least four signaling mechanisms: reduced TOR signaling (likely to regulate autophagy), DAF-16/FOXO regulation, increased steroid signaling via the DAF-36/DAF-9/DAF-12 pathway (which regulates various genes), and increased NHR-80/HNF-4 signaling (which enhances fatty-acid desaturation). Of these mechanisms, the most-studied mechanism is DAF-16/FOXO regulation. Germline-less animals require specific cofactors (not required in daf-2/InR mutants) for activation of DAF-16/FOXO in the nucleus of intestinal cells. DAF-12/FXR, TCER-1/TCERG1, KRI-1/KRIT-1 and PHI-62 all function specifically in germline-less animals, whereas DAF-18/PTEN, SMK-1/SMEK and HSF-1 are common between the insulin/IGF-1 and germline signaling pathways. Cell-nonautonomous regulation of DAF-16/FOXO is also mediated through a microRNA, mir-71, which is produced in neurons.
Factors with longevity-promoting effects are in green and those with lifespan-limiting effects are in red. TOR effects are listed in grey to indicate that these links are presently inferred. Transcription factors are in boxes. See text for details.
A lipophilic-hormone/steroid signaling pathway also affects DAF-16/FOXO localization and is required for longevity in germline-less animals. This pathway includes three key components, DAF-9, DAF-36 and DAF-12. DAF-9 is an ortholog of the mammalian cytochrome P450 enzyme, CYP27 and is expressed in the hypodermis, the spermatheca, and two neuron-like cells. DAF-36 is a Rieske-like oxygenase primarily expressed in the intestine [11]. DAF-9/CYP27 and DAF-36/oxygenase are involved in the synthesis or modification of ligands for the nuclear hormone receptor DAF-12/FXR. Consistent with this idea, supplementation with the DAF-12 ligand delta-4 dafachronic acid restores lifespan extension in germline-less animals deficient in daf-9/CYP27 or daf-36/oxygenase [70]. DAF-12/FXR is likely to interact in a complex manner with DAF-16 DAF-9/CYP27, and the two proteins may directly bind each other [71]. Thus, in response to germline removal, steroid signaling mediates specific changes in the intestine that stimulate DAF-16/FOXO activity to extend lifespan (Figure 2).
Target genes
Conveniently, C. elegans lifespan extension induced by germline ablation can be mimicked genetically. For instance, animals carrying mutations in the Notch-1 receptor (glp-1/NotchR), a mediator of germline development, have extended lifespans [72]. These mutants were employed in a recent microarray analysis to identify DAF-16/FOXO and DAF-12/FXR target genes [68]. This detailed analysis revealed a significant overlap in DAF-16/FOXO–regulated genes in the daf-2/InR mutants and in germline-less animals, and a small but overlapping set of genes were regulated by both daf-16/FOXO and daf-12/FXR in germline-less animals. Of note, some of these genes are involved in lipid metabolism, as described below.
Cellular effector mechanisms
Lipid metabolism
Germline-less animals display notable changes in lipid metabolism. These include increased lipolysis, resulting in large part from DAF-16/FOXO–mediated increases in the lipase lipl-4 expression [32], and concomitant increases in lipid stores [73], similar to daf-2/InR and tor mutants. Moreover, other lipid genes such as the lipase lips-17 and the fatty acyl reductase fard-17 are upregulated and required for the long lifespan of germline-less glp-1/NotchR mutants, further supporting a role for lipid remodeling in longevity [68]. In addition, a recent study showed that the nuclear hormone receptor NHR-80/HNF-4 regulates oleic acid synthesis via the fatty acid desaturases fat-5, -6 and -7 and mutation of fat-6 and fat-7 together shortens the lifespan of germline-less animals, an effect that can be reversed by adding oleic acid to the media [74]. Taken together, these results are consistent with a key role for fatty acid desaturation in germline signaling. It will be interesting to determine if products of these lipid-modifying genes could serve as signaling molecules to coordinate metabolism and longevity regulation of the entire organism.
Autophagy
As observed for both daf-2/InR and tor mutants, germline-less glp-1/NotchR animals display an increase in autophagy, which is required for their long lifespan [44]. Autophagy induction in glp-1/NotchR animals is at least partly mediated by a PHA-4/FOXA–dependent increase in expression of autophagy genes [44]. Autophagy is predominantly induced in hypodermal and intestinal cells, suggesting that these tissues are particularly important for longevity. In particular, overexpression of the lipase LIPL-4 in the intestine [32], or under its endogenous promoter [44] is sufficient to extend lifespan, which occurs in part through PHA-4/FOXA–mediated induction of autophagy [44]. On the other hand, autophagy is required for the increased lipase activity in germline-less animals, strengthening the hypothesis that a functional link exists between autophagy and lipid breakdown. Further investigation will be necessary to determine how autophagy functions as a possible regulator of lipid metabolism in germline-less animals.
Overlap with insulin/IGF-1 and TOR signaling
Germline-less animals and daf-2/InR mutants have several mechanistic similarities, including neuron-to-intestine communication for longevity, increased autophagy, nuclear-localized DAF-16/FOXO in the intestine, and significant overlap in DAF-16/FOXO–regulated targets [68]. However, notable differences also exist; for example, KRI-1/KRIT1 and TCER-1/TCERG1 regulate germline-dependent but not daf-2/InR–mediated longevity. Moreover, germline removal in daf-2/InR mutants leads to extreme longevity, suggesting that parallel pathways determine longevity in daf-2/InR and germline-less animals [75].
The lifespan of germline-less glp-1/NotchR animals is not further extended by TOR inhibition [44], suggesting that overlap exists between TOR and germline signaling. This is supported by the observation that TOR levels are reduced in glp-1/NotchR animals [44], and both pathways share common transcriptional targets (e.g., the lipase lipl-4) and regulators (e.g., DAF-16/FOXO). However, since inhibition of raga-1 (i.e., TORC1) does extend the lifespan of germline-less animals [53], this overlap is likely to be complex and awaits further investigation.
Summary
The effects of germline signaling on aging in C. elegans have received much attention in recent years, and this pathway is now known to share common effector mechanisms with insulin/IGF-1 and TOR signaling, most notably lipid metabolism and autophagy. Among the three longevity pathways discussed here, germline signaling is associated with the largest number of transcription factors known to modulate longevity, namely daf-16/FOXO, daf-12/FXR, hsf-1, nhr-80/HNF4, and pha-4/FOXA. Future experiments should evaluate how the function of these factors is coordinated, both intracellularly and at the intercellular level, to extend lifespan in germline-less animals.
Concluding Remarks and Future Perspectives
The involvement of the insulin/IGF-1, TOR, and germline signaling pathways in lifespan determination of C. elegans is the subject of intense investigation. Each pathway regulates downstream effector mechanisms, including lipid metabolism and autophagy, through overlapping sets of transcription factors. All three pathways converge on DAF-16/FOXO, whereas SKN-1/Nrf1, PHA-4/FOXA and HSF-1 are employed in specific combinations. Notably, some transcriptional target genes are known to be regulated by both DAF-16/FOXO and HSF-1, whereas others are specifically controlled by one factor [18]. It will be crucial to identify the targets of each transcription factor, and to determine how they are coordinately regulated in the different longevity models.
At least two cellular processes are regulated in common by insulin/IGF-1, TOR, and germline signaling to affect longevity; namely, autophagy and lipolysis. The interplay between autophagy and lipolysis may be critical for longevity as LIPL-4–mediated lipase activity and autophagy function interdependently [44]. Moreover, concomitant changes in fatty acid desaturation may be necessary for somatic maintenance [76]. As such, increased activity in lipid remodeling pathways, such as fatty acid desaturation observed in germline signaling, may stimulate membrane biogenesis and maintain functional autophagy [77]. The significance of lipid breakdown in aging appears to be conserved, because in humans, the capacity to maintain dynamic lipid turnover in adipocytes correlates with better health status [78]. Paradoxically, mutants of all three longevity pathways have excessive lipid stores [76]. Future biochemical experiments in C. elegans may reveal how aging is modulated by different metabolic routes governing energy and lipid homeostasis.
As in the past decades, future research in C. elegans will likely help clarify how endocrine and metabolic signaling between different tissues modulate organismal aging. Such studies could uncover novel genetic factors with effects on survival, which may help develop therapeutic solutions against human age-related diseases and aging. The nematode is also emerging as a practical platform to test compounds and rapidly validate their bioactivity in different disease models [79,80]. In sum, research using the model organism C. elegans will continue to accelerate discoveries, and will likely provide a better molecular and cellular understanding relevant to treatment of age-related disorders and metabolic diseases in humans.
Outstanding Questions.
How are energy and lipid homeostasis changed to ensure survival?
What is the cargo(s) degraded by autophagy that leads to longer lifespan?
How do TOR complexes differ in their ability to mediate longevity?
What are the mechanisms by which multiple transcription factors coordinate transcription of distinct subset of genes in a longevity pathway?
Acknowledgments
We would like to thank Drs. H. Aguilaniu, A. Ghazi and A. O’Rourke for feedback on the manuscript. We also thank Jamie Simon from the Salk Institute for Biological Studies for help with figures. LRL is funded by a pilot grant from NIH/NIA (3 P50 AG005131-28), and MH is funded by two R01s from NIH/NIA (R01 AG038664 and R01 AG039756). MH is also an Ellison Medical Foundation New Scholar in Aging (AG-NS-0481-08).
Glossary of C. elegans gene orthologs
- aak-2
alpha-subunit of AMP-activated kinase (AMPK)
- age-1
phosphatidylinosytol 3-kinase
- akt-1,2
serine/threonine kinase Akt/PKB
- daf-2
insulin/IGF-1 receptor
- daf-9
cytochrome P450 of CYP27 subfamily
- daf-12
nuclear receptor FXR
- daf-15
TORC1-binding partner Raptor
- daf-16
transcription factor FOXO3A
- daf-18
phosphatidylinositol 3,4,5-triphosphate 3-phosphatase, PTEN
- daf-36
catalytic subunit of Rieske-like oxygenase
- fat-5
palmitoyl-CoA- Δ9-Desaturase
- fat-6-7
stearoyl-CoA- Δ9-Desaturase SCD
- ftt-2
14-3-3 (FOXO-interacting) protein
- glp-1
Notch receptor
- hcf-1
Host cell factor 1
- hsf-1
Heat shock factor 1
- ins-7
one of 40 insulin-like peptides in C. elegans and encodes an insulin/IGF-1-like peptide that likely functions as an agonist for DAF-2
- jnk-1
c-Jun N-terminal kinase
- kri-1
ankyrin-repeat protein KRIT-1
- lipl-4
lipase with high homology to human lysosomal acid lipase
- MAPK
mitogen-activated protein kinase
- mir-71
one of hundreds of microRNAs with a specific effect on longevity germline-less animals
- nhr-80
nuclear hormone receptor HNF-4
- pdk-1
3-phosphoinositide-dependent kinase 1
- pha-4
transcription factor FOXA
- phi-62
Putative RNAse-binding protein
- prmt-1
PRotein arginine MethylTransferase
- raga-1
Ras-related GTPase RagA
- ragc-1
Ras-related GTPase RagC
- rheb-1
Rheb GTPase
- rict-1
TORC2-binding partner Rictor (also referred to as lpo-6)
- rle-1
E3 ubiquitin ligase
- rsks-1
p70 ribosomal S6 kinase
- skn-1
transcription factor Nrf
- smk-1
suppressor of MEK
- tcer-1
TransCription Elongation Regulator homolog 1
- TOR
Target-of-rapamycin (also referred to as let-363)
- TORC1
TOR complex 1
- TORC2
TOR complex 2
- unc-51
ULK-1 (Unc-51-Like Kinase)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Panowski SH, Dillin A. Signals of youth: endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:259–264. doi: 10.1016/j.tem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Ni Z, Lee SS. RNAi screens to identify components of gene networks that modulate aging in Caenorhabditis elegans. Brief Funct Genomics. 2010;9:53–64. doi: 10.1093/bfgp/elp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type [see comments] Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 4.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 6.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans [see comments] Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 7.Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone EA, Inoue T, Thomas JH. Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics. 1996;143:1193–1205. doi: 10.1093/genetics/143.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy CT, Lee SJ, Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 12.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 13.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 14.Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y, Daitoku H, Hirota K, Tamiya H, Yokoyama A, et al. Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 2011;13:505–516. doi: 10.1016/j.cmet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Wolff S, Ma H, Burch D, Maciel GA, Hunter T, et al. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 18.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 19.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 Regulators DDL-1/2 Link Insulin-like Signaling to Heat-Shock Responses and Modulation of Longevity. Cell. 2012;148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 24.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 25.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 26.Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 28.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 31.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 34.Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendez A, Levine B. Autophagy in C. elegans. WormBook. 2009:1–26. doi: 10.1895/wormbook.1.147.1. [DOI] [PubMed] [Google Scholar]
- 38.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 39.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 40.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapierre LR, Gelino SR, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate aging in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 52.Long X, Spycher C, Han ZS, Rose AM, Muller F, et al. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- 53.Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, et al. TOR Signaling and Rapamycin Influence Longevity by Regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, McIntire SL. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. 2010;6:e1000972. doi: 10.1371/journal.pgen.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- 58.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 60.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 61.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 63.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 64.Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- 65.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 66.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000639. doi: 10.1371/journal.pgen.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulias K, Horvitz HR. The C. elegans MicroRNA mir-71 Acts in Neurons to Promote Germline-Mediated Longevity through Regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 72.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 73.O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, et al. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- 76.Ackerman D, Gems D. The mystery of C. elegans aging: An emerging role for fat: Distant parallels between C. elegans aging and metabolic syndrome? Bioessays. 2012;34:466–471. doi: 10.1002/bies.201100189. [DOI] [PubMed] [Google Scholar]
- 77.Girardi JP, Pereira L, Bakovic M. De novo synthesis of phospholipids is coupled with autophagosome formation. Med Hypotheses. 2011;77:1083–1087. doi: 10.1016/j.mehy.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 78.Arner P, Bernard S, Salehpour M, Possnert G, Liebl J, et al. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature. 2011;478:110–113. doi: 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petrascheck M, Ye X, Buck LB. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature. 2007;450:553–556. doi: 10.1038/nature05991. [DOI] [PubMed] [Google Scholar]
- 80.Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]