Abstract
Unilamellar phospholipid vesicles prepared using the polymerizable lipid bis-sorbylphosphatidylcholine (bis-SorbPC) yield three-dimensional nanoarchitectures that are highly permeable to small molecules. The resulting porous phospholipid nanoshells (PPNs) are potentially useful for a range of biomedical applications including nanosensors and nanodelivery vehicles for cellular assays and manipulations. The uniformity and size distribution of the pores, key properties for sensor design and utilization, has not previously been reported. Fluorophore-assisted carbohydrate electrophoresis (FACE) was utilized to assess the nominal molecular weight cutoff limit (NMCL) of the PPN via analysis of retained dextran with single monomer resolution. The NMCL of PPNs prepared from pure bis-SorbPC was equivalent to a 1800 Da linear dextran, corresponding to a maximum pore diameter of 2.6 nm. Further investigation of PPNs prepared using binary mixtures of bis-SorbPC and dioleylphosphatidylcholine (DOPC) revealed a similar NMCL when the bis-SorbPC content exceeded 30 mol %, whereas different size-dependent permeation was observed below this composition. Below 30 mol % bis-SorbPC, dextran retention provided insufficient mass resolution (162 Da) to observe porosity on the experimental time scale; however, proton permeability showed a marked enhancement for bis-SorbPC ≥ 10 mol %. Combined these data suggest that the NMCL for native pores in bis-SorbPC PPNs results from an inherent property within the lipid assembly that can be partially disrupted by dilution of bis-SorbPC below a critical value for domain formation. Additionally, the analytical method described herein should prove useful for the challenging task of elucidating porosity in a range of three-dimensional nanomaterials.
INTRODUCTION
The advent of biocompatible nanoarchitectures has contributed significantly to a broad range of biomedical applications including biosensing,1–8 drug delivery,1,7,9–16 and diagnostic imaging.1,12,13,17–24 A highly diverse catalog of nanomaterials has been prepared from inorganic, polymeric and hybrid materials in a multitude of geometric configurations. Among the most promising geometries for intracellular sensing and delivery is the recent advent of silica and polymer nanoshells.10,18–21,25–36 The nanoshell architecture provides an aqueous interior free of diffusional restrictions and non-covalent interactions with the nanoparticle matrix which may degrade the activity of encapsulated cargo, e.g. enzymes, aptamers, etc. Simultaneously, the shell provides a protective barrier which minimizes deleterious interactions between the cargo and the exterior biological system. Combined, these properties provide nanomaterials capable of new paradigms in molecular delivery applications where large molecular weight species, e.g. enzymes, are retained in a protected environment within the biological system, yet still retain the capability to modulate the chemical composition of the surroundings via size dependent mass transport across the membrane. Thus, these nanoarchitectures should prove useful for a wide range of biological applications including sensing, therapeutics, and many others.
A key factor that limits many potential nanoshell applications is restricted mass transport across the nanoshell surface to the encapsulated cargo. To overcome this limitation, porous nanoshell architectures have been developed.10,21,24,25,28,30–33,35,37–41 The resultant thin porous nanoshells function similar to a dialysis membrane where the size exclusion limit is determined by the pore size within the nanoshell surface. Porous silica nanoshells are prepared via selective etching of condensed silica nanoparticles to yield a tunable nanoshell diameter and shell thickness.29–33,42 A very promising approach for nanoshell synthesis relies upon self-assembly of natural or synthetic materials into a three dimensional spherical nanoshell,21,26–28,34–36 similar to a liposome, followed by pore formation using pore forming proteins,27,34,37,38,43 bilayer disrupting compounds44 or skeletonization10. Self-assembly is particularly attractive for encapsulating water-soluble, large molecular weight species that may be damaged during sol-gel polymerization and the subsequent chemical etching. We recently prepared porous nanoshell materials utilizing the polymerizable lipid bis-SorbPC.35 The resultant porous phospholipid nanoshells (PPNs), were resistant to non-specific protein adsorption, could readily encapsulate large molecular weight species while allowing smaller molecules to traverse the nanoshell membrane irrespective of molecular charge and exhibited very high chemical and physical stability, facilitating intracellular loading.35
A key property of porous nanoshell materials is the molecular weight cutoff, which must be sufficiently small to permanently include cargo, yet large enough to facilitate transport of desired species across the shell surface. Further, when enzymes and other cargos that interact with charged species are employed, the transport must be insensitive to molecular charge. While a number of porous nanoshell architectures exist, detailed characterization and subsequent control of pore diameters and pore density remain challenging.
Characterization of nanoshell porosity has typically relied upon permeation and exclusion assays or high resolution imaging. Estimations of pore sizes in silica nanoshell were performed with N2 permeation assays 32,33 and TEM,30–33 with pore diameters of ca. 3.5 nm. Pore diameters smaller than this are difficult to directly visualize in silica due to the nature of the sol-gel network. Further, these approaches are less amenable to pore size determination in flexible, porous polymer nanoshells, including PPNs. AFM has been used to investigate defects in bis-SorbPC planar supported lipid bilayers (PSLB),45 though the resolution was insufficient to detect the nanoscale defects that would lead to leakage of low molecular weight species. Further, this approach is less amenable to pore size determination in fully-enclosed three-dimensional nanometer-sized architectures. Porosity in polymeric bis-SorbPC lipid bilayers has been explored using a number of approaches;35,46–49 though to date the NMCL of bis-SorbPC PPNs has not been precisely measured. Electrophysiology47 and fluorescence35 studies have revealed enhanced leakage of low molecular weight species in bis-SorbPC bilayers compared to natural lipid membranes; however, AFM45 and TEM35 of poly(bis-SorbPC) PSLBs and PPNs, respectively, showed no obvious nanoscale level defects. Thus, new approaches are necessary to address this parameter in self-assembled porous nanoarchitectures.
Intrinsic and induced nanoshell permeability has been evaluated via liposome retention of species of varying molecular weight. Encapsulated, fluorescently-labeled dextrans were used to explore broad permeability of nanoporous liposome preparations; however, the mass resolution was limited to ca. 5,000 Da.10,37,38 The mass resolution could be significantly increased using the native monomer distribution of an unlabeled dextran population where the mass difference of oligomers is 162 Da, if sufficient resolution in the analysis of the retained species can be achieved. For example, a pre-labeled dextran population was previously utilized to investigate cellular junctions with single monomer resolution.50 In this work, we utilized a high-sensitivity dextran retention assay wherein a dextran distribution was encapsulated within self-assembled bis-SorbPC PPNs and the retained fraction was collected. The retained dextran was subsequently derivatized with 1-aminopyrene-3,6,8-trisulfonate (APTS) and analyzed by fluorophore assisted carbohydrate electrophoresis (FACE).51–54 This approach provides the first approximation of pore diameters in PPNs and is amenable to pore size determination in other self-assembled porous nanoshell architectures.
EXPERIMENTAL
Materials
1-aminopyrene-3,6,8-trisulfonate (APTS) was obtained from Biotium (Hayward, CA). 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt) (Rh-DPPE), Mini-Extruder and 200 nm nominal pore size nucleopore membranes were purchased from Avanti Polar Lipids (Alabaster, AL). Bis-Sorbyl phosphatidylcholine (Bis-SorbPC) was prepared and purified as previously described.48 Dextran (6,000 Da) and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Dialysis tubing was obtained from Spectrapor (Houston, TX) (1,000 Da and 12,000 – 14,000 Da NMCL) and Pierce (Rockford, IL) (3,500 Da NMCL). All chemicals were used as received. All solutions were prepared using deionized water (18 MΩ cm).
PPN preparation, polymerization and recovery of retained dextran
PPNs were prepared by lipid film rehydration followed by freeze-thaw extrusion techniques. A 2 mg aliquot of total lipid (bis-SorbPC or bis-SorbPC/DOPC mixtures) dissolved in chloroform was added to a glass vial and dried overnight under vacuum. A 200 μL aliquot of dextran (100 mg/mL in H2O; 6,000 Da) was added to the dried lipid films for rehydration. Liposome suspensions were subjected to 10 freeze-thaw cycles and extruded 21 times through two 200 nm pore size membranes using a miniextruder. Separation of non-encapsulated materials was performed using a Sepharose CL-4B column (Sigma-Aldrich). Quasi-elastic dynamic light scattering (BI-200 with a BI-DS detector and BI-800 AT autocorrelator software, Brookhaven Instruments, Holtsville, NY) was used for size determination. Where indicated, polymerization was performed by 15 min exposure to a UV polymerization apparatus consisting of a 100 W Hg arc lamp (Newport model 6281), a UV band pass filter (Edmund U-330) and a custom-built water IR filter. Importantly, these polymerization conditions generate partially polymerized PPNs that retain sufficient stability for the present experiments yet can still be dissociated via surfactant dissolution to allow dextran retrieval. Polymerized samples were placed in a dialysis bag (12,000 – 14,000 Da NMCL) and dialyzed for 18 hours against H2O at 4 °C. To recover retained dextran from PPNs and DOPC vesicles, Triton X-100 was added at 10x molar excess compared to the total lipid content and the mixture was vortexed. Following lysis, dextran was isolated from lipids and detergents by two consecutive chloroform extractions. The recovered dextran was frozen and lyophilized prior to derivatization with APTS.
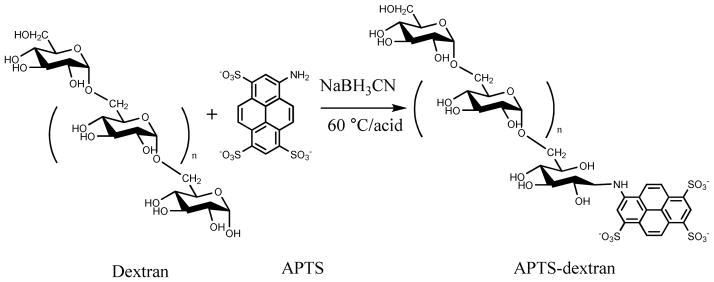
Derivatization of dextran with APTS
Dextran and other sugars (100 μg or less) were derivatized similarly to published protocols.51–55 Briefly, 2 μL of 0.01M APTS in 1M citric acid and 1 μL of 1M sodium cyanoborohydride in DMSO were added to dried dextran samples (Scheme 1). The samples were heated to 60 °C for 4 hr and diluted with FACE running buffer (20 mM phosphate at pH 7.0). Occasionally, APTS-derivatized samples contained insoluble matter indicating inefficient extraction of lipids and detergent before dextran derivatization. Thus, chloroform extraction of derivatized sample was performed prior to FACE analysis. Fluorescein was added as a migration marker to a final concentration of 50 nM. Samples were stored at −20 °C after derivatization with no observable degradation up to 3 months.
Scheme I.
Derivatization of dextran with APTS via reductive amination.
FACE instrumentation
FACE instrumentation was built in-house and data was acquired using a custom Lab View (National Instruments) program.56 Fused silica capillaries (25 μm i.d., 360 μm o.d.) (Innovaquartz, Phoenix, AZ) with a 32 cm separation distance and 42 cm total length were coated with 0.2 % polyethylene oxide (PEO).57 A 24 kV potential was applied using a Spellman CZE 1000R power supply (Hauppauge, NY). Samples were injected using gravity flow for 4–6 s at a height of 10 cm. Capillaries were rinsed with buffer between runs and a freshly coated capillary was used each day. Peak analysis was performed using Cutter 7.0 (beta).58 Fractional peak areas were calculated as follows:
| Equation 1 |
where peaks 4–20 in the resulting electropherograms were utilized (see below for explanation). The relative retention ratio of dextrans in PPNs was calculated as follows:
| Equation 2 |
where the peak area in the original distribution was determined prior to encapsulation.
Estimation of PPN pore size
The hydrodynamic pore radius (rp) was estimated by calculating the molecular weight of the smallest dextran oligomers with 90% retention in PPNs:59
| Equation 3 |
Preparation of PPNs for proton permeability assays
H+-sensitive PPNs were prepared by encapsulating 10,000 MW FITC-dextran in 10 mM phosphate buffer (pH =7, supplemented with 100 mM NaCl), into PPNs that were doped with 1 mol % Rh-DPPE. Rh-DPPE-labeled liposomes were prepared using binary bis-SorbPC/DOPC compositions ranging from 0 mol % to 20 mol % bis-SorbPC. Residual unencapsulated material was separated using a Sepharose CL-4B (Sigma, St. Louis, MI) packed column equilibrated with the same buffer. After initial recording, pH was lowered by adding 100 mM citrate buffer (pH =5.5) with 100 mM NaCl. Fluorescence was recorded for both FITC and rhodamine using a Fluorolog 3 spectrofluorometer (Jobin Yvon, Edison, NJ).
RESULTS AND DISCUSSION
Characterization of porosity in three-dimensional, polymeric nanostructures presents a number of physical and chemical challenges. The primary goal of this work was to determine the NMCL within the PPN membrane using dextran retention, an approach that is amenable to a range of other porous nanoarchitectures. For these experiments, we encapsulated unlabeled linear dextrans from a 6,000 Da dextran sample within PPNs and allowed them to leak from the porous nanoarchitecture to establish an equilibrium distribution of retained material. Linear dextrans were chosen to meet three primary requirements of the encapsulated material: a) water soluble; b) minimal shape and conformation variability over the size range; and c) quantized differences in molecular size. Dextrans were subjected to post-labeling with APTS (MW = 523.40) following recovery to decrease the minimum size that could be resolved. We hypothesized that dextran oligomers with hydrodynamic radii that were smaller than the inherent pores within the PPN membrane would leak from the PPN, whereas larger dextran oligomers would be retained. Comparison of the equilibrium encapsulated dextran oligomers to the original dextran solution would therefore reveal the approximate NMCL within the PPN membrane.
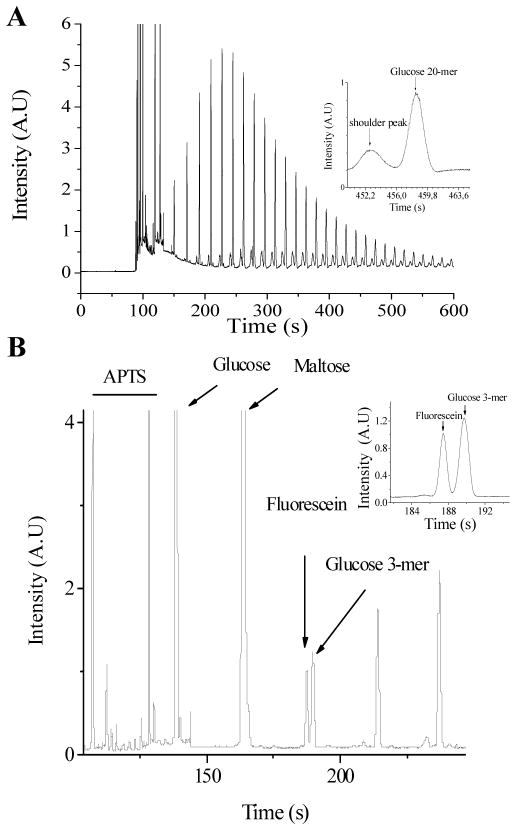
To determine the maximal size-dependent resolution that could be obtained from the dextran population, high resolution FACE separation of APTS-labeled dextrans was performed under EOF-suppressed conditions using polyethylene oxide (PEO)-coated capillaries.57 APTS derivatization yields fluorescent dextrans that contain only one fluorophore with no size-dependent labeling biases within this size distribution range, thus providing a clear measure of the dextran size distribution.51,53,54 Under EOF suppressed conditions, migration of APTS-labeled dextran oligomers is dependent on the viscous drag on the molecule, which is directly proportional to the hydrodynamic radius and thus number of glucose monomers. The apparent electrophoretic mobility (μapp) was inversely proportional to the number of glucose monomers, supporting the size dependent separation (Supporting Information).60 A representative electropherogram for APTS-labeled dextrans is shown in Figure 1A. A 6,000 Da dextran distribution was chosen based on preliminary observations that NMCL of PPNs was > 1,000 Da, but likely < 3,000 Da. The inset represents the peaks obtained for the glucose 20-mer from the 6,000 Da dextran distribution, which yielded N = 460,000 plates/m. The low abundance peak that corresponds to branched oligomers55 was omitted in all calculations. Unambiguous identification of the number of glucose units in derivatized dextran oligomers is required to assign molecular weight cutoff values to PPN pores. To obtain this information, we utilized fluorescein in combination with APTS-labeled glucose and maltose (glucose disaccharide) standards.55 Fluorescein reproducibly migrated immediately before the glucose 3-mer, and was thus added to all derivatized dextran samples to serve as a migration marker for convenient peak identification (Figure 1B). Thus, optimal separation resolution was obtained between 4 and 20 glucose monomers (MW = 666 to 3258 Da in the unlabeled dextran sample) and peaks corresponding to this size range were used for all further analyses.
Figure 1.
FACE of APTS-dextran. (A) Electropherogram of APTS-labeled 6000 MW dextran sample. Inset: Peaks corresponding to a glucose 20-mer. (B) Peak identification for dextran oligomers using APTS-labeled glucose, maltose and fluorescein standards. Inset: Enhanced resolution of the glucose trimer and fluorescein peaks. All separations were performed using a 25μm i.d., PEO-coated capillary at 570 V/cm in 20 mM phosphate buffer at pH 7.0.
Post-labeling of single dextran distribution coupled with FACE separation has not previously been utilized for determination of NMCL in porous membranes. To evaluate the feasibility of this approach, we used dialysis membranes with NMCL = 1,000 Da and NMCL = 3,500 Da. The dextran sample was divided into two fractions: the first fraction was dialyzed overnight in the dialysis membrane against H2O at 4 °C and the retained dextran was collected whereas the second fraction was stored under identical conditions with no dialysis. The samples were then lyophilized, APTS-labeled, and analyzed by FACE.
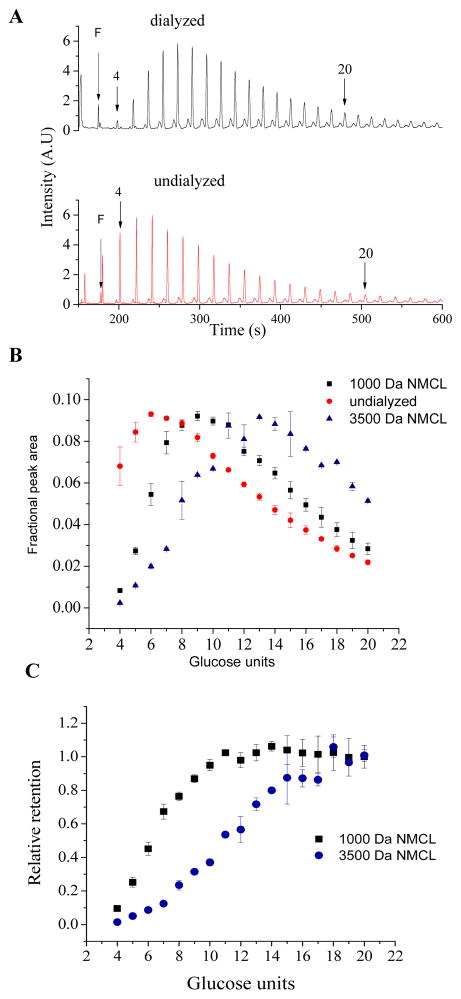
The APTS-labeled samples show two distinct peak distributions, with the dialyzed dextran yielding decreased peak areas for peaks that correspond to lower molecular weight (MW) oligomers compared to the non-dialyzed dextran (Figure 2A). The fraction of dextran oligomers retained in 1,000 Da NMCL membranes was compared to the non-dialyzed dextran and used to estimate the 50 % (MW50) and 90 % (NMCL) cutoff of the membrane. The fractional contribution of each peak to the total peak area (for peaks 4–20) calculated using Equation 1 is shown in Figure 2B where the contribution from lower MW oligomers is observed to decrease with increasing NMCL of the dialysis membrane. Further, the relative retention of oligomers was determined by calculating the ratio of fractional peak areas for individual oligomers. The relative retention of oligomers with > 14 glucose units was constant indicating that these oligomers did not permeate the dialysis membrane (Figure 2C). The 1,000 Da NMCL dialysis membrane yielded MW50 and NMCL corresponding to 6 (990 Da) and 10 (1638 Da) glucose units, respectively. The 3,500 Da NMCL dialysis membrane yielded MW50 corresponding to 1,300 Da (11 glucose units) and a less-defined NMCL corresponding to ca. 2,600 Da (ca. 22 glucose units). These results vary slightly from the manufacturers’ stated values, which are determined using globular proteins, suggesting that the observed variations may be due to shape effects. The results obtained by the dextran retention FACE assay lie within a factor of 2 of the company provided values, suggesting this analytical method is suitable for assessing the NMCL of porous nanoarchitectures based on differential dextran retention.
Figure 2.
Evaluation of APTS-dextran retention using dialysis membranes. (A) Electropherograms of APTS-labeled 6000 MW dextran sample before and after dialysis using a 1000 Da NMCL membrane. Peaks corresponding to dextrans comprised of 4 and 20 glucose units are denoted by arrows. (B) Fractional peak areas corresponding to dextran oligomers retained in 1000 Da NMCL membrane (black squares), 3500 Da NMCL membrane (blue triangles) and undialyzed 6000 Da MW dextran (red circles). (C) Relative retention of dextran by 1000 Da NMCL membrane (squares) and 3500 Da NMCL membrane (circles).
NMCL determination in PPNs
PPNs were used to encapsulate a population of dextran oligomers that were then allowed to leak from the PPN. PPN leakage was performed in a dialysis bag (NMCL = 12,000 – 14,000 Da) at 4°C for 18 h to ensure complete size-dependent removal of low MW species and to remove leaked contents that may result from premature vesicle disruption. Analysis of retained compounds compared to leaked compounds avoided dilution effects during the leakage assay and minimized contamination from vesicles that were inadvertently disrupted. Retained dextran was then recovered from PPNs via surfactant lysis and solvent extraction followed by APTS-labeling. Dextran retention in DOPC liposomes was used as a control for assessing PPN permeability since DOPC liposomes exhibit low permeability towards hydrophilic species.35
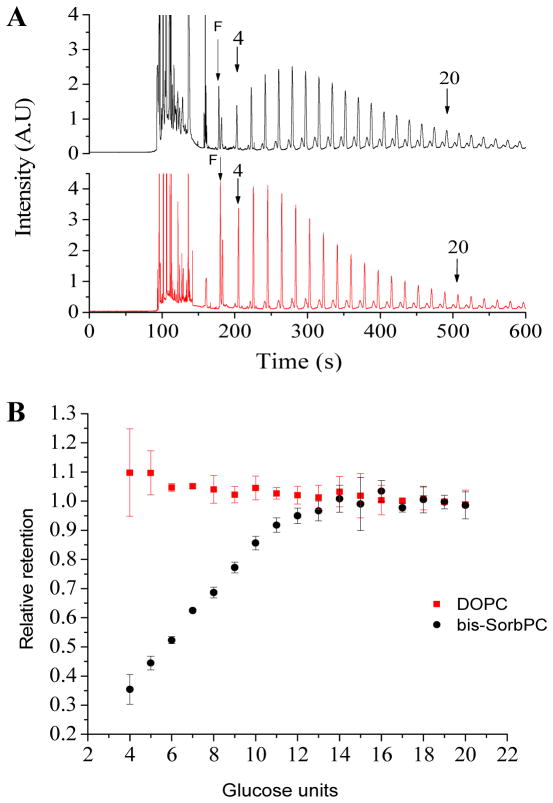
When analyzed by FACE, dextran recovered from DOPC liposomes exhibited a distinctly different oligomer distribution compared to dextran recovered from PPNs (Figure 3A). Specifically, a larger fraction of lower MW (< 7 glucose units) species were retained within the DOPC liposomes compared to PPNs. The MW50 of PPNs was determined to be 990 Da, similar to 1,000 Da NMCL dialysis membrane whereas the NMCL corresponded to ca. 11 glucose units (1800 Da) (Figure 3B). No distinct NMCL was observed for DOPC, further supporting the integrity of the DOPC membrane, relative to the inherently porous PPN.
Figure 3.
Size-based retention of dextrans in PPNs. (A) Electropherograms of APTS-labeled dextran retained within PPNs (top) and DOPC vesicles (bottom). Peaks corresponding to dextrans comprised of 4 and 20 glucose units are denoted by arrows. (B) Relative retention as a function of size for dextrans retained in DOPC vesicles (squares) and PPNs (circles).
These dextran retention data were then used to estimate rp of the pores found within the PPN membrane. Dextran has been modeled as oblate cylinders and the dextran radius (rD) was calculated using DOSY-NMR61 and gel permeation chromatography.59 For 2,000 Da dextran, rD was estimated to be 1.1 nm using the relationship derived from DOSY-NMR.61 Conversely, permeation chromatography tends to overestimate rD compared to equivalent-molecular weight spherical proteins and other spherical molecules such as Ficoll, though this effect is minimal (< 25%) in the lower molecular weight regime.62,63,64 Since the persistence length of dextrans ranges from 6–13 Å,63 the possibility of adapting multiple molecular configurations is also minimal in this size regime. Thus, rD should approximate the pore radius, rp.
These previously-derived relationships were therefore used to predict maximal rp of PPN pores. For NMCL = 1800, rp was calculated to be 1.3 nm using Equation 3 assuming the temperature and concentration of dextran has minimal effect on rD. Thus, the maximal pore diameter is estimated to be 2.6 nm for PPNs. These data provide the highest MW resolution to date for pore size and membrane transport in bis-SorbPC and provide a quantitative explanation for prior empirical observations of lower encapsulation efficiency of low molecular weight species in bis-SorbPC liposomes.65,66 Interestingly, assuming a cross-sectional area of 65 Å2 per phosphorylcholine lipid,67 the largest pores occupy an area of only ca. eight lipid molecules.
Permeability of binary lipid mixtures
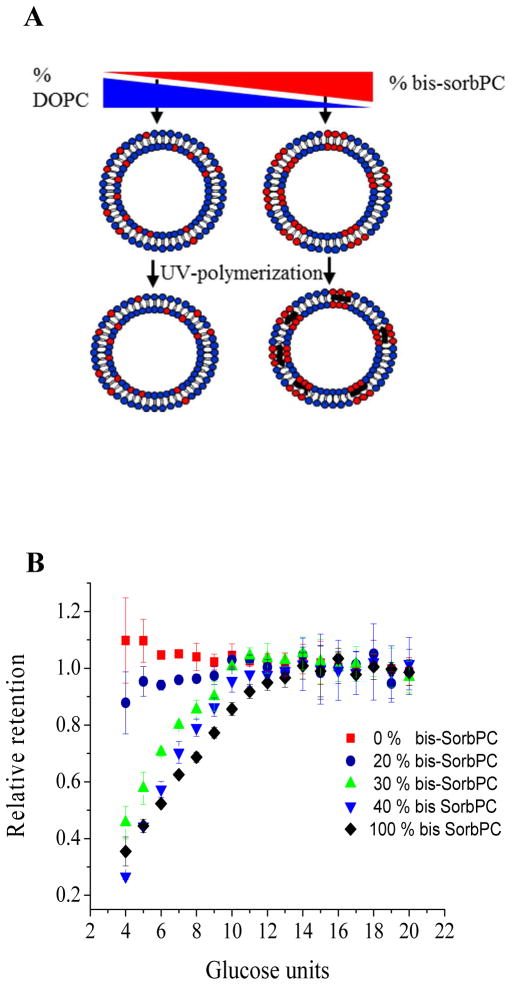
We hypothesized that the two distinct permeability characteristics of liposomes prepared from DOPC or bis-SorbPC may be utilized to control the relative porosity, and thus the membrane transport properties, of PPNs via preparation of PPNs with binary lipid compositions. Previous studies had shown for bis-SorbPC:DOPC mixtures that the membrane permeability was similar to that of pure DOPC when bis-SorbPC ≤ 30 mol%.46,48 Binary membrane compositions with higher bis-SorbPC content have not been reported. Specifically, an increase in bis-SorbPC content was hypothesized to increase pore density without affecting NMCL, since NMCL is primarily dependent on the largest pore diameter within the membrane, a property inherent to the molecular structure of bis-SorbPC as revealed in pure bis-SorbPC PPNs. To test this hypothesis, we prepared binary PPNs with varying compositions of DOPC and bis-SorbPC (Figure 4A). Size determination at different stages of the experiment confirmed that a substantial fraction of the binary PPNs were intact until detergent lysis (Supporting Information). Further, PPNs were collected immediately off of the Sepharose column after a defined leakage time, thus separating lysed vesicles from the analyzed population. Combined, these factors indicate that leakage occurred through the pores and not via PPN disruption.
Figure 4.
Dextran retention in mixed lipid PPNs. (A) Schematic representation of PPNs comprised of binary mixtures of bis-SorbPC and DOPC. (B) Relative retention of 6,000 MW dextran sample in PPNs prepared with 100 mol % bis-SorbPC (black diamonds), 40 mol % bis-SorbPC (blue triangles), 30 mol % bis-SorbPC (green triangles), 20 mol % bis-SorbPC (blue circles) and 100mol % DOPC (red squares).
PPNs prepared with 40 mol % bis-SorbPC have similar permeability characteristics to that of 100 mol % bis-SorbPC (Figure 4B), suggesting that the size of the pores in the bilayer is not significantly altered compared to the pore structure found in PPNs comprised of 100 mol % bis-SorbPC. Thus, during the 18 h time course of the experiment, membrane transport was similar in both populations. Unfortunately, the dextran retention assay is not amenable to time resolved measurement of leakage kinetics, which may reveal more subtle differences in rapid transport dynamics.
In contrast, permeability is drastically reduced when bis-SorbPC content is reduced to 20 mol %, where dextran leakage is only slightly higher than that observed from DOPC liposomes. Interestingly, at 30 mol % bis-SorbPC the permeability properties approach that observed for ≥ 40 mol % bis-SorbPC (Figure 4B). Prior studies have shown that for binary lipid membranes comprised solely of phosphorylcholine containing lipids, there was no significant difference in the lipid arrangement between polymerized and unpolymerized domains.46,48 Interestingly, the inherent pore formation within pure bis-SorbPC suggests that pore formation should occur within binary PPNs as well if the DOPC and bis-SorbPC domains are completely phase segregated upon polymerization as previously suggested. Thus, the data observed here suggest and alternate arrangement within the membrane that leads to lower permeability at low bis-SorbPC compositions. The significant change in PPN permeability observed above 30 mol % bis-SorbPC may be understood by considering the molecular arrangement of lipid mixtures at the gel point. At the gel point, the miscibility of lipids is greatly reduced, leading to the formation of lipid domains. The net permeability of the structure is governed by the lipid domain with the higher permeability. Thus below the gel point, DOPC and bis-SorbPC are evenly dispersed, minimizing the formation of packing defects found for bis-SorbPC. In fact, the gel point of bis-SorbPC is ca. 30 mol % bis-SorbPC in a binary membrane,68 where a significant shift in the liposome stability and solubility is attributed to an increase in cross-linking. The reduced NMCL observed for <30 mol % bis-SorbPC likely results from increased lipid miscibility which minimizes pore formation. Further elucidation of the underlying mechanism will require an assay with higher temporal resolution.
Proton permeability into binary lipid mixtures
The low permeability of dextrans observed for bis-SorbPC compositions < 30 mol%, combined with previous observations of low bilayer permeability in this size regime poses a series of interesting questions. Most importantly, if bis-SorbPC forms domains upon polymerization, and if these domains are intrinsically more permeable than unpolymerized lipids, then why do the membranes appear impermeable to low molecular weight species such as glucose and calcein?46,48 To probe for altered permeability below 30 mol % bis-SorbPC, where dextran retention lacked resolution, we used a proton permeability assay pH-sensitive PPNs were prepared by encapsulating dextran conjugated FITC and doping rhodamine-conjugated lipids into the membrane. The result is a pH-sensitive PPN that exhibits a change in fluorescein emission intensity as a function of pH, whereas the rhodamine intensity is unchanged.
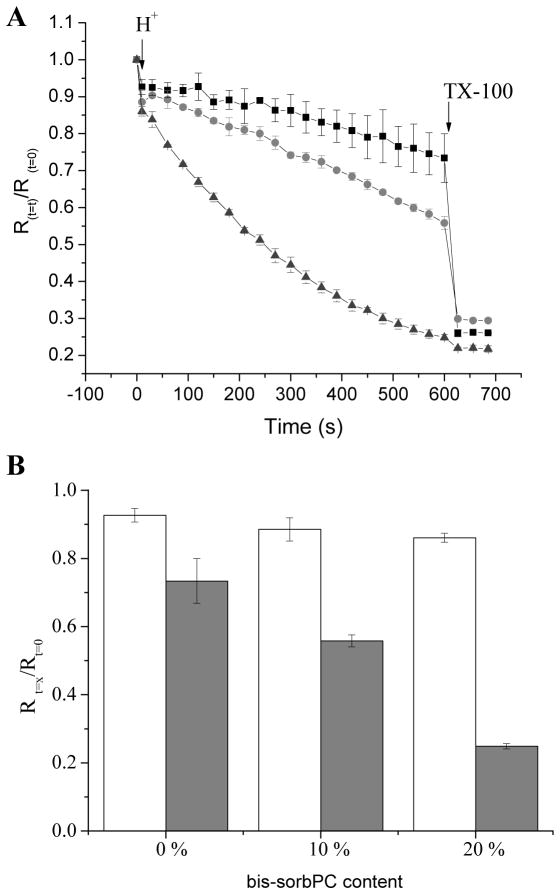
When pH-sensitive binary PPNs were subjected to a step change from pH 7.0 to 5.5, the fluorescence ratio of FITC/rhodamine decreased as a function of time (Figure 5A). The rate of change increased with increasing molar ratio (0 – 20 mol %) of bis-SorbPC indicative of more rapid proton access to the encapsulated FITC. At 600 s, PPNs were disrupted using surfactant to facilitate complete proton access to the encapsulated FITC. Importantly, the H+ permeability observed for DOPC liposomes is consistent with previous reports.69 For 20 mol % bis-SorbPC, the encapsulated FITC experienced the maximal pH change prior to disruption, whereas only partial leakage was observed for 0 – 10 mol % bis-SorbPC (Figure 5B). Combined with the dextran retention, these data suggest that even at 20 mol % bis-SorbPC, higher permeability is observed in PPNs compared to natural lipid liposomes, though the size dependence and mechanism for the enhanced permeability requires further exploration.
Figure 5.
H+-permeability of binary lipid composition PPNs. (A) Normalized fluorescence ratio of FITC/rhodamine for PPNs comprised of 0 mol % bis-SorbPC (black squares), 10 mol % bis-SorbPC (red circles) and 20 mol % bis-SorbPC (green triangles). DOPC was used as the other primary lipid component. The pH was decreased from 7.0 to 5.5 at t = 0 s. (B) The normalized fluorescence ratio compared at 10 s (open) and 600 s (gray) after H+ spike with the three different compositions. Triton X-100 was added at t = 600s to lyse PPNs.
The capability to tune the pore size and the relative porosity in PPNs, and other nanostructures facilitates a wide range of applications in chemical sensing and delivery. For example, encapsulation of enzymes within the PPN necessitates sufficiently small membrane pores for retention of the enzyme, yet sufficiently large pores to allow unobstructed transport of enzyme substrates and products. Conversely, tuning pore density may facilitate control over mass transport kinetics, enabling optimization of individual PPN properties for specific enzyme combinations. The data presented here support the hypothesis that, above 30 mol % bis-SorbPC, the pore size is an inherent property of the bis-SorbPC, though techniques with higher MW resolution may reveal subtle differences in the future. A limitation of the approach presented herein is the lack of temporally resolved data for dextran retention, which restricts the ability to compare pore density and pore size as primary mechanisms of mass transport near the gel point.
CONCLUSIONS
PPNs composed of bis-SorbPC yield NMCL of ca. 1800 Da as assessed by FACE of retained dextran with a mass resolution of 162 Da. This NMCL corresponds to a maximum pore diameter of 2.6 nm. Modulation of relative porosity via preparation of binary lipid PPNs revealed that NMCL does not change significantly down to 30 mol % bis-SorbPC. However, a significant reduction in permeability to larger molecular weight species was observed when bis-SorbPC content was decreased to 20 mol %, though proton permeability remained significantly higher compared to non-porous liposomes. The differences in permeability as a function of bis-SorbPC composition are likely due to the alterations in lipid distribution. Improved temporal resolution in future studies should further clarify the dynamic, size-dependent permeability of PPN membranes. Finally, this approach is amenable to size determination in other nanoshell systems and the information garnered should markedly impact the rational design and synthesis of novel porous nanoshell architectures.
Supplementary Material
Acknowledgments
We thank Dr. Yehia Mechref for useful comments on dextran derivatization. We also thank Dr. Femina Rauf and Elyssia Steinwinter for their assistance with the FACE instrumentation. Funding was provided by NIH (EB007047) and NSF (CHE-0548167).
References
- 1.Janczak CM, Aspinwall CA. Anal Bioanal Chem. 2012;402(1):83–89. doi: 10.1007/s00216-011-5482-5. [DOI] [PubMed] [Google Scholar]
- 2.Cheng ZL, Aspinwall CA. Analyst. 2006;131(2):236–243. doi: 10.1039/b511083a. [DOI] [PubMed] [Google Scholar]
- 3.Aylott JW. Analyst. 2003;128(4):309–312. doi: 10.1039/b302174m. [DOI] [PubMed] [Google Scholar]
- 4.Tapec R, Zhao XJJ, Tan WH. J Nanosci Nanotech. 2002;2(3–4):405–409. doi: 10.1166/jnn.2002.114. [DOI] [PubMed] [Google Scholar]
- 5.Qhobosheane M, Santra S, Zhang P, Tan WH. Analyst. 2001;126(8):1274–1278. doi: 10.1039/b101489g. [DOI] [PubMed] [Google Scholar]
- 6.Lee YEK, Smith R, Kopelman R. Ann Rev Anal Chem. 2009;2:57–76. doi: 10.1146/annurev.anchem.1.031207.112823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jesorka A, Orwar O. Ann Rev Anal Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- 8.Clark HA, Hoyer M, Philbert MA, Kopelman R. Anal Chem. 1999;71(21):4831–4836. doi: 10.1021/ac990629o. [DOI] [PubMed] [Google Scholar]
- 9.Gao D, Xu H, Philbert MA, Kopelman R. Nano Letters. 2008;8(10):3320–3324. doi: 10.1021/nl8017274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeoka S, Sakai H, Ohno H, Tsuchida E. Macromolecules. 1991;24(6):1279–1283. [Google Scholar]
- 11.Yamada T, Iwasaki Y, Tada H, Iwabuki H, Chuah MKL, VandenDriessche T, Fukuda H, Kondo A, Ueda M, Seno M, Tanizawa K, Kuroda S. Nature Biotechn. 2003;21(8):885–890. doi: 10.1038/nbt843. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Barnes JC, Bosoy A, Stoddart J, Zink JI. Chem Soc Rev. 2012;41(7):2590–2605. doi: 10.1039/c1cs15246g. [DOI] [PubMed] [Google Scholar]
- 13.Tassa C, Shaw SY, Weissleder R. Acc Chem Res. 2011;44(10):842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones CF, Grainger DW. Adv Drug Delivery Rev. 2009;61(6):438–456. doi: 10.1016/j.addr.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puri A, Loomis K, Smith B, Lee JH, Yavlovich A, Heldman E, Blumenthal R. Crit Rev Therapeutic Drug Carrier Systems. 2009;26(6):523–580. doi: 10.1615/critrevtherdrugcarriersyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian T, Ho RJY. J Pharmaceutical Sci. 2001;90(6):667–680. doi: 10.1002/jps.1023. [DOI] [PubMed] [Google Scholar]
- 17.Vivero-Escoto JL, Huxford-Phillips RC, Lin W. Chem Soc Rev. 2012;41(7):2673–2685. doi: 10.1039/c2cs15229k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardhan R, Lal S, Joshi A, Halas NJ. Acc Chem Res. 2011;44(10):936–946. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanson C, Diou O, Thevenot J, Ibarboure E, Soum A, Brulet A, Miraux S, Thiaudiere E, Tan S, Brisson A, Dupuis V, Sandre O, Lecommandoux S. ACS Nano. 2011;5(2):1122–1140. doi: 10.1021/nn102762f. [DOI] [PubMed] [Google Scholar]
- 20.Su CH, Sheu HS, Lin CY, Huang CC, Lo YW, Pu YC, Weng JC, Shieh DB, Chen JH, Yeh CS. J Am Chem Soc. 2007;129(7):2139–2146. doi: 10.1021/ja0672066. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Z, Tsourkas A. Langmuir. 2008;24(15):8169–8173. doi: 10.1021/la801027q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z, Thorek DL, Tsourkas A. Angew Chem-Int Ed. 2010;49(2):346–350. doi: 10.1002/anie.200905133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Z, Elias DR, Kamat NP, Johnston ED, Poloukhtine A, Popik V, Hammer DA, Tsourkas A. Bioconj Chem. 2011;22(10):2021–2029. doi: 10.1021/bc200214g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z, Thorek DL, Tsourkas A. Adv Func Mat. 2009;19(23):3753–3759. doi: 10.1002/adfm.200901253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreft O, Prevot M, Mohwald H, Sukhorukov GB. Angew Chem Int Ed. 2007;46(29):5605–5608. doi: 10.1002/anie.200701173. [DOI] [PubMed] [Google Scholar]
- 26.Hotz J, Meier W. Langmuir. 1998;14(5):1031–1036. [Google Scholar]
- 27.Graff A, Sauer M, Van Gelder P, Meier W. Proc Nat Acad Sci USA. 2002;99(8):5064–5068. doi: 10.1073/pnas.062654499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuiper SM, Nallani M, Vriezema DM, Cornelissen JJ, van Hest JC, Nolte RJ, Rowan AE. Organic & Biomolecular Chemistry. 2008;6(23):4315–4318. doi: 10.1039/b811196k. [DOI] [PubMed] [Google Scholar]
- 29.Zhang T, Ge J, Hu Y, Zhang Q, Aloni S, Yin Y. Angew Chem-Int Ed. 2008;47(31):5806–5811. doi: 10.1002/anie.200800927. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Zhang T, Ge J, Yin Y. Nano Letters. 2008;8(9):2867–2871. doi: 10.1021/nl8016187. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Zhang Q, Goebl J, Zhang T, Yin Y. Phys Chem Chem Phys. 2010;12(38):11836–11842. doi: 10.1039/c0cp00031k. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Chen H, Guo L, He Q, Chen F, Zhou J, Feng J, Shi J. ACS Nano. 2010;4(1):529–539. doi: 10.1021/nn901398j. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Zhou Y, Li Y, Bandosz TJ, Akins DL. J Coll Interf Sci. 2012;375:106–111. doi: 10.1016/j.jcis.2012.02.046. [DOI] [PubMed] [Google Scholar]
- 34.Graff A, Winterhalter M, Meier W. Langmuir. 2001;17(3):919–923. [Google Scholar]
- 35.Cheng Z, D’Ambruoso GD, Aspinwall CA. Langmuir. 2006;22(23):9507–9511. doi: 10.1021/la061542i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts DL, Ma Y, Bowles SE, Janczak CM, Pyun J, Saavedra SS, Aspinwall CA. Langmuir. 2009;25(4):1908–1910. doi: 10.1021/la803358m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan L, Miao Q, Sun Y, Yang FY. FEBS Letters. 2003;555(3):545–550. doi: 10.1016/s0014-5793(03)01346-2. [DOI] [PubMed] [Google Scholar]
- 38.Ladokhin AS, Selsted ME, White SH. Biophys J. 1997;72(4):1762–1766. doi: 10.1016/S0006-3495(97)78822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueno M. Biochim Biophys Acta. 1987;904(1):140–144. doi: 10.1016/0005-2736(87)90095-2. [DOI] [PubMed] [Google Scholar]
- 40.Park J, McShane MJ. ACS Appl Mat Inter. 2010;2(4):991–997. doi: 10.1021/am900673r. [DOI] [PubMed] [Google Scholar]
- 41.Stein EW, Grant PS, Zhu H, McShane MJ. Anal Chem. 2007;79(4):1339–1348. doi: 10.1021/ac061414z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siuti P, Retterer ST, Doktycz MJ. Lab on A Chip. 2011;11(20):3523–3529. doi: 10.1039/c1lc20462a. [DOI] [PubMed] [Google Scholar]
- 43.Lindemann M, Winterhalter M. IEE Proc-Nanobiotechnology. 2006;153(4):107–111. doi: 10.1049/ip-nbt:20050027. [DOI] [PubMed] [Google Scholar]
- 44.Scrimin P, Tecilla P, Moss RA, Bracken K. J Am Chem Soc. 1998;120(6):1179–1185. [Google Scholar]
- 45.Ross EE, Rozanski LJ, Spratt T, Liu SC, O’Brien DF, Saavedra SS. Langmuir. 2003;19(5):1752–1765. [Google Scholar]
- 46.Frankel DA, Lamparski H, Liman U, Obrien DF. J Am Chem Soc. 1989;111(26):9262–9263. [Google Scholar]
- 47.Heitz BA, Xu J, Jones IW, Keogh JP, Comi TJ, Hall HK, Aspinwall CA, Saavedra SS. Langmuir. 2011;27(5):1882–1890. doi: 10.1021/la1025944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamparski H, Liman U, Barry JA, Frankel DA, Ramaswami V, Brown MF, O’Brien DF. Biochemistry. 1992;31(3):685–694. doi: 10.1021/bi00118a008. [DOI] [PubMed] [Google Scholar]
- 49.Ross EE, Rozanski LJ, Spratt T, Liu SC, O’Brien DF, Saavedra SS. Langmuir. 2003;19(5):1752–1765. [Google Scholar]
- 50.Neuhaus W, Bogner E, Wirth M, Trzeciak J, Lachmann B, Gabor F, Noe CR. Pharmaceutical Res. 2006;23(7):1491–1501. doi: 10.1007/s11095-006-0256-z. [DOI] [PubMed] [Google Scholar]
- 51.Evangelista RA, Guttman A, Chen FTA. Electrophoresis. 1996;17(2):347–351. doi: 10.1002/elps.1150170210. [DOI] [PubMed] [Google Scholar]
- 52.Guttman A, Chen FTA, Evangelista RA, Cooke N. Anal Biochem. 1996;233(2):234–242. doi: 10.1006/abio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 53.Evangelista RA, Liu MS, Chen FTA. Anal Chem. 1995;67(13):2239–2245. [Google Scholar]
- 54.O’Shea MG, Samuel MS, Konik CM, Morell MK. Carbohydrate Research. 1998;307(1–2):1–12. [Google Scholar]
- 55.Chen F-TA. Characterization of Oligosaccharides from Starch, Dextran, Cellulose, and Glycoproteins by Capillary Electrophoresis. In: Thibault P, Honda S, editors. Capillary Electrophoresis of Carbohydrates. Humana Press; Totowa, N.J: 2003. pp. 105–120. [DOI] [PubMed] [Google Scholar]
- 56.Rauf F, Huang Y, Muhandiramlage TP, Aspinwall CA. Anal Bioanal Chem. 2010;397(8):3359–3367. doi: 10.1007/s00216-010-3776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preisler J, Yeung ES. Anal Chem. 1996;68(17):2885–2889. doi: 10.1021/ac960260s. [DOI] [PubMed] [Google Scholar]
- 58.Shackman JG, Watson CJ, Kennedy RT. J Chrom A. 2004;1040(2):273–282. doi: 10.1016/j.chroma.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Venturoli D, Rippe B. Am J Physiol-Renal Physiol. 2005;288(4):F605–F613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- 60.Stefansson M, Novotny M. Anal Chem. 1994;66(7):1134–1140. doi: 10.1021/ac00079a031. [DOI] [PubMed] [Google Scholar]
- 61.Viel S, Capitani D, Mannina L, Segre A. Biomacromolecules. 2003;4(6):1843–1847. doi: 10.1021/bm0342638. [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen KE, Moller JV. Am J Physiol. 1979;236(2):F103–F111. doi: 10.1152/ajprenal.1979.236.2.F103. [DOI] [PubMed] [Google Scholar]
- 63.Bohrer MP, Patterson GD, Carroll PJ. Macromolecules. 1984;17(6):1170–1173. [Google Scholar]
- 64.Oliver JD, Anderson S, Troy JL, Brenner BM, Deen WH. J Am Soc Nephrology. 1992;3(2):214–228. doi: 10.1681/ASN.V32214. [DOI] [PubMed] [Google Scholar]
- 65.Armitage BA, Bennett DE, Lamparski HG, O’Brien DF. Adv Poly Sci. 1996;126:53–84. [Google Scholar]
- 66.Bondurant B, Mueller A, O’Brien DF. Biochim Biophys Acta. 2001;1511:113–121. doi: 10.1016/s0005-2736(00)00388-6. [DOI] [PubMed] [Google Scholar]
- 67.Nagle JF. Biophys l J. 1993;64(5):1476–1481. doi: 10.1016/S0006-3495(93)81514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sisson TM, Lamparski HG, Kolchens S, Elayadi A, O’Brien DF. Macromolecules. 1996;29(26):8321–8329. [Google Scholar]
- 69.Norris FA, Powell GL. Biochim Biophys Acta. 1990;1030(1):165–171. doi: 10.1016/0005-2736(90)90252-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.