Abstract
The diverse spatial and temporal expression of alternatively spliced transcript isoforms shapes neurodevelopment and plays a major role in neuronal adaptability. Although alternative splicing is extremely common in the brain, its role in mental illnesses such as schizophrenia has received little attention. To examine this relationship, postmortem brain tissue was obtained from 20 individuals with schizophrenia (SZ) and 20 neuropsychiatrically normal comparison subjects. Gray matter samples were extracted from two brain regions implicated in the disorder: Brodmann area 10 and caudate. Affymetrix Human Gene 1.0 ST arrays were used on four subjects per group to attain an initial profile of differential expression of transcribed elements within and across brain regions in SZ. Numerous genes of interest with altered mRNA transcripts were identified by microarray through the differential expression of particular exons and 3′ untranslated regions (UTRs) between diagnostic groups. Select microarray results—including dysregulation of ENAH exon 11a and CPNE3 3′UTR—were verified by qRTPCR and replicated in the remaining independent sample of 16 SZ patients and 16 normal comparison subjects. These results, if further replicated, clearly illustrate the importance of Identifying transcriptomic variants in expression studies, and implicate novel candidate genes in the disorder.
INTRODUCTION
One method commonly employed to unravel the underlying biology of schizophrenia (SZ) is transcriptomic profiling of postmortem brain tissue (Glatt et al., 2005; Horvath et al., 2011; Mirnics et al., 2006). Most prior transcriptomic studies of the brain in SZ overlooked a potentially important source of phenotypic diversity: the expression of alternatively spliced variants (ASVs). Alternative splicing is a major mechanism by which eukaryotes create enormous proteomic diversity from a smaller-than-expected number of genes. The manner in which elements of a particular gene are spliced has a significant impact on protein function. In fact, the discrete ASVs of some genes have diametrically opposed physiological functions (Clark et al., 2007); thus, traditional discussions about a the function of a given gene may be moot unless a particular ASV of the gene is invoked.
Transcriptome-wide sequence analysis has detected splicing events in up to 95% of multi-exon genes (Pan et al., 2008). Alternative splicing events are tissue-specific, and can be regionally specific within a tissue, including the brain (Twine et al., 2011), resulting in an extremely complex expression profile. These diverse splicing patterns dictate important regulatory decisions in many steps of neuronal development, including axon guidance (Schmucker et al., 2000; Zipursky et al., 2006), cell-fate determination (Dho et al., 1999; Dho et al., 2006; Reugels et al., 2006), and synaptogenesis (Li et al., 2007). Notably, a small handful of candidate genes for SZ have been found to exhibit abnormal splicing patterns in individual regions of postmortem brain (Glatt et al., 2011), including CTNNA2 (Mexal et al., 2008), DISC1 (Nakata et al., 2009), ERBB4 (Law et al., 2007), ESR1 (Weickert et al., 2008), GRM3 (Sartorius et al., 2008), and NRG1 (Tan et al., 2007). It is not clear if these splicing patterns are abnormal only in those specific brain regions or if other regions are similarly affected.
The key technological advance provided by the Affymetrix Human Gene 1.0 ST arrays is measurement of the expression levels of individual exons and UTRs (hereafter collectively referred to as transcribed elements, or TEs). The aim of the current study was to use this technology to compare the expression profiles of TEs across the whole transcriptome in postmortem brain of individuals with SZ and control subjects in two brain regions previously implicated in the disorder (Brodmann Area 10 [BA10] and caudate head) (Ellison-Wright et al., 2008; Goghari, 2010; Yu et al., 2010) in order to identify alternative splicing abnormalities in SZ and to estimate their regional specificity.
METHODS
Samples
Postmortem brain samples were obtained from 20 SZ and 20 neuropsychiatrically normal comparison (NC) subjects in the Harvard Brain Tissue Resource Center. All tissues were collected with the full consent of the family or next of kin of the deceased. A “discovery sample” was constructed of four SZ and four NC subjects matched on age, sex, postmortem interval, and RNA integrity Number (RIN) (Table 1). The SZ discovery sample was selected to include two subjects on antipsychotic medication at the time of death and two who were not, allowing identification and exclusion of some medication effects from further validation steps (we note here that this contrast of small samples did not have adequate power to detect all medication-related effects, but it does provide one basis for filtering out genes that were strongly regulated by medication). Subsequent to verification, select observations from microarray analysis of the discovery sample were tested for replication in an independent, larger, and less homogeneous group of SZ and NC subjects (the “replication sample”).
Table 1.
Demographic and Agonal Characteristics of the SZ and NC Discovery and Replication Samples
| Discovery Samples | Replication Samples | |||
|---|---|---|---|---|
|
| ||||
| SZ | NC | SZ | NC | |
| Sample Size: n | 4 | 4 | 16 | 16 |
| Sex: % male (n) | 100 (4) | 100 (4) | 37.5 (6) | 31.2 (5) |
| Age: mean years±S.D. | 59.0±3.7 | 58.3±2.1 | 68.2±22.7 | 66.5±20.7 |
| Postmortem Interval: mean hours±S.D. | 24.9±4.1 | 24.0±3.5 | 18.8±7.0 | 21.9±5.3 |
| RNA Integrity Number (RIN): mean±S.D. | 8.0±1.2 | 7.8±1.3 | 7.9±0.5 | 8.3±1.3 |
No significant differences on any factor were observed between the SZ and NC discovery samples (all p>0.737) or between the SZ and NC replication samples (all p>0.183).
RNA Extraction and Purification
Gray matter tissue samples were dissected from two brain regions (BA10 and CAUD). All brains had a pH between 6 and 7. Samples remained frozen over dry ice until placed in RLT buffer with β-mercaptoethanol and homogenized using Qiashredder columns for subsequent nucleic acid isolation using Qiagen AllPrep DNA/RNA Mini Kits (see Supplemental Methods for further details). Using a Bioanalyzer 2100 and an RNA Nanochip, purity of each sample was determined by the A260:280 ratio, with acceptable values ranging from 1.8–2.2. The quality of each sample was assured by a RIN≥6 and visual confirmation of clear, distinct 28S and 18S rRNA peaks.
Microarray Procedures
Reverse-transcription, hybridization, and scanning were performed according to well-established protocols. Ambion WT Expression Kits were used for amplification and Affymetrix WT Terminal Labeling Kits were used for labeling. Total RNA was reverse-transcribed and hybridized onto GeneChip Human Gene 1.0 ST Arrays (Affymetrix) and scanned on a GeneChip 7G/4C scanner (Affymetrix). The Human Gene 1.0ST Array improves upon previous generations of Affymetrix arrays by probing the entire length of each transcript, rather than the 3′ end only. The Gene 1.0 ST Array interrogates 28,869 well-annotated genes with 764,885 distinct probes, with an average of one probe per exon and 27 probes per transcript. The array has greater than 99 percent coverage of NM sequences present in the November 3, 2006, RefSeq database (Affymetrix, 2007).
Microarray Data Preparation
Partek Genomics Suite software was utilized for all analyses of microarray scan data. Corrections for background signal were made by robust multi-array average (RMA) (Irizarry et al., 2003). The set of eight GeneChips was standardized using quantile normalization, and expression levels of each probe underwent log-2 transformation to yield data distributions more closely approximating normality. Summarization of redundant probesets was obtained by median polish. Probesets with a signal:noise ratio of less than 3.0 were excluded from subsequent analyses (Handran et al., 2002); this led to the exclusion of six probes from the BA10 dataset and seven probes from the CAUD dataset.
Microarray Data Analyses
Primary analyses were designed to detect genes with significantly different TE expression between diagnostic groups, potentially indicating different alternative splicing events between them. SZ and NC groups were compared on mean expression levels of all TEs in each gene on a gene-by-gene basis through ANCOVAs and inspection of interaction terms, as described previously (Glatt et al., 2009; Partek Incorporated, 2008) and in more detail in the Supplemental Methods. The key term for the present analyses was the interaction of TE identity (ID) with diagnostic group, which allowed for the detection of differences in the expression of one or more TEs in a gene between diagnostic groups (Partek Incorporated, 2008). The significance of these interaction terms (one per gene) was judged against a stringent Bonferroni-corrected threshold of p=2.47e−06, and post-hoc F-tests were used to identify the specific dysregulated TE(s) in the genes showing significant interactions.
Genes influenced by a significant interaction of diagnosis and TE ID in one or more brain regions were subjected to the DAVID algorithm (Dennis et al., 2003) to determine if the set was enriched with genes mapping to a particular biological pathway, or genes containing particular protein domains, which might indicate that the corresponding exonic sequences shared by these genes might provide the basis for their targeting by a common alternative-splicing regulator(s).
Reverse Transcription and Quantitative PCR
Quantitative reverse transcription (qRT) PCR was performed to confirm and validate select microarray results. Concentrations of total RNA isolates were adjusted prior to RT, and the same amount of total RNA was used in each reaction (20 ng/ul). QuantiTect RT kits (Qiagen) were used according to the manufacturer’s protocol. Each subject’s cDNA was run by qRTPCR in duplicate for each primer set (Supplemental Tables). Additional details are provided in Supplemental Methods.
Expression of each dysregulated TE was evaluated and compared with a non-dysregulated TE of the same gene. The primer set amplifying the non-dysregulated region of the transcript was considered the “control” primer set. The expression value from the primer set that amplified the dysregulated region (considered the “experimental” primer set) was then compared to that of the control primer set using the ΔΔCT method (Supplemental Methods). TE dysregulation was confirmed using linear regression models with diagnosis, age, sex, PMI, and RIN as independent predictors. The significance of the effect of diagnosis was determined with a 1-tailed p<0.05 for this term after backward removal of clearly non-significant terms (p<0.20) from each regression model.
RESULTS
Discovery Analyses
In BA10 from four SZ subjects and four well-matched NC subjects, 43 transcripts were influenced by a Bonferroni-corrected significant interaction of diagnosis and TE ID, indicative of differential expression of one or some (but not all) TEs between diagnostic groups (Table 2). DAVID analysis found no particular biological pathways or protein domains enriched among these 43 genes.
Table 2.
Transcripts Exhibiting a Bonferroni-Corrected Significant Interaction of Diagnosis (SZ vs. NC) and Transcript Element ID in Brodmann Area 10 (BA10)
| Gene Symbol | Gene Product | Reference Sequence | Transcript Cluster ID | Probe Sets (n) | F | p | Direction of Dysregulation# |
|---|---|---|---|---|---|---|---|
| CPNE3 | Copine III | NM_003909 | 8147172 | 21 | 15.003 | 6.6e−24 | ↓ |
| SCIN | Scinderin | NM_001112706 | 8131550 | 22 | 6.992 | 5.4e−13 | ↑ |
| SAMHD1 | SAM domain and HD domain 1 | NM_015474 | 8066117 | 18 | 8.354 | 7.6e−13 | ↑ |
| C4A | Complement component 4A (Rodgers blood group) | NM_007293 | 8118409 | 42 | 3.830 | 2.9e−11 | ↑ |
| IARS | Isoleucyl-tRNA synthetase | NM_013417 | 8162313 | 38 | 4.012 | 5.0e−11 | ↑ |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | NM_001276 | 7923547 | 14 | 9.026 | 5.7e−11 | ↑ |
| LPPR4 | Plasticity related gene 1 | NM_014839 | 7903214 | 12 | 9.775 | 3.3e−10 | ↑ |
| C4A | Complement component 4A (Rodgers blood group) | NM_007293 | 8118455 | 42 | 3.458 | 9.8e−10 | ↑ |
| SERPINA3 | Serpin peptidase inhibitor, clade A (alpha-1 antiprotein) | NM_001085 | 7976496 | 9 | 12.930 | 1.0e−9 | ↑ |
| C4A | Complement component 4A (Rodgers blood group) | NM_007293 | 8179399 | 40 | 3.507 | 1.5e−9 | ↑ |
| ALOX5 | Arachidonate 5-lipoxygenase | NM_000698 | 7927215 | 15 | 7.011 | 2.6e−9 | ↑ |
| CSDE1 | Cold shock domain containing E1, RNA-binding | NM_001007553 | 7918825 | 21 | 5.214 | 3.5e−9 | ↑ |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, a | NM_000944 | 8101971 | 18 | 5.734 | 6.3e−9 | ↑ |
| TLN1 | Talin 1 | NM_006289 | 8161056 | 57 | 2.791 | 6.7e−9 | ↑ |
| IFI16 | Interferon, gamma-inducible protein 16 | NM_005531 | 7906400 | 16 | 6.209 | 9.3e−9 | ↑ |
| RNASET2 | Ribonuclease T2 | NM_003730 | 8130768 | 14 | 6.923 | 1.1e−8 | ↑ |
| ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | NM_000690 | 7958784 | 15 | 6.330 | 1.9e−8 | ↓ |
| CD86 | CD86 molecule | NM_175862 | 8082035 | 10 | 9.325 | 2.2e−8 | ↑ |
| CUX1 | Cut-like homeobox 1 | NM_181552 | 8135114 | 34 | 3.424 | 4.5e−8 | ↑ |
| MAGI2 | Membrane associated guanylate kinase, WW and PDZ domain containing 2 | NM_012301 | 8140504 | 25 | 4.003 | 9.1e−8 | ↑ |
| LAT2 | Linker for activation of T cells family, member 2 | NM_032464 | 8133442 | 16 | 5.454 | 1.1e−7 | ↑ |
| CFDP1 | Craniofacial development protein 1 | NM_006324 | 8002865 | 12 | 6.858 | 1.5e−7 | ↓ |
| RAB8B | RAB8B, member RAS oncogene family | NM_016530 | 7984112 | 11 | 7.240 | 2.3e−7 | ↑ |
| ERCC5 | Excision repair cross-complementing 5 | NM_000123 | 7969935 | 19 | 4.556 | 2.8e−7 | ↑ |
| MYO5A | Myosin VA (heavy chain 12, myoxin) | NM_000259 | 7988921 | 41 | 2.895 | 2.9e−7 | ↑ |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | NM_002838 | 7908553 | 37 | 3.042 | 3.0e−7 | ↑ |
| C5orf15 | Chromosome 5 open reading frame 15 | NM_020199 | 8114138 | 6 | 14.126 | 3.9e−7 | ↑ |
| LPCAT2 | Lysophosphatidylcholine acyltransferase 2 | NM_017839 | 7995697 | 14 | 5.525 | 5.4e−7 | ↑ |
| ENAH | Enabled homolog (Drosophila) | NM_001008493 | 7924619 | 16 | 4.976 | 5.5e−7 | ↓ |
| LAPTM5 | Lysosomal protein transmembrane 5 | NM_006762 | 7914270 | 11 | 6.802 | 5.6e−7 | ↑ |
| UHRF2 | Ubiquitin-like with PHD and ring finger domains 2 | NM_152896 | 8154316 | 16 | 4.970 | 5.6e−7 | ↑ |
| VTI1A | Vesicle transport through interaction with t-SNAREs homolog | NM_145206 | 7930524 | 8 | 9.380 | 6.0e−7 | ↑ |
| RAPGEF4 | Rap guanine nucleotide exchange factor (GEF) 4 | NM_007023 | 8046428 | 32 | 3.159 | 7.5e−7 | ↓ |
| LGALS9C | Lectin, galactoside-binding, soluble, 9C | NM_001040078 | 8005458 | 12 | 6.102 | 8.6e−7 | ↑ |
| CPVL | Carboxypeptidase, vitellogenic-like | NM_019029 | 8138805 | 16 | 4.779 | 1.1e−6 | ↑ |
| RBM33 | RNA binding motif protein 33 | NM_053043 | 8137558 | 7 | 10.236 | 1.3e−6 | ↓ |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma | NM_004106 | 7906720 | 7 | 9.985 | 1.7e−6 | ↑ |
| ZMYND8 | Zinc finger, MYND-type containing 8 | NM_183047 | 8066786 | 31 | 3.091 | 1.8e−6 | ↑ |
| SAMSN1 | SAM domain, SH3 domain and nuclear localization signals 1 | NM_022136 | 8069541 | 11 | 6.252 | 1.8e−6 | ↑ |
| DOCK8 | Dedicator of cytokinesis 8 | NM_203447 | 8153959 | 52 | 2.422 | 2.0e−6 | ↑ |
| MUM1 | Melanoma associated antigen (mutated) 1 | NR_024247 | 8024255 | 17 | 4.409 | 2.0e−6 | ↑ |
| IGSF6 | Immunoglobulin superfamily, member 6 | NM_005849 | 8000184 | 7 | 9.837 | 2.0e−6 | ↑ |
| ITGB2 | Integrin, beta 2 (complement component 3 receptor 3 and 4) | NM_000211 | 8070826 | 20 | 3.943 | 2.1e−6 | ↑ |
Rows are sorted by p-value in ascending order.
of the single most differentially expressed transcribed element per gene.
In CAUD, 31 results surpassed the Bonferroni-adjusted significance threshold (Table 3). These 31 transcripts were significantly enriched with genes sharing common protein domains, suggesting a possible basis for their common exonic dysregulation. The most significantly enriched protein-domain annotations were spectrin repeats (p=5.07e−04) and actinin-type actin-binding domains (p=6.09e−04), both of which were present in the same three transcripts. This ratio (3/31) represents an approximately 80-fold enrichment of both annotations compared to chance expectation. One biological pathway (Agrin in Postsynaptic Differentiation) was also over-represented at a Bonferroni-corrected level of significance (p=0.019). The appearance of two genes in this pathway on a list of 31 transcripts represents an approximately 52-fold enrichment compared to chance expectation.
Table 3.
Transcripts Exhibiting a Bonferroni-Corrected Significant Interaction of Diagnosis (SZ vs. NC) and Transcript Element ID in Caudate Head (CAUD)
| Gene Symbol | Gene Product | Reference Sequence | Transcript Cluster ID | Probe Sets (n) | F | p* | Direction of Dysregulation# |
|---|---|---|---|---|---|---|---|
| CPNE3 | Copine III | NM_003909 | 8147172 | 21 | 18.518 | 1.0e−27 | ↓ |
| UTRNa | Utrophin | NM_007124 | 8122464 | 72 | 4.946 | 7.1e−26 | ↑ |
| RXFP2 | Relaxin | NM_130806 | 7968389 | 21 | 8.816 | 1.6e−15 | ↑ |
| DMDa | Dystrophin | NM_000109 | 8171921 | 95 | 2.540 | 2.0e−11 | ↓ |
| GPR98 | G protein-coupled receptor 98 | NM_032119 | 8106827 | 91 | 2.409 | 6.6e−10 | ↓ |
| RGPD6 | Ranbp2-like and grip domain containing 6 | NM_001123363 | 8044304 | 28 | 4.286 | 2.8e−9 | ↓ |
| CIT | Citron (rho-interacting, serine) | NM_007174 | 7966878 | 50 | 3.008 | 4.6e−9 | ↓ |
| SYNE1a | Spectrin repeat containing, nuclear envelope 1 | NM_182961 | 8130211 | 158 | 1.873 | 1.3e−8 | ↑ |
| C6orf174 | Chromosome 6 open reading frame 174 | NM_001012279 | 8129392 | 17 | 5.689 | 1.9e−8 | ↑ |
| Unknown | Unknown | Unknown | 8054557 | 22 | 4.447 | 5.8e−8 | ↑ |
| NME7 | Non-metastatic cells 7 | NM_013330 | 7922137 | 14 | 6.201 | 8.0e−8 | ↑ |
| CEP192 | Centrosomal protein 192Kda | NM_032142 | 8020267 | 40 | 3.050 | 9.7e−8 | ↑ |
| KIAA0319L | KIAA0319-Like | NM_024874 | 7914809 | 24 | 4.042 | 1.3e−7 | ↓ |
| DGKI | Diacylglycerol kinase, iota | NM_004717 | 8143154 | 33 | 3.333 | 1.4e−7 | ↑ |
| KCNK2 | Potassium channel, subfamily k, member 2 | NM_001017425 | 7909730 | 14 | 5.970 | 1.5e−7 | ↑ |
| MYT1L | Myelin transcription factor 1-like | NM_015025 | 8050031 | 28 | 3.585 | 2.3e−7 | ↑ |
| SEC31A | Sec31 Homolog A (S. Cerevisiae) | NM_014933 | 8101376 | 34 | 3.193 | 2.7e−7 | ↑ |
| HSPA4 | Heat shock 70kda protein 4 | NM_002154 | 8108015 | 24 | 3.849 | 3.7e−7 | ↑ |
| SLC4A1AP | solute carrier family 4 (anion exchanger), member 1, adaptor protein | NM_018158 | 8041015 | 11 | 6.701 | 6.9e−7 | ↓ |
| EPB49 | Erythrocyte membrane protein band 4.9 (dematin) | NM_001978 | 8145005 | 21 | 4.022 | 8.3e−7 | ↓ |
| STXBP2 | Syntaxin binding protein 2 | NM_006949 | 8025255 | 22 | 3.902 | 8.4e−7 | ↑ |
| RABEP1 | Rabaptin, rab gtpase binding effector protein 1 | NM_004703 | 8004111 | 21 | 4.011 | 8.8e−7 | ↓ |
| FBXO44 | F-box protein 44 | NM_001014765 | 7897714 | 13 | 5.689 | 8.9e−7 | ↑ |
| EPRS | Glutamyl-prolyl-trna synthetase | NM_004446 | 7924351 | 34 | 3.021 | 1.0e−6 | ↑ |
| C17orf44 | Chromosome 17 open reading frame 44 | NR_026951 | 8012416 | 6 | 12.700 | 1.1e−6 | ↓ |
| PSMA5 | Proteasome (prosome, macropain) subunit, alpha type, 5 | NM_002790 | 7918345 | 13 | 5.622 | 1.1e−6 | ↓ |
| PSD3 | Pleckstrin and sec7 domain containing 3 | NM_015310 | 8149555 | 18 | 4.342 | 1.3e−6 | ↑ |
| TNKS2 | TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | NM_025235 | 7929168 | 32 | 3.078 | 1.4e−6 | ↑ |
| ADORA3 | Adenosine a3 receptor | NM_020683 | 7918533 | 14 | 5.180 | 1.5e−6 | ↑ |
| VWA3A | Von Willebrand factor a domain containing 3a | NM_173615 | 7993898 | 36 | 2.837 | 2.2e−6 | ↓ |
| OVOS | Ovostatin | BX647938 | 7961026 | 4 | 22.882 | 2.3e−6 | ↓ |
Rows are sorted by p-value in ascending order.
of the single most differentially expressed transcribed element per gene.
Genes encoding proteins with actinin-type actin-binding domains.
CPNE3, was the lone gene whose modulation by a diagnosis-x-TE ID interaction surpassed Bonferroni-corrected significance in both brain regions; however, 340 additional transcripts were influenced by at least a nominally significant interaction of diagnosis and TE ID in both brain regions (Table 4). This list of 341 transcripts was also significantly enriched with genes with one or more common protein domains, including a 9.4-fold enrichment of genes with actinin-type actin-binding domains (p=8.45e−03). The most significant enrichment was for genes containing one or more fibronectin type-III-like fold (p=7.14e−03), with ten out of 341 genes containing this domain, representing a 2.9-fold enrichment beyond chance expectation. Other protein domains enriched among the genes on this list included calponin-like actin-binding (six genes; 4.6-fold enrichment; p=9.55e−03), peptidase C1A, papain C-terminal (three genes; 11.5-fold enrichment; p=2.68e−02), and histone core (four genes; 5.0-fold enrichment; p=4.50e−02). No biological pathways were enriched at a Bonferroni-corrected level of significance within the list of 341 transcripts.
Table 4.
Transcripts Exhibiting a Significant Interaction of Diagnosis (SZ vs. NC) and Transcript Element ID in both Brodmann Area 10 (BA10) and Caudate (CAUD)
| Gene Symbol | Gene Product | Reference Sequence | Transcript Cluster ID | Probe Sets (n) | BA10 | CAUD | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| F | p* | F | p* | |||||
| CPNE3 | Copine III | NM_003909 | 8147172 | 21 | 15.003 | 6.60e−24 | 18.518 | 1.00e−27 |
| CEP192 | Centrosomal protein 192kDa | NM_032142 | 8020267 | 40 | 2.587 | 6.20e−06 | 3.050 | 9.70e−08 |
| SCIN | Scinderin | NM_001112706 | 8131550 | 22 | 6.992 | 5.40e−13 | 3.055 | 5.80e−05 |
| OVOS | Ovostatin | BX647938 | 7953873 | 4 | 12.265 | 1.30e−04 | 17.412 | 1.50e−05 |
| NCKAP1L | NCK-associated protein 1-like | NM_005337 | 7955908 | 34 | 2.359 | 1.50e−04 | 2.642 | 1.80e−05 |
| LCP1a,c | Lymphocyte cytosolic protein 1 (L-plastin) | NM_002298 | 7971461 | 20 | 3.062 | 1.20e−04 | 3.169 | 7.20e−05 |
| OVOS | Ovostatin | BX647938 | 7961026 | 4 | 11.274 | 2.10e−04 | 22.882 | 2.30e−06 |
| KLHL5 | Kelch-like 5 (Drosophila) | NM_015990 | 8094625 | 12 | 4.170 | 1.10e−04 | 3.891 | 2.40e−04 |
| NME7 | Non-metastatic cells 7, protein expressed in (nucleoside) | NM_013330 | 7922137 | 14 | 3.348 | 4.30e−04 | 6.201 | 8.00e−08 |
| LAPTM5 | Lysosomal protein transmembrane 5 | NM_006762 | 7914270 | 11 | 6.802 | 5.60e−07 | 3.864 | 4.50e−04 |
| LAT2 | Linker for activation of T cells family, member 2 | NM_032464 | 8133442 | 16 | 5.454 | 1.10e−07 | 3.064 | 5.10e−04 |
| ST6GAL1 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | NM_173216 | 8084717 | 8 | 4.984 | 3.60e−04 | 5.319 | 2.10e−04 |
| APOC2 | Apolipoprotein C-II | NM_000483 | 8029551 | 8 | 6.543 | 3.00e−05 | 4.739 | 5.40e−04 |
| RPL13A | Ribosomal protein L13a | NM_012423 | 8030351 | 8 | 4.700 | 5.80e−04 | 7.204 | 1.10e−05 |
| HSD17B4 | Hydroxysteroid (17-beta) dehydrogenase 4 | NM_000414 | 8107532 | 21 | 2.677 | 5.10e−04 | 2.917 | 1.60e−04 |
| RHOV | Ras homolog gene family, member V | NM_133639 | 7987574 | 9 | 5.332 | 8.20e−05 | 4.281 | 6.10e−04 |
| NACA | Nascent polypeptide-associated complex alpha subunit | NM_001113201 | 7964262 | 8 | 4.838 | 4.60e−04 | 5.239 | 2.40e−04 |
| SMOX | Spermine oxidase | NM_175839 | 8060745 | 12 | 4.805 | 2.10e−05 | 3.504 | 6.80e−04 |
| APOL1 | Apolipoprotein L, 1 | NM_145343 | 8072735 | 8 | 7.687 | 5.80e−06 | 4.299 | 1.20e−03 |
| EFNA5 | Ephrin-A5 | NM_001962 | 8113433 | 9 | 6.592 | 8.80e−06 | 3.951 | 1.20e−03 |
| JTB | Jumping translocation breakpoint | NM_006694 | 7920409 | 12 | 4.304 | 7.90e−05 | 3.326 | 1.10e−03 |
| ADORA3 | Adenosine A3 receptor | NM_020683 | 7918533 | 14 | 3.019 | 1.20e−03 | 5.180 | 1.50e−06 |
| LEPRb | Leptin receptor | NM_002303 | 7902074 | 27 | 2.369 | 6.10e−04 | 2.347 | 6.90e−04 |
| CPVL | Carboxypeptidase, vitellogenic-like | NM_019029 | 8138805 | 16 | 4.779 | 1.10e−06 | 2.805 | 1.30e−03 |
| RNASET2 | Ribonuclease T2 | NM_003730 | 8130768 | 14 | 6.923 | 1.10e−08 | 2.985 | 1.40e−03 |
| Unknown | Unknown | Unknown | 8179731 | 17 | 2.967 | 5.10e−04 | 2.800 | 9.80e−04 |
| NDUFS1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (Na+) | NM_005006 | 8058428 | 21 | 2.448 | 1.50e−03 | 2.963 | 1.30e−04 |
| ALDH3A2 | Aldehyde dehydrogenase 3 family, member A2 | NM_001031806 | 8005638 | 16 | 2.760 | 1.50e−03 | 3.459 | 1.20e−04 |
| HLA-E | Major histocompatibility complex, class I, E | NM_005516 | 8117890 | 9 | 4.056 | 9.50e−04 | 4.130 | 8.20e−04 |
| TBXAS1 | Thromboxane A synthase 1 (platelet) | NM_001130966 | 8136557 | 18 | 3.700 | 1.80e−05 | 2.567 | 1.90e−03 |
| DNAJC11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | NM_018198 | 7912112 | 20 | 2.614 | 9.00e−04 | 2.567 | 1.10e−03 |
| DDRGK1 | DDRGK domain containing 1 | NM_023935 | 8064601 | 11 | 3.206 | 2.30e−03 | 4.249 | 1.70e−04 |
| IRF8 | Interferon regulatory factor 8 | NM_002163 | 7997712 | 11 | 5.891 | 3.90e−06 | 3.163 | 2.60e−03 |
| IARS | Isoleucyl-tRNA synthetase | NM_013417 | 8162313 | 38 | 4.012 | 5.00e−11 | 1.888 | 2.80e−03 |
| SPTLC1 | Serine palmitoyltransferase, long chain base subunit 1 | NM_006415 | 8162294 | 16 | 2.669 | 2.20e−03 | 2.983 | 6.80e−04 |
| LST1 | Leukocyte specific transcript 1 | NM_007161 | 8118149 | 8 | 4.695 | 5.90e−04 | 3.896 | 2.40e−03 |
| AKR1CL2 | Aldo-keto reductase family 1, member C-like 2 | NM_001040177 | 7925904 | 13 | 4.268 | 4.50e−05 | 2.834 | 3.10e−03 |
| PARVGc | Parvin, gamma | NM_022141 | 8073682 | 15 | 2.889 | 1.30e−03 | 2.795 | 1.90e−03 |
| COL6A2 | Collagen, type VI, alpha 2 | NM_001849 | 8069301 | 30 | 2.069 | 2.20e−03 | 2.184 | 1.10e−03 |
| CTSSd | Cathepsin S | NM_004079 | 7919800 | 14 | 3.283 | 5.30e−04 | 2.753 | 2.90e−03 |
| HS6ST2 | Heparan sulfate 6-O-sulfotransferase 2 | NM_001077188 | 8175195 | 10 | 3.282 | 3.00e−03 | 4.068 | 5.00e−04 |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | NM_001276 | 7923547 | 14 | 9.026 | 5.70e−11 | 2.693 | 3.50e−03 |
| PSD3 | Pleckstrin and Sec7 domain containing 3 | NM_015310 | 8149555 | 18 | 2.395 | 3.70e−03 | 4.342 | 1.30e−06 |
| TGFBR1 | Transforming growth factor, beta receptor 1 | NM_004612 | 8156826 | 11 | 4.710 | 5.80e−05 | 2.999 | 3.90e−03 |
| PPHLN1 | Periphilin 1 | NM_016488 | 7954940 | 19 | 2.470 | 2.20e−03 | 2.426 | 2.10e−03 |
| MS4A7 | Membrane-spanning 4-domains, subfamily A, member 7 | NM_021201 | 7940259 | 19 | 3.330 | 5.30e−05 | 2.304 | 4.40e−03 |
| FAM118A | Family with sequence similarity 118, member A/B | NM_001104595 | 8073752 | 13 | 2.812 | 3.30e−03 | 3.165 | 1.20e−03 |
| ITGAV | Integrin, alpha V (vitronectin receptor, alpha polypeptide | NM_002210 | 8046861 | 33 | 1.917 | 4.00e−03 | 2.085 | 1.30e−03 |
| ERBB4 | V-erb-a erythroblastic leukemia viral oncogene homolog 4 | NM_005235 | 8058627 | 30 | 2.049 | 2.50e−03 | 2.027 | 2.90e−03 |
| ZC3H14 | Zinc finger CCCH-type containing 14 | NM_024824 | 7976101 | 24 | 3.043 | 3.00e−05 | 2.064 | 5.60e−03 |
| SYK | Spleen tyrosine kinase | NM_003177 | 8156321 | 15 | 2.628 | 3.30e−03 | 2.680 | 2.80e−03 |
| RRP1B | Ribosomal RNA processing 1 homolog B (S. cerevisiae) | NM_015056 | 8068902 | 16 | 2.656 | 2.30e−03 | 2.514 | 3.80e−03 |
| SLC6A18 | Solute carrier family 6, member 18 | NM_182632 | 8104281 | 16 | 2.565 | 3.10e−03 | 2.581 | 3.00e−03 |
| CIT | Citron (rho-interacting, serine) | NM_007174 | 7966878 | 50 | 1.649 | 6.60e−03 | 3.008 | 4.60e−09 |
| C3 | Complement component 3 | NM_000064 | 8033257 | 42 | 2.629 | 2.50e−06 | 1.720 | 6.70e−03 |
| SIK1 | Salt-inducible kinase 1 | NM_173354 | 8070665 | 15 | 2.408 | 7.00e−03 | 3.524 | 1.50e−04 |
| UHRF2 | Ubiquitin-like with PHD and ring finger domains 2 | NM_152896 | 8154316 | 16 | 4.970 | 5.60e−07 | 2.323 | 7.60e−03 |
| FCGBP | Fc fragment of IgG binding protein | NM_003890 | 8036787 | 20 | 2.370 | 2.70e−03 | 2.229 | 5.00e−03 |
| CRHR1 | Corticotropin releasing hormone receptor 1 | NM_001145146 | 8007808 | 17 | 2.730 | 1.30e−03 | 2.312 | 6.40e−03 |
| ALDOA | Aldolase A, fructose-bisphosphate | NM_000034 | 7994737 | 18 | 2.354 | 4.40e−03 | 2.418 | 3.40e−03 |
| ATP5SL | ATP5S-like | NM_018035 | 8037037 | 5 | 4.702 | 6.00e−03 | 5.856 | 2.00e−03 |
| MCM6 | Minichromosome maintenance complex component 6 | NM_005915 | 8055426 | 18 | 3.304 | 9.00e−05 | 2.206 | 7.90e−03 |
| ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | NM_001776 | 7929511 | 13 | 2.696 | 4.70e−03 | 2.793 | 3.50e−03 |
| NCF4 | Neutrophil cytosolic factor 4, 40kDa | NM_013416 | 8072744 | 12 | 3.347 | 1.00e−03 | 2.649 | 7.20e−03 |
| ERCC8 | Excision repair cross-complementing rodent repair deficiency | NM_000082 | 8112285 | 16 | 3.321 | 2.00e−04 | 2.305 | 8.00e−03 |
| HNRNPAB | Heterogeneous nuclear ribonucleoprotein A | NM_031266 | 8110450 | 10 | 2.958 | 6.30e−03 | 3.404 | 2.20e−03 |
| SLC30A10 | Solute carrier family 30, member 10 | NM_018713 | 7924342 | 8 | 3.763 | 3.00e−03 | 3.388 | 5.90e−03 |
| FYB | FYN binding protein (FYB-120) | NM_001465 | 8111739 | 17 | 2.763 | 1.10e−03 | 2.260 | 7.80e−03 |
| SCAMP2 | Secretory carrier membrane protein 2 | NM_005697 | 7990417 | 11 | 4.307 | 1.50e−04 | 2.682 | 8.80e−03 |
| TYROBP | TYRO protein tyrosine kinase binding protein | NM_003332 | 8036224 | 7 | 5.079 | 7.30e−04 | 3.458 | 8.40e−03 |
| PDIA2 | Protein disulfide isomerase family A, member 2 | NM_006849 | 7991815 | 12 | 3.062 | 2.30e−03 | 2.662 | 6.90e−03 |
| ARPC1B | Actin related protein 2 | NM_005720 | 8134552 | 11 | 2.669 | 9.10e−03 | 4.392 | 1.00e−04 |
| CD68 | CD68 molecule | NM_001251 | 8004510 | 10 | 5.239 | 4.10e−05 | 2.791 | 9.20e−03 |
| H2AFJe | H2A histone family, member J | NM_177925 | 7954124 | 7 | 3.541 | 7.40e−03 | 4.369 | 2.10e−03 |
| SECISBP2L | SECIS binding protein 2-like | NM_014701 | 7988581 | 19 | 2.197 | 6.90e−03 | 2.415 | 2.70e−03 |
| RNF7 | Ring finger protein 7 | NM_014245 | 8083119 | 9 | 3.456 | 3.20e−03 | 3.107 | 6.60e−03 |
| BIRC6 | Baculoviral IAP repeat-containing 6 | NM_016252 | 8041283 | 76 | 1.561 | 3.50e−03 | 1.510 | 6.30e−03 |
| RTKN2 | Rhotekin 2 | NM_145307 | 7933855 | 16 | 2.650 | 2.30e−03 | 2.314 | 7.80e−03 |
| IL13RA1b | Interleukin 13 receptor, alpha 1 | NM_001560 | 8169580 | 17 | 2.254 | 8.00e−03 | 2.570 | 2.40e−03 |
| C1QC | Complement component 1, q subcomponent, C chain | NM_001114101 | 7898799 | 5 | 13.897 | 5.30e−06 | 4.158 | 1.07e−02 |
| BTF3 | Basic transcription factor 3 | NM_001037637 | 8106181 | 7 | 3.305 | 1.08e−02 | 6.077 | 1.80e−04 |
| WDR74 | WD repeat domain 74 | NM_018093 | 7948881 | 12 | 3.360 | 1.00e−03 | 2.525 | 1.01e−02 |
| CD37 | CD37 molecule | NM_001774 | 8030277 | 14 | 3.460 | 3.00e−04 | 2.340 | 1.09e−02 |
| OSBP | Oxysterol binding protein | NM_002556 | 7948379 | 17 | 2.940 | 5.70e−04 | 2.174 | 1.07e−02 |
| AVPR2 | Arginine vasopressin receptor 2 | NM_001146151 | 8170794 | 8 | 3.072 | 1.05e−02 | 4.186 | 1.40e−03 |
| SAFB2 | Scaffold attachment factor B2 | NM_014649 | 8032974 | 21 | 2.003 | 1.15e−02 | 2.652 | 5.70e−04 |
| SYNPO | Synaptopodin | NM_007286 | 8109305 | 9 | 2.907 | 1.00e−02 | 3.661 | 2.10e−03 |
| TCIRG1 | T-cell, immune regulator 1, ATPase, H+ transporting | NM_006019 | 7941985 | 21 | 2.165 | 5.60e−03 | 2.111 | 7.10e−03 |
| RPS28 | Ribosomal protein S28 | NM_001031 | 8025395 | 6 | 3.525 | 1.26e−02 | 7.811 | 8.30e−05 |
| LGALS9C | Lectin, galactoside-binding, soluble, 9C | NM_001040078 | 8005458 | 12 | 6.102 | 8.60e−07 | 2.430 | 1.31e−02 |
| CDC42BPA | CDC42 binding protein kinase alpha (DMPK-like) | NM_003607 | 7924773 | 43 | 2.006 | 5.80e−04 | 1.627 | 1.27e−02 |
| NUDT9 | Nudix (nucleoside diphosphate linked moiety X)-type motif | NM_024047 | 8096251 | 13 | 3.337 | 6.90e−04 | 2.361 | 1.27e−02 |
| CYBB | Cytochrome b-245, beta polypeptide | NM_000397 | 8166730 | 14 | 3.664 | 1.60e−04 | 2.272 | 1.35e−02 |
| FGR | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | NM_005248 | 7914112 | 14 | 2.645 | 4.10e−03 | 2.380 | 9.60e−03 |
| GNPTAB | N-acetylglucosamine-1-phosphate transferase, alpha and beta | NM_024312 | 7965812 | 23 | 1.920 | 1.30e−02 | 2.478 | 8.10e−04 |
| ELK1 | ELK1, member of ETS oncogene family | NM_001114123 | 8172345 | 12 | 3.579 | 5.50e−04 | 2.425 | 1.33e−02 |
| RGPD1 | RANBP2-like and GRIP domain containing 1 | NM_001024457 | 8053622 | 23 | 3.369 | 7.80e−06 | 1.902 | 1.41e−02 |
| CYTL1 | Cytokine-like 1 | NM_018659 | 8099132 | 7 | 3.145 | 1.39e−02 | 5.361 | 4.90e−04 |
| LGALS9B | Lectin, galactoside-binding, soluble, 9B | NM_001042685 | 8013450 | 14 | 4.636 | 7.60e−06 | 2.249 | 1.45e−02 |
| BSG | Basigin (Ok blood group) | NM_001728 | 8023955 | 12 | 2.420 | 1.35e−02 | 3.196 | 1.60e−03 |
| PPP2R5B | Protein phosphatase 2, regulatory subunit B′, beta isoform | NM_006244 | 7941087 | 16 | 2.124 | 1.53e−02 | 3.889 | 2.50e−05 |
| NR1D2 | Nuclear receptor subfamily 1, group D, member 2 | NM_005126 | 8078272 | 11 | 2.640 | 9.80e−03 | 2.856 | 5.70e−03 |
| ITGB2 | Integrin, beta 2 (complement component 3 receptor 3 and 4) | NM_000211 | 8070826 | 20 | 3.943 | 2.10e−06 | 1.962 | 1.58e−02 |
| BEX4 | Brain expressed, X-linked 4 | NM_001080425 | 8169009 | 5 | 5.476 | 2.80e−03 | 3.973 | 1.30e−02 |
| SERPINA1 | Serpin peptidase inhibitor, clade A (alpha-1) | NM_001002236 | 7981068 | 9 | 6.057 | 2.20e−05 | 2.684 | 1.60e−02 |
| ALOX5 | Arachidonate 5-lipoxygenase | NM_000698 | 7927215 | 15 | 7.011 | 2.60e−09 | 2.161 | 1.60e−02 |
| MORC2 | MORC family CW-type zinc finger 2 | NM_014941 | 8075430 | 27 | 2.332 | 7.60e−04 | 1.798 | 1.54e−02 |
| PASK | PAS domain containing serine | NM_015148 | 8060205 | 19 | 2.946 | 2.80e−04 | 1.988 | 1.62e−02 |
| IFNGR1b | Interferon gamma receptor 1 | NM_000416 | 8129861 | 10 | 2.933 | 7.00e−03 | 2.756 | 1.00e−02 |
| ACTN1a,c | Actinin, alpha 1 | NM_001130004 | 7979824 | 24 | 1.978 | 9.00e−03 | 1.987 | 8.20e−03 |
| FAM50B | Family with sequence similarity 50, member B | NM_012135 | 8116658 | 3 | 6.831 | 1.00e−02 | 7.868 | 6.60e−03 |
| LPAR5 | Lysophosphatidic acid receptor 5 | NM_020400 | 7960637 | 4 | 5.092 | 1.00e−02 | 5.541 | 7.10e−03 |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase | NM_001127605 | 7934920 | 15 | 2.567 | 4.00e−03 | 2.216 | 1.34e−02 |
| SAMHD1 | SAM domain and HD domain 1 | NM_015474 | 8066117 | 18 | 8.354 | 7.60e−13 | 2.002 | 1.76e−02 |
| CD53 | CD53 molecule | NM_000560 | 7903893 | 12 | 2.329 | 1.70e−02 | 3.636 | 4.70e−04 |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | NM_004396 | 8017634 | 16 | 2.088 | 1.70e−02 | 3.002 | 6.40e−04 |
| DCLK1 | Doublecortin-like kinase 1 | NM_004734 | 7970954 | 18 | 2.446 | 3.00e−03 | 2.044 | 1.50e−02 |
| EPRS | Glutamyl-prolyl-tRNA synthetase | NM_004446 | 7924351 | 34 | 1.661 | 1.90e−02 | 3.021 | 1.00e−06 |
| HSPA4 | Heat shock 70kDa protein 4 | NM_002154 | 8108015 | 24 | 1.819 | 1.87e−02 | 3.849 | 3.70e−07 |
| GSTM1 | Glutathione S-transferase mu 1 | NM_000561 | 7903765 | 9 | 2.603 | 1.89e−02 | 6.225 | 1.70e−05 |
| S100A11 | S100 calcium binding protein A11 | NM_005620 | 7920128 | 6 | 4.134 | 5.60e−03 | 3.481 | 1.34e−02 |
| ARHGAP26 | Rho GTPase activating protein 26 | NM_015071 | 8108873 | 26 | 2.376 | 7.20e−04 | 1.781 | 1.86e−02 |
| BAIAP3 | BAI1-associated protein 3 | NM_003933 | 7992219 | 35 | 1.655 | 1.81e−02 | 2.040 | 1.30e−03 |
| FAM111B | Family with sequence similarity 111, member B | NM_198947 | 7940147 | 5 | 4.244 | 9.70e−03 | 4.211 | 1.01e−02 |
| LGMN | Legumain | NM_005606 | 7980958 | 11 | 4.797 | 4.70e−05 | 2.357 | 2.01e−02 |
| CSF2RAb | Colony stimulating factor 2 receptor, alpha, low-affinity | NM_001161531 | 8165735 | 16 | 2.081 | 1.80e−02 | 2.636 | 2.40e−03 |
| CSF2RAb | Colony stimulating factor 2 receptor, alpha, low-affinity | NM_001161531 | 8176306 | 16 | 2.081 | 1.80e−02 | 2.636 | 2.40e−03 |
| KDM5B | Lysine (K)-specific demethylase 5B | NM_006618 | 7923453 | 29 | 1.851 | 9.00e−03 | 1.821 | 1.11e−02 |
| PPP2R2A | Protein phosphatase 2 (formerly 2A), regulatory subunit | NM_002717 | 8145440 | 13 | 2.204 | 2.00e−02 | 3.410 | 5.60e−04 |
| UTRNa,c | Utrophin | NM_007124 | 8122464 | 72 | 1.415 | 2.10e−02 | 4.946 | 7.10e−26 |
| BAX | BCL2-associated X protein | NR_027882 | 8030158 | 12 | 2.659 | 7.00e−03 | 2.410 | 1.39e−02 |
| MUM1 | Melanoma associated antigen (mutated) 1 | NR_024247 | 8024255 | 17 | 4.409 | 2.00e−06 | 1.986 | 2.15e−02 |
| ZNF509 | Zinc finger protein 509 | NM_145291 | 8093829 | 9 | 2.619 | 1.83e−02 | 3.417 | 3.50e−03 |
| GABARAPL2 | GABA(A) receptor-associated protein-like 2 | NM_007285 | 7997272 | 8 | 2.694 | 2.12e−02 | 4.776 | 5.10e−04 |
| CHST15 | Carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) | NM_015892 | 7936856 | 14 | 2.139 | 2.05e−02 | 3.019 | 1.20e−03 |
| CD247 | CD247 molecule | NM_198053 | 7922040 | 10 | 3.484 | 1.90e−03 | 2.457 | 2.00e−02 |
| SBF2 | SET binding factor 2 | NM_030962 | 7946516 | 42 | 1.740 | 5.80e−03 | 1.599 | 1.66e−02 |
| SEC13 | SEC13 homolog (S. cerevisiae) | NR_024272 | 8085300 | 10 | 3.295 | 2.90e−03 | 2.465 | 1.96e−02 |
| PWP2 | PWP2 periodic tryptophan protein homolog (yeast) | NM_005049 | 8069003 | 22 | 1.965 | 1.19e−02 | 1.986 | 1.08e−02 |
| ELMOD3 | ELMO/CED-12 domain containing 3 | NM_001135021 | 8043131 | 21 | 2.138 | 6.30e−03 | 1.920 | 1.66e−02 |
| ZFP82 | Zinc finger protein 82 homolog (mouse) | NM_133466 | 8036309 | 8 | 2.874 | 1.52e−02 | 3.222 | 8.00e−03 |
| APBB1IP | Amyloid beta (A4) precursor protein-binding, family B, m | NM_019043 | 7926786 | 18 | 1.969 | 2.00e−02 | 2.374 | 4.10e−03 |
| IGSF6 | Immunoglobulin superfamily, member 6 | NM_005849 | 8000184 | 7 | 9.837 | 2.00e−06 | 2.800 | 2.44e−02 |
| HDDC2 | HD domain containing 2 | NM_016063 | 8129363 | 11 | 2.423 | 1.70e−02 | 2.736 | 7.70e−03 |
| GRK1 | G protein-coupled receptor kinase 1 | NM_002929 | 7970325 | 3 | 12.093 | 1.30e−03 | 5.219 | 2.34e−02 |
| C6orf114 | Chromosome 6 open reading frame 114 | AF264036 | 8123981 | 3 | 8.242 | 5.60e−03 | 5.597 | 1.92e−02 |
| PEX10 | Peroxisomal biogenesis factor 10 | NM_153818 | 7911720 | 9 | 2.496 | 2.37e−02 | 3.854 | 1.40e−03 |
| RBM12B | RNA binding motif protein 12B | NM_203390 | 8151788 | 6 | 3.592 | 1.15e−02 | 3.472 | 1.36e−02 |
| TRPM3 | Transient receptor potential cation channel, subfamily M | NM_206946 | 8161654 | 38 | 1.574 | 2.51e−02 | 2.281 | 1.30e−04 |
| PHKG2 | Phosphorylase kinase, gamma 2 (testis) | NM_000294 | 7994928 | 10 | 2.654 | 1.27e−02 | 2.651 | 1.27e−02 |
| C20orf11 | Chromosome 20 open reading frame 11 | NM_017896 | 8064007 | 6 | 4.198 | 5.20e−03 | 3.173 | 2.04e−02 |
| GFM2 | G elongation factor, mitochondrial 2 | NM_032380 | 8112622 | 23 | 1.770 | 2.61e−02 | 2.695 | 2.70e−04 |
| RUNX1T1 | Runt-related transcription factor 1; translocated to, 1 | NM_175634 | 8151768 | 15 | 2.030 | 2.47e−02 | 2.813 | 1.70e−03 |
| CHAF1B | Chromatin assembly factor 1, subunit B (p60) | NM_005441 | 8068478 | 15 | 2.618 | 3.40e−03 | 2.047 | 2.33e−02 |
| ZNF483 | Zinc finger protein 483 | NM_133464 | 8157193 | 9 | 2.486 | 2.42e−02 | 3.560 | 2.60e−03 |
| C6orf25 | Chromosome 6 open reading frame 25 | NM_138277 | 8178074 | 5 | 3.752 | 1.65e−02 | 4.142 | 1.09e−02 |
| TSPY1 | Testis specific protein, Y-linked 1 | NM_003308 | 8176524 | 7 | 5.534 | 3.80e−04 | 2.735 | 2.71e−02 |
| ZBTB8B | Zinc finger and BTB domain containing 8B | NM_001145720 | 7899797 | 13 | 2.231 | 1.86e−02 | 2.472 | 9.10e−03 |
| STXBP2 | Syntaxin binding protein 2 | NM_006949 | 8025255 | 22 | 1.776 | 2.79e−02 | 3.902 | 8.40e−07 |
| ME3 | Malic enzyme 3, NADP(+)-dependent, mitochondrial | NM_001014811 | 7950864 | 18 | 3.910 | 7.60e−06 | 1.876 | 2.84e−02 |
| HECW2 | HECT, C2 and WW domain containing E3 ubiquitin protein | NM_020760 | 8057898 | 30 | 1.869 | 7.60e−03 | 1.694 | 2.09e−02 |
| ZNF827 | Zinc finger protein 827 | NM_178835 | 8103025 | 15 | 2.204 | 1.39e−02 | 2.177 | 1.52e−02 |
| RPS28 | Ribosomal protein S28 | NM_001031 | 7942824 | 5 | 3.249 | 2.90e−02 | 8.974 | 1.40e−04 |
| FLJ46300 | FLJ46300 protein | eNST00000341866 | 7937073 | 5 | 5.286 | 3.40e−03 | 3.345 | 2.60e−02 |
| RRAGD | Ras-related GTP binding D | NM_021244 | 8128123 | 9 | 2.511 | 2.29e−02 | 3.100 | 6.70e−03 |
| BRP44 | Brain protein 44 | NR_026550 | 7922095 | 8 | 4.073 | 1.70e−03 | 2.528 | 2.90e−02 |
| ANGEL2 | Angel homolog 2 (Drosophila) | NM_144567 | 7924190 | 12 | 2.190 | 2.53e−02 | 2.744 | 5.50e−03 |
| PRRG4 | Proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) | NM_024081 | 7939150 | 7 | 2.888 | 2.11e−02 | 3.344 | 1.01e−02 |
| IL1B | Interleukin 1, beta | NM_000576 | 8054722 | 8 | 2.764 | 1.86e−02 | 2.955 | 1.30e−02 |
| ADAR | Adenosine deaminase, RNA-specific | NM_001111 | 7920531 | 20 | 1.803 | 3.04e−02 | 2.495 | 1.50e−03 |
| SYNE2a,c | Spectrin repeat containing, nuclear envelope 2 | NM_182914 | 7974920 | 124 | 1.461 | 1.80e−03 | 1.280 | 3.02e−02 |
| CXCL16 | Chemokine (C-X-C motif) ligand 16 | NM_022059 | 8011713 | 10 | 2.254 | 3.19e−02 | 4.472 | 2.10e−04 |
| CSF1 | Colony stimulating factor 1 (macrophage) | NM_000757 | 7903786 | 16 | 2.060 | 1.91e−02 | 2.168 | 1.31e−02 |
| PTPRE | Protein tyrosine phosphatase, receptor type, E | NM_006504 | 7931353 | 25 | 1.775 | 2.10e−02 | 1.923 | 1.13e−02 |
| ZNF540 | Zinc finger protein 540 | NM_152606 | 8028266 | 8 | 2.510 | 2.99e−02 | 3.854 | 2.50e−03 |
| QSOX1 | Quiescin Q6 sulfhydryl oxidase 1 | NM_002826 | 7907830 | 16 | 1.925 | 3.05e−02 | 2.686 | 2.00e−03 |
| RYR2 | Ryanodine receptor 2 (cardiac) | NM_001035 | 7910792 | 104 | 1.364 | 1.49e−02 | 1.351 | 1.76e−02 |
| ABCD4 | ATP-binding cassette, sub-family D (ALD), member 4 | NM_005050 | 7980115 | 24 | 1.983 | 8.40e−03 | 1.764 | 2.44e−02 |
| SPPL2B | Signal peptide peptidase-like 2B | NM_001077238 | 8024446 | 20 | 1.789 | 3.22e−02 | 2.682 | 6.60e−04 |
| CTSD | Cathepsin D | NM_001909 | 7945666 | 11 | 2.184 | 3.11e−02 | 3.281 | 1.90e−03 |
| SLC9A6 | Solute carrier family 9 (sodium | NM_001042537 | 8170097 | 19 | 2.046 | 1.28e−02 | 1.933 | 2.02e−02 |
| ARID1A | AT rich interactive domain 1A (SWI-like) | NM_006015 | 7899220 | 23 | 1.720 | 3.27e−02 | 2.524 | 6.40e−04 |
| RAPGEF4 | Rap guanine nucleotide exchange factor (GEF) 4 | NM_007023 | 8046428 | 32 | 3.159 | 7.50e−07 | 1.585 | 3.35e−02 |
| VAPB | VAMP (vesicle-associated membrane protein)-associated protein | NM_004738 | 8063620 | 13 | 2.075 | 2.94e−02 | 2.733 | 4.20e−03 |
| SLC1A1 | Solute carrier family 1 (neuronal) | NM_004170 | 8154135 | 15 | 1.932 | 3.40e−02 | 4.279 | 1.20e−05 |
| CIRH1A | Cirrhosis, autosomal recessive 1A (cirhin) | NM_032830 | 7996891 | 16 | 2.323 | 7.50e−03 | 1.966 | 2.65e−02 |
| NCRNA00169 | Non-protein coding RNA 169 | NR_026675 | 7993821 | 3 | 4.867 | 2.83e−02 | 8.058 | 6.00e−03 |
| ETF1 | Eukaryotic translation termination factor 1 | NM_004730 | 8114443 | 11 | 2.412 | 1.75e−02 | 2.416 | 1.73e−02 |
| CDH5 | Cadherin 5, type 2 (vascular endothelium) | NM_001795 | 7996264 | 16 | 1.984 | 2.50e−02 | 2.229 | 1.05e−02 |
| ACSL4 | Acyl-CoA synthetase long-chain family member 4 | NM_022977 | 8174474 | 19 | 2.901 | 3.40e−04 | 1.788 | 3.57e−02 |
| LRFN2b | Leucine rich repeat and fibronectin type III domain | NM_020737 | 8119390 | 3 | 14.282 | 6.70e−04 | 4.468 | 3.55e−02 |
| FLJ43315 | Similar to Asparagine synthetase [glutamine-hydrolyzing] | BC057848 | 8077198 | 5 | 5.599 | 2.50e−03 | 3.117 | 3.37e−02 |
| RGPD1 | RANBP2-like and GRIP domain containing 1 | NM_001024457 | 8043324 | 22 | 1.747 | 3.18e−02 | 2.158 | 4.80e−03 |
| CPSF3 | Cleavage and polyadenylation specific factor 3, 73kDa | NM_016207 | 8040142 | 20 | 1.789 | 3.21e−02 | 2.242 | 4.70e−03 |
| B4GALT1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, poly | NM_001497 | 8160637 | 9 | 3.330 | 4.20e−03 | 2.336 | 3.31e−02 |
| NPTXR | Neuronal pentraxin receptor | NM_014293 | 8076169 | 6 | 3.733 | 9.60e−03 | 2.941 | 2.82e−02 |
| CYFIP1 | Cytoplasmic FMR1 interacting protein 1 | NM_014608 | 7981824 | 34 | 2.359 | 1.50e−04 | 1.542 | 3.82e−02 |
| C9orf78 | Chromosome 9 open reading frame 78 | NM_016520 | 8164596 | 10 | 2.619 | 1.40e−02 | 2.362 | 2.49e−02 |
| KLHL7 | Kelch-like 7 (Drosophila) | NM_018846 | 8131815 | 15 | 1.979 | 2.90e−02 | 2.319 | 9.40e−03 |
| OCRL | Oculocerebrorenal syndrome of Lowe | NM_000276 | 8169811 | 24 | 1.664 | 3.90e−02 | 2.881 | 7.30e−05 |
| KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | NM_002266 | 8019737 | 13 | 2.241 | 1.80e−02 | 2.193 | 2.08e−02 |
| RPGR | Retinitis pigmentosa GTPase regulator | NM_000328 | 8172056 | 25 | 1.648 | 3.90e−02 | 2.646 | 2.00e−04 |
| ABAT | 4-aminobutyrate aminotransferase | NM_020686 | 7993126 | 19 | 1.988 | 1.60e−02 | 1.900 | 2.30e−02 |
| ZNF852 | Zinc finger protein 852 | AK296954 | 8086494 | 3 | 6.607 | 1.20e−02 | 4.900 | 2.78e−02 |
| KIAA1324L | KIAA1324-like | NM_001142749 | 8140709 | 20 | 2.502 | 1.00e−03 | 1.747 | 3.81e−02 |
| ATP4A | ATPase, H+ | NM_000704 | 8036110 | 22 | 2.147 | 5.00e−03 | 1.727 | 3.46e−02 |
| JAKMIP2 | Janus kinase and microtubule interacting protein 2 | NM_014790 | 8114938 | 25 | 1.651 | 3.80e−02 | 2.268 | 1.60e−03 |
| PRG2 | Plasticity-related gene 2 | NM_024888 | 8032094 | 11 | 2.426 | 1.70e−02 | 2.305 | 2.29e−02 |
| ZMYND8 | Zinc finger, MYND-type containing 8 | NM_183047 | 8066786 | 31 | 3.091 | 1.80e−06 | 1.565 | 3.98e−02 |
| FCGR1B | Fc fragment of IgG, high affinity Ib, receptor (CD64) | NM_001017986 | 7919133 | 5 | 15.021 | 2.80e−06 | 2.970 | 3.99e−02 |
| PTBP2 | Polypyrimidine tract binding protein 2 | NM_021190 | 7903188 | 14 | 3.029 | 1.20e−03 | 1.931 | 3.88e−02 |
| PAF1 | Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) | NM_019088 | 8036720 | 16 | 3.231 | 2.80e−04 | 1.847 | 3.98e−02 |
| CCDC90B | Coiled-coil domain containing 90B | NM_021825 | 7950753 | 8 | 2.743 | 1.90e−02 | 2.700 | 2.10e−02 |
| TSPY1 | Testis specific protein, Y-linked 1 | NM_003308 | 8176508 | 8 | 3.347 | 6.00e−03 | 2.437 | 3.43e−02 |
| CTSZd | Cathepsin Z | NM_001336 | 8067279 | 8 | 2.353 | 4.00e−02 | 4.688 | 5.90e−04 |
| ZDHHC16 | Zinc finger, DHHC-type containing 16 | NM_198046 | 7929634 | 14 | 2.485 | 7.00e−03 | 1.975 | 3.41e−02 |
| SMA5 | Glucuronidase, beta pseudogene | AK289851 | 8177544 | 9 | 2.797 | 1.30e−02 | 2.410 | 2.83e−02 |
| CTPS | CTP synthase | NM_001905 | 7900510 | 20 | 1.815 | 2.90e−02 | 2.020 | 1.23e−02 |
| TUBB4Q | Tubulin, beta polypeptide 4, member Q | NM_020040 | 8021919 | 4 | 4.782 | 1.30e−02 | 3.797 | 2.86e−02 |
| ANXA6 | Annexin A6 | NM_001155 | 8115234 | 26 | 2.174 | 2.00e−03 | 1.630 | 3.93e−02 |
| SMARCA1 | SWI/SNF related, matrix associated, actin dependent | NM_003069 | 8174985 | 30 | 2.980 | 5.40e−06 | 1.568 | 4.15e−02 |
| TMCO3 | Transmembrane and coiled-coil domains 3 | NM_017905 | 7970301 | 15 | 2.377 | 7.70e−03 | 1.934 | 3.38e−02 |
| ULK4 | Unc-51-like kinase 4 (C. elegans) | NM_017886 | 8086352 | 15 | 2.540 | 4.40e−03 | 1.899 | 3.77e−02 |
| ZNF341 | Zinc finger protein 341 | NM_032819 | 8061946 | 11 | 4.044 | 2.90e−04 | 2.061 | 4.22e−02 |
| TSPAN7 | Tetraspanin 7 | NM_004615 | 8166784 | 10 | 2.134 | 4.21e−02 | 3.962 | 6.40e−04 |
| TPD52L1 | Tumor protein D52-like 1 | NM_001003395 | 8121838 | 11 | 3.000 | 3.90e−03 | 2.091 | 3.92e−02 |
| ATP6V0E1 | ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 | NM_003945 | 8110022 | 9 | 2.487 | 2.41e−02 | 2.588 | 1.95e−02 |
| SREBF1 | Sterol regulatory element binding transcription factor | NM_001005291 | 8013135 | 21 | 1.784 | 2.97e−02 | 1.958 | 1.41e−02 |
| AQP1 | Aquaporin 1 (Colton blood group) | NM_198098 | 8132118 | 11 | 2.545 | 1.25e−02 | 2.176 | 3.17e−02 |
| DPY19L4 | Dpy-19-like 4 (C. elegans) | NM_181787 | 8147375 | 20 | 1.731 | 4.06e−02 | 2.306 | 3.60e−03 |
| ZWINT | ZW10 interactor | NM_032997 | 7933707 | 15 | 2.432 | 6.40e−03 | 1.895 | 3.83e−02 |
| C3orf1 | Chromosome 3 open reading frame 1 | NM_016589 | 8081867 | 10 | 2.802 | 9.00e−03 | 2.203 | 3.59e−02 |
| CCR10 | Chemokine (C-C motif) receptor 10 | NM_016602 | 8015681 | 3 | 6.185 | 1.43e−02 | 4.721 | 3.07e−02 |
| ITGA8 | Integrin, alpha 8 | NM_003638 | 7932254 | 30 | 1.599 | 3.52e−02 | 1.823 | 9.90e−03 |
| BRD7 | Bromodomain containing 7 | NM_013263 | 8001350 | 20 | 1.704 | 4.52e−02 | 3.622 | 9.10e−06 |
| MPHOSPH9 | M-phase phosphoprotein 9 | NM_022782 | 7967386 | 25 | 2.763 | 1.00e−04 | 1.615 | 4.52e−02 |
| MKLN1 | Muskelin 1, intracellular mediator containing kelch motifs | NM_013255 | 8136259 | 21 | 1.724 | 3.83e−02 | 2.112 | 7.10e−03 |
| UGGT1 | UDP-glucose glycoprotein glucosyltransferase 1 | NM_020120 | 8045090 | 43 | 1.496 | 3.27e−02 | 1.617 | 1.36e−02 |
| ZCCHC2 | Zinc finger, CCHC domain containing 2 | NM_017742 | 8021546 | 16 | 2.057 | 1.93e−02 | 1.961 | 2.70e−02 |
| STK19 | Serine/threonine kinase 19 | NR_026717 | 8118395 | 12 | 5.450 | 4.20e−06 | 1.965 | 4.64e−02 |
| STK19 | Serine/threonine kinase 19 | NR_026717 | 8178164 | 12 | 5.450 | 4.20e−06 | 1.965 | 4.64e−02 |
| CEMP1 | Cementum protein 1 | NM_001048212 | 7998817 | 2 | 11.348 | 1.51e−02 | 7.819 | 3.13e−02 |
| TRPC4 | Transient receptor potential cation channel, subfamily C, | NM_016179 | 7971104 | 17 | 1.854 | 3.46e−02 | 2.148 | 1.18e−02 |
| PLA1A | Phospholipase A1 member A | NM_015900 | 8081890 | 12 | 2.641 | 7.30e−03 | 2.029 | 3.91e−02 |
| PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G | NM_000962 | 8157650 | 17 | 1.773 | 4.61e−02 | 2.951 | 5.40e−04 |
| POLM | Polymerase (DNA directed), mu | NM_013284 | 8139281 | 17 | 1.960 | 2.36e−02 | 1.966 | 2.32e−02 |
| PPM1D | Protein phosphatase 1D magnesium-dependent, delta isoform | NM_003620 | 8008922 | 10 | 2.872 | 7.60e−03 | 2.164 | 3.93e−02 |
| COBLL1 | COBL-like 1 | NM_014900 | 8056343 | 15 | 2.081 | 2.09e−02 | 2.014 | 2.61e−02 |
| CTSL2d | Cathepsin L2 | NM_001333 | 8162652 | 11 | 2.613 | 1.05e−02 | 2.119 | 3.66e−02 |
| TRIM22 | Tripartite motif-containing 22 | NM_006074 | 7938035 | 11 | 2.867 | 5.50e−03 | 2.063 | 4.20e−02 |
| FLII | Flightless I homolog (Drosophila) | NM_002018 | 8013191 | 30 | 2.052 | 2.50e−03 | 1.552 | 4.53e−02 |
| EEF1A2 | Eukaryotic translation elongation factor 1 alpha 2 | NM_001958 | 8067652 | 9 | 2.183 | 4.55e−02 | 3.504 | 2.90e−03 |
| AGFG1 | ArfGAP with FG repeats 1 | NM_001135187 | 8048847 | 16 | 1.791 | 4.80e−02 | 2.931 | 8.20e−04 |
| GMDS | GDP-mannose 4,6-dehydratase | NM_001500 | 8123562 | 16 | 1.804 | 4.60e−02 | 2.597 | 2.80e−03 |
| TMC3 | Transmembrane channel-like 3 | NM_001080532 | 7990848 | 24 | 1.660 | 3.94e−02 | 1.987 | 9.40e−03 |
| USP18 | Ubiquitin specific peptidase 18 | NM_017414 | 8071155 | 5 | 2.875 | 4.45e−02 | 5.041 | 4.30e−03 |
| PTPRSb | Protein tyrosine phosphatase, receptor type, S | NM_002850 | 8032926 | 38 | 2.614 | 8.10e−06 | 1.469 | 4.89e−02 |
| ATP13A2 | ATPase type 13A2 | NM_022089 | 7912898 | 29 | 1.745 | 1.71e−02 | 1.630 | 3.19e−02 |
| ARX | Aristaless related homeobox | NM_139058 | 8171867 | 6 | 2.693 | 3.99e−02 | 3.754 | 9.30e−03 |
| AP2A1 | Adaptor-related protein complex 2, alpha 1 subunit | NM_014203 | 8030470 | 27 | 2.989 | 1.40e−05 | 1.569 | 4.95e−02 |
| LAT | Linker for activation of T cells | NM_014387 | 7994541 | 17 | 2.383 | 4.90e−03 | 1.782 | 4.47e−02 |
| PLVAP | Plasmalemma vesicle associated protein | NM_031310 | 8035297 | 6 | 2.663 | 4.16e−02 | 3.832 | 8.40e−03 |
| SLC25A46 | Solute carrier family 25, member 46 | NM_138773 | 8107259 | 10 | 2.503 | 1.80e−02 | 2.251 | 3.21e−02 |
| DPF1 | D4, zinc and double PHD fingers family 1 | NM_004647 | 8036460 | 12 | 1.949 | 4.85e−02 | 3.183 | 1.60e−03 |
| PDGFD | Platelet derived growth factor D | NM_025208 | 7951351 | 11 | 2.718 | 8.00e−03 | 2.057 | 4.27e−02 |
| C1orf175 | Chromosome 1 open reading frame 175 | NR_026782 | 7901634 | 27 | 1.727 | 2.23e−02 | 1.680 | 2.85e−02 |
| POM121 | POM121 membrane glycoprotein (rat) | NM_172020 | 8133275 | 17 | 1.804 | 4.14e−02 | 2.204 | 9.60e−03 |
| SUGT1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | NM_001130912 | 7969271 | 14 | 2.474 | 7.10e−03 | 1.891 | 4.39e−02 |
| TSPY1 | Testis specific protein, Y-linked 1 | NM_003308 | 8176544 | 8 | 2.657 | 2.28e−02 | 2.537 | 2.85e−02 |
| TBC1D21 | TBC1 domain family, member 21 | NM_153356 | 7984759 | 11 | 2.383 | 1.88e−02 | 2.164 | 3.26e−02 |
| CMTM4 | CKLF-like MARVEL transmembrane domain containing 4 | NM_181521 | 8001830 | 10 | 2.095 | 4.60e−02 | 3.016 | 5.50e−03 |
| LST1 | Leukocyte specific transcript 1 | NM_007161 | 8177988 | 7 | 2.367 | 4.97e−02 | 4.478 | 1.70e−03 |
| LST1 | Leukocyte specific transcript 1 | NM_007161 | 8179268 | 7 | 2.367 | 4.97e−02 | 4.478 | 1.70e−03 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71 kDa | NM_002535 | 7958913 | 17 | 1.759 | 4.84e−02 | 2.399 | 4.60e−03 |
| TNS1 | Tensin 1 | NM_022648 | 8058869 | 35 | 1.578 | 2.92e−02 | 1.609 | 2.41e−02 |
| ECSIT | ECSIT homolog (Drosophila) | NM_016581 | 8034286 | 10 | 2.653 | 1.27e−02 | 2.146 | 4.09e−02 |
| PCMTD1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase | NM_052937 | 8150714 | 7 | 2.696 | 2.89e−02 | 2.785 | 2.50e−02 |
| RBAK | RB-associated KRAB zinc finger | NM_021163 | 8131292 | 3 | 5.417 | 2.11e−02 | 4.573 | 3.34e−02 |
| RBM33 | RNA binding motif protein 33 | NM_053043 | 8137542 | 15 | 2.154 | 1.64e−02 | 1.896 | 3.82e−02 |
| GCN1L1 | GCN1 general control of amino-acid synthesis 1-like 1 | NM_006836 | 7966938 | 57 | 1.620 | 5.50e−03 | 1.372 | 4.91e−02 |
| SSH2 | Slingshot homolog 2 (Drosophila) | NM_033389 | 8013965 | 19 | 2.277 | 4.90e−03 | 1.702 | 4.97e−02 |
| ZPLD1 | Zona pellucida-like domain containing 1 | NM_175056 | 8081407 | 19 | 1.806 | 3.33e−02 | 1.919 | 2.13e−02 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | NM_001827 | 8156290 | 4 | 3.169 | 4.96e−02 | 5.916 | 5.40e−03 |
| EGFLAMb | EGF-like, fibronectin type III and laminin G domains | NM_152403 | 8105013 | 26 | 1.853 | 1.28e−02 | 1.614 | 4.25e−02 |
| UROD | Uroporphyrinogen decarboxylase | NM_000374 | 7901073 | 13 | 2.013 | 3.52e−02 | 2.202 | 2.03e−02 |
| CWH43 | Cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) | AK300495 | 8095005 | 3 | 4.689 | 3.13e−02 | 5.063 | 2.55e−02 |
| REL | V-rel reticuloendotheliosis viral oncogene homolog (avian) | NM_002908 | 8042144 | 12 | 1.948 | 4.86e−02 | 2.579 | 8.70e−03 |
| EPB41 | Erythrocyte membrane protein band 4.1 (elliptocytosis 1, R) | NM_203342 | 7899534 | 25 | 1.605 | 4.73e−02 | 1.914 | 1.05e−02 |
| TSPY1 | Testis specific protein, Y-linked 1 | NM_003308 | 8176484 | 9 | 2.411 | 2.83e−02 | 2.383 | 3.00e−02 |
| TM2D2 | TM2 domain containing 2 | NM_031940 | 8150364 | 8 | 2.444 | 3.39e−02 | 2.620 | 2.44e−02 |
| CLSPN | Claspin homolog (Xenopus laevis) | NM_022111 | 7914851 | 26 | 1.691 | 2.92e−02 | 1.690 | 2.93e−02 |
| CKMT1A | Creatine kinase, mitochondrial 1A | NM_001015001 | 7983256 | 13 | 1.890 | 4.98e−02 | 2.488 | 8.70e−03 |
| THADA | Thyroid adenoma associated | NM_022065 | 8051820 | 41 | 1.652 | 1.21e−02 | 1.455 | 4.68e−02 |
| GPR179 | G protein-coupled receptor 179 | NM_001004334 | 8014666 | 12 | 2.309 | 1.83e−02 | 2.014 | 4.07e−02 |
| PNPLA8 | Patatin-like phospholipase domain containing 8 | NM_015723 | 8142307 | 16 | 1.784 | 4.92e−02 | 2.237 | 1.03e−02 |
| IGDCC3b | Immunoglobulin superfamily, DCC subclass, member 3 | NM_004884 | 7989770 | 14 | 2.267 | 1.37e−02 | 1.877 | 4.58e−02 |
| ADAM9 | ADAM metallopeptidase domain 9 (meltrin gamma) | NM_003816 | 8146000 | 23 | 1.714 | 3.35e−02 | 1.769 | 2.61e−02 |
| PNKP | Polynucleotide kinase 3′-phosphatase | NM_007254 | 8038458 | 18 | 1.813 | 3.60e−02 | 1.924 | 2.38e−02 |
| IFITM2 | Interferon induced transmembrane protein 2 (1–8D) | NM_006435 | 7937330 | 4 | 3.691 | 3.13e−02 | 3.793 | 2.87e−02 |
| KCNG2 | Potassium voltage-gated channel, subfamily G, member 2 | NM_012283 | 8021900 | 2 | 8.880 | 2.46e−02 | 7.247 | 3.60e−02 |
| LHFPL2 | Lipoma HMGIC fusion partner-like 2 | NM_005779 | 8112803 | 3 | 4.391 | 3.71e−02 | 5.200 | 2.36e−02 |
| PDCD11 | Programmed cell death 11 | NM_014976 | 7930226 | 37 | 1.615 | 2.04e−02 | 1.503 | 4.15e−02 |
| HIST1H4He | Histone cluster 1, H4h | NM_003543 | 8124448 | 4 | 4.697 | 1.36e−02 | 3.188 | 4.88e−02 |
| SDCBP | Syndecan binding protein (syntenin) | NM_005625 | 8146550 | 8 | 2.390 | 3.75e−02 | 2.603 | 2.52e−02 |
| MGC42105 | Serine | NM_153361 | 8105146 | 4 | 3.781 | 2.90e−02 | 3.602 | 3.38e−02 |
| DIAPH1 | Diaphanous homolog 1 (Drosophila) | NM_005219 | 8114658 | 32 | 1.522 | 4.75e−02 | 1.716 | 1.56e−02 |
| IL17RA | Interleukin 17 receptor A | NM_014339 | 8071069 | 14 | 2.100 | 2.32e−02 | 1.921 | 4.00e−02 |
| MYO18A | Myosin XVIIIA | NM_078471 | 8013860 | 45 | 1.474 | 3.49e−02 | 1.502 | 2.85e−02 |
| ACLY | ATP citrate lyase | NM_001096 | 8015460 | 29 | 1.687 | 2.35e−02 | 1.587 | 4.00e−02 |
| KCNH3 | Potassium voltage-gated channel, subfamily H (eag-related) | NM_012284 | 7955231 | 16 | 1.794 | 4.76e−02 | 2.090 | 1.73e−02 |
| CAMTA2 | Calmodulin binding transcription activator 2 | NM_015099 | 8011774 | 22 | 1.642 | 4.96e−02 | 1.908 | 1.55e−02 |
| CD4 | CD4 molecule | NM_000616 | 7953428 | 13 | 2.282 | 1.60e−02 | 1.896 | 4.91e−02 |
| PLXNB1 | Plexin B1 | NM_001130082 | 8086908 | 40 | 1.452 | 4.98e−02 | 1.625 | 1.57e−02 |
| WISP1 | WNT1 inducible signaling pathway protein 1 | NM_003882 | 8148435 | 10 | 2.379 | 2.39e−02 | 2.138 | 4.17e−02 |
| SLC18A2 | Solute carrier family 18 (vesicular monoamine), member 2 | NM_003054 | 7930837 | 19 | 1.942 | 1.95e−02 | 1.720 | 4.64e−02 |
| KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | NM_002266 | 8009417 | 12 | 2.332 | 1.72e−02 | 1.940 | 4.96e−02 |
| MAPRE3c | Microtubule-associated protein, RP | NM_012326 | 8040742 | 10 | 2.330 | 2.68e−02 | 2.148 | 4.07e−02 |
| RPL9 | Ribosomal protein L9 | NM_001024921 | 8099887 | 9 | 2.281 | 3.71e−02 | 2.346 | 3.24e−02 |
| FOXD4L6 | Forkhead box D4-like 6 | NM_001085476 | 8161533 | 3 | 4.580 | 3.33e−02 | 4.399 | 3.69e−02 |
| TGM2b | Transglutaminase 2 (C polypeptide, protein-glutamine-gamma) | NM_004613 | 8066214 | 16 | 1.858 | 3.83e−02 | 1.908 | 3.23e−02 |
| ZDHHC17 | Zinc finger, DHHC-type containing 17 | NM_015336 | 7957277 | 20 | 1.838 | 2.63e−02 | 1.708 | 4.46e−02 |
| ATP9A | ATPase, class II, type 9A | NM_006045 | 8067055 | 31 | 1.553 | 4.25e−02 | 1.623 | 2.88e−02 |
| ZNF382 | Zinc finger protein 382 | NM_032825 | 8028194 | 5 | 3.392 | 2.46e−02 | 2.834 | 4.67e−02 |
| HIST1H3De | Histone cluster 1, H3d | NM_003530 | 8124416 | 6 | 2.717 | 3.86e−02 | 2.824 | 3.32e−02 |
| SLC25A3 | Solute carrier family 25 (mitochondrial carrier) | NM_213611 | 7957746 | 12 | 2.217 | 2.35e−02 | 1.946 | 4.88e−02 |
| C12orf56 | Chromosome 12 open reading frame 56 | NM_001099676 | 7964687 | 11 | 2.185 | 3.10e−02 | 2.051 | 4.32e−02 |
| AEBP1 | AE binding protein 1 | NM_001129 | 8132557 | 22 | 1.699 | 3.91e−02 | 1.714 | 3.65e−02 |
| TNPO1 | Transportin 1 | NM_002270 | 8106122 | 18 | 1.748 | 4.58e−02 | 1.863 | 2.99e−02 |
| ACOT2 | Acyl-CoA thioesterase 2 | NM_006821 | 7975602 | 4 | 3.878 | 2.66e−02 | 3.175 | 4.93e−02 |
| POLR3E | Polymerase (RNA) III (DNA directed) polypeptide E (80kDa) | NM_018119 | 7993973 | 25 | 1.693 | 3.14e−02 | 1.613 | 4.56e−02 |
| STK32A | Serine/threonine kinase | NM_001112724 | 8108981 | 13 | 1.971 | 3.96e−02 | 1.954 | 4.16e−02 |
| PARP14 | Poly (ADP-ribose) polymerase family, member 14 | NM_017554 | 8082100 | 17 | 1.864 | 3.35e−02 | 1.755 | 4.91e−02 |
| FOLR3 | Folate receptor 3 (gamma) | NM_000804 | 7942328 | 3 | 4.042 | 4.55e−02 | 4.389 | 3.71e−02 |
| HIST1H2BDe | Histone cluster 1, H2bd | NM_021063 | 8117382 | 6 | 2.783 | 3.51e−02 | 2.808 | 4.82e−02 |
| CRLF3b | Cytokine receptor-like factor 3 | NM_015986 | 8014037 | 9 | 2.313 | 3.47e−02 | 2.141 | 4.97e−02 |
| MLC1 | Megalencephalic leukoencephalopathy with subcortical cysts | NM_015166 | 8076894 | 14 | 1.915 | 4.08e−02 | 1.881 | 4.53e−02 |
| DNAJC9 | DnaJ (Hsp40) homolog, subfamily C, member 9 | NM_015190 | 7934320 | 5 | 2.882 | 4.42e−02 | 2.912 | 4.27e−02 |
| MTSS1 | Metastasis suppressor 1 | NM_014751 | 8152764 | 17 | 1.780 | 4.51e−02 | 1.799 | 4.21e−02 |
| GLRX3 | Glutaredoxin 3 | NM_006541 | 7931393 | 9 | 2.197 | 4.42e−02 | 2.140 | 4.98e−02 |
| ACSL5 | Acyl-CoA synthetase long-chain family member 5 | NM_016234 | 7930498 | 25 | 1.610 | 4.64e−02 | 1.602 | 4.79e−02 |
| NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | NM_002135 | 7955589 | 16 | 1.787 | 4.87e−02 | 1.785 | 4.90e−02 |
Rows are sorted in ascending order by the average p-value across both brain regions.
Genes encoding proteins with actinin-type actin-binding domains
Genes encoding proteins with fibronectin type-III-folds
Genes encoding proteins with calponin-like actin-binding domains
Genes encoding proteins with peptidase C1A, papain C-terminals
Genes encoding histone core proteins
Confirmation and Replication Analyses
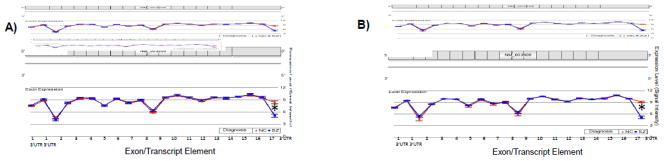
We selected three genes for confirmation and replication analyses. First was CPNE3, which exhibited a Bonferroni-corrected significant interaction in both BA10 (F=15.00, p=6.6e−24) and CAUD (F=18.52, p=1.0e−27). As shown in Figure 1, the significant difference in expression detected between diagnostic groups by microarray in both brain regions was relegated to the most distal end of the gene’s 3′UTR (panel A: BA10, F=35.89, p=9.7e−4; panel B: CAUD, F=113.56, p=4.0e−5), suggesting that SZ subjects expressed transcripts with relatively shorter 3′UTRs. In the same discovery sample used for the microarray analyses described above, qRTPCR confirmed differences between diagnostic groups in the expression levels of this segment of the 3′UTR of CPNE3 relative to a control region of the gene (upstream in the 3′UTR). This difference attained statistical significance in both CAUD (p=0.001) and BA10 (p=0.041). We next sought to replicate the decreased expression level of the distal 3′UTR segment of CPNE3 (again, relative to the non-differentially expressed proximal 3′UTR segment) in the remaining 16 SZ and 16 NC postmortem brain tissue samples from the Harvard Brain Tissue Resource Center. In this fully independent sample, we successfully replicated a highly significant decrease in 3′UTR expression in BA10 in SZ (p<0.001), and in CAUD SZ samples as well (p=0.034), indicating that this result generalizes across samples and brain regions.
Figure 1. Exonic Expression and Alternative 3′UTR Usage of CPNE3in: A) BA10; and B) CAUD.
Microarray results of differential probe expression of Copine3 (CPNE3) in Brodman Area 10 (A) and Caudate (B) in postmortem brain tissue samples from normal control subjects (NC) (n=4) and individuals with schizophrenia (SZ) (n=4). The interaction of diagnosis and exon ID was highly significant in both brain regions; in fact, it was the only gene for which a Bonferroni-corrected threshold for significance of this term was met. *There was a statistically significant difference in probe-level expression between diagnostic groups at the most distal end of the 3′UTR in both BA10 (p=9.7e-4) and CAUD (p=4.0e-5), indicating that a truncated transcript may be expressed in SZ patients.
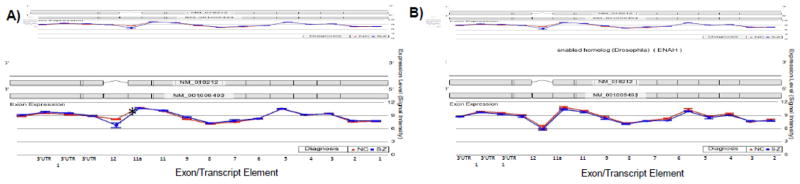
The second gene followed-up by qRTPCR was ENAH, which was chosen based on the appearance of exonic dysregulation in the vicinity of a known splice site. Initial analyses of the small discovery sample by microarray showed that ENAH expression levels were influenced by a Bonferroni-corrected significant interaction of diagnosis and TE ID in BA10 (F=4.98, p=5.5e−07) but not in CAUD (F=0.57, p=0.890). As shown in Figure 2, SZ and NC subjects had equivalent levels of TE expression in most gene regions in both BA10 (panel A) and CAUD (panel B); however, there was a significant decrease in exon 11a expression in SZ in BA10 (F=15.96, p=0.007). This precise pattern was recapitulated by qRTPCR, with a significant SZ-associated decrease in ENAH exon 11a expression (relative to a non-dysregulated region of the gene: the boundary of exons 2 and 3) confirmed in BA10 (p=0.037), and no significant difference observed in CAUD (p=0.141) in the discovery sample. When extending this evaluation to the replication sample, the significant dysregulation of ENAH exon 11a was again detected in BA10 in SZ (p=0.035); unexpectedly, we also detected a significant decrease in exon 11a expression in CAUD in the replication sample by qRTPCR (p=0.001).
Figure 2. Exonic Expression and Alternative Splicing of ENAH in: A) BA10; and B) CAUD.
Microarray results of differential exon expression of Enabled Homolog (ENAH) in Brodman Area 10 (A) and Caudate (B) in postmortem brain tissue samples from normal control subjects (NC) (n=4) and individuals with schizophrenia (SZ) (n=4). The interaction of diagnosis and exon ID was significant only in BA10, not CAUD, indicating the possibility of regionally specific differential splice-variant expression between the groups. *There was a statistically significant difference in expression between diagnostic groups at exon 11a in BA10 (p=0.047) but not CAUD (p=0.489), indicating increased expression of the short (11a) isoform in SZ patients in BA10 only.
The last result selected for confirmation by qRTPCR was the up-regulation of KLHL5 exon 10 in SZ. This gene’s expression levels showed highly significant evidence of a diagnosis by TE ID interaction in both BA10 (F=4.17; p=1.1e−4) and CAUD (F=3.89; p=2.4e−4) by microarray. Interestingly, the strongest difference in expression in both brain regions did not involve the use of the known alternate transcription start-sites in the 5′ end of the gene, but was found at exon 10 (BA10: F=5.96, p=0.050; CAUD: F=8.79 p=0.025), where SZ subjects exhibited higher expression on average than NC subjects. Reanalysis of the discovery samples by qRTPCR did not directly verify the microarray results, as expression levels of the exon 10 and 11 boundary (relative to a non-dysregulated region of the gene: the boundary of exons 6 and 7) were not significantly higher in SZ (BA10: p=0.229; CAUD: p=1.000). Yet, when we examined the expression levels of this exon by qRTPCR in the independent replication sample, we again observed a significant increase in exon 10 expression in SZ in BA10 (p=0.011), though not in CAUD (p=1.000). Our inability to consistently verify the precise results observed by microarray in the discovery sample nor confirm them in the replication sample is perhaps not surprising given the relatively lax criteria used in selecting this gene for follow-up study relative to the Bonferroni-corrected significant results that drove our follow-up work on CPNE3 and ENAH. However, further investigation of KLHL5’s transcriptomic profile by direct sequencing or qRTPCR using primers with greater coverage may resolve this uncertainty in the future.
DISCUSSION
The main objective of this study was to assess how common and widespread exonic expression abnormalities are in postmortem brain in SZ. Our work suggests they are not uncommon, and are more complex than initially conceived, involving not only alternative splicing of traditional cassette exons, but also selection of mutually exclusive exons, alternate promoter selection, and 3′UTR constitution. By comparing brain regions linked to the disorder, as well as medicated and unmedicated patients (although in just a small, initial discovery sample), we were able to identify some expression abnormalities that generalized across regions and which may reflect stable traits associated with the disorder. Further work could evaluate whether such expression abnormalities arise from inherited mutations or biological insults acquired early in development. In contrast, most TE expression abnormalities observed in this study were restricted to either BA10 or CAUD. Such regionally specific abnormalities may be less likely attributable to regulatory effects of “splicing quantitative trait loci” (sQTLs) than are ubiquitous expression abnormalities; however, this does not preclude the possibility that sQTLs could be differentially regulated in different cells or brain regions. Such region-specific effects might reflect the diverse developmental influences governing maturation of those brain regions, or their differential sensitivity to schizophrenogenic environmental insults. Alternatively, these region-specific effects may result from differential expression of facilitators or inhibitors of splicing (i.e., alternative splicing regulators) in a similarly region-specific manner. In support of this possibility, we found nominally significant evidence of dysregulation of full-length splicing-regulatory transcripts such as HNRNPH1 (p=0.004), HNRNPH3 (p=0.039), HNRNPC (p=0.040), and SFRS16 (p=0.018) in BA10 but not CAUD in the same samples examined here (unpublished data). The systematic follow-up of our results and evaluation of these contributory possibilities should be a high priority for future work.
Our analyses of microarray-derived TE expression data from a small sample of well-matched SZ and NC samples generated many leads, two of which we validated (either perfectly or partially) using another, more sensitive analytic technique (qRTPCR). These two results also replicated in a larger, more heterogeneous, and fully independent sample of SZ and NC subjects. One of these genes (CPNE3, a calcium-dependent membrane-binding protein that co-localizes with phosphorylated focal adhesion kinase at the leading edge of migrating cells (Heinrich et al., 2010)) has never before been implicated in SZ, and the other (ENAH, an actin-associated protein involved in cytoskeleton remodeling and neuronal projection) was targeted just once among a panel of 18 target genes (Kahler et al., 2008), highlighting the advantage of a transcriptome-wide approach that can generate new candidate genes and hypotheses regarding this complex multifactorial disorder.
Aside from generating new hypotheses, this work validates our prior blood-based biomarker study of TE expression in SZ (Glatt et al., 2009). For example, 44 (28%) of the 156 genes that we previously found to exhibit Bonferroni-corrected significant abnormal expression of a TE in peripheral blood cells in psychotic subjects (SZ plus psychotic bipolar disorder) were also found to have at least nominally significant abnormal TE expression in either BA10 or CAUD in the present study of postmortem brain. Further, eight of those 156 genes (ADAR, ARHGAP26, BIRC6, MAPK14, STXBP2, SYNE2, UTRN, and ZDHHC17) had TEs that were Bonferroni-corrected significant in blood and at least nominally significantly dysregulated in both brain regions, a result very unlikely to occur by chance (binomial test, p<0.0001). This type of convergence suggests two areas for future work; the pursuit of factors that may be capable of disrupting the expression of particular TEs regardless of tissue type, and the validation of blood-based biomarkers for these disorders.
This study also lends support to hypotheses generated by the prior work of others. In particular, the dysregulation of ERBB4 exons we observed in both BA10 and CAUD confirmed prior postmortem work linking particular splice variants of this neuregulin-1 cofactor to risk for the disorder (Kao et al., 2010; Law et al., 2007; Silberberg et al., 2006). We could not confirm in our small microarray sample the differential splicing of other previously observed results for SZ candidate genes, such as CTNNA2, DISC1, ESR1, GRM3, and NRG1; however, because our microarray sample was very small it is possible that these effects may have eluded detection in our sample due primarily to lack of power. This seems particularly likely for DISC1 and NRG1, as we did observe patterns of TE-expression data suggestive of differential splicing of known alternatively spliced TEs of these genes that did not attain statistical significance due to relatively large variance. It is also possible that the prior results are specific for regions of the brain (e.g., BA9) we did not have available for analysis. Yet, several other genes for which functionally distinct and neurodevelopmentally important splice variants are known (including NUMBL, DSCAM, FGF, SNAP25, and Neuroligins 2 and 4X) were also found in our study to have SZ-associated dysregulation of one or more exons in postmortem BA10, but not CAUD, reinforcing the importance of pursuing both ubiquitous and regionally specific alterations in exonic expression.
This study, like all human postmortem studies of brain disorders, is subject to several limitations. Primarily, because postmortem brain tissue from schizophrenia patients is an extremely rare and highly prized commodity, the sample size available for this study was quite small. Our design, which included carving both discovery and replication samples from the same small primary sample, further exacerbated this problem; however, this approach was taken to foster more confidence in those results that did survive replication. This anonymized sample was also not ideal because we were unable to determine the ancestry of each subject, which might relate to either genetically or environmentally mediated differences in TE expression. Thus, if the SZ and NC groups differed systematically in ancestry and if the represented ancestral groups differed systematically in expression levels of particular mRNA isoforms, then it is possible we would have falsely attributed some of these ancestry-related differences to diagnosis. Another limitation is that, because we did not have access to brain tissue from first-episode or prodromal patients, we are unable to rule out the possibility that the observed results are a function of (rather than cause of or contributor to) having SZ, such as treatment, hospitalization, medical and psychiatric comorbidity, and substance use disorders, all of which are more common among SZ patients than NC subjects. In an attempt to control for these factors, we followed-up by qRTPCR only those genes that did not differ in TE expression between the treated and untreated subgroups; however, power to detect such differences was quite low due to the small number of subjects in each group and the fact that all patients had been medicated at some time, even if not at the time of death. Future in vitro studies will be instrumental for validating these splicing abnormalities and more strongly attributing them to sequence variation rather than personal, clinical, agonal, or other factors.
In conclusion, we demonstrated the utility of examining the functional genomic output of the human brain in SZ using TE- and brain-region-specific profiling of expression intensity by microarray. The SZ-associated TE-expression abnormalities validated and replicated in this study would not have been detected using earlier generations of “whole-transcript” expression arrays. Looking back, this might explain in part why prior microarray-based studies of postmortem brain tissue in SZ have not routinely observed identical patterns of transcript dysregulation. Looking ahead, the use of RNA sequencing should facilitate the detection of additional subtle splicing (and other RNA-processing) variations that might characterize particular brain regions (or the whole organism) in SZ. Additional work is needed to extend our results into other samples and other implicated brain regions, as well as to uncover the factors (genetic or otherwise) that influence the observed expression abnormalities in brain and in blood. Ultimately the generation of region-specific transcriptome profiles that include information on the relative abundance of each splice variant may prove essential for gaining a better understanding of the biological basis of SZ and the development of better biomarkers and treatments for the disorder.
Supplementary Material
Acknowledgments
This work was supported in part by grants R21MH075027 (M.T.T.), P50MH081755-0003 (S.J.G.), and R01MH085521 (S.J.G.) from the U.S. National Institutes of Health, a Young Investigator Award and the Sidney R. Baer, Jr. Prize for Schizophrenia Research (S.J.G.) from NARSAD: The Brain and Behavior Research Fund, and A Research Grant from The Gerber Foundation (S.J.G.). The authors wish to thank Dr. Francine M. Benes, George Tejada, and the staff of the Harvard Brain Tissue Resource Center for providing postmortem brain tissue samples, Dr. George H. Trksak for determining medication status at the time of death of the SZ patients, and those subjects and family members who made this study possible.
The funding source had no role in designing the study, analyzing the data, or interpreting the results.
Footnotes
The authors report no conflicts of interest.
Ori S. Cohen designed and performed the experiments and wrote the manuscript.
Sarah Y. Mccoy, Sean Bialosuknia, Lu Liu, and Yanli Zhang-James assisted with experiments and contributed sections of text to the manuscript.
Frank A. Middleton participated in the design and execution of the neuroanatomical and molecular biological aspects of the experiments.
Ming T. Tsuang, Stephen V. Faraone, & Stephen J. Glatt designed the experiments and wrote and edited portions of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affymetrix, Inc. Data Sheet: GeneChip® Gene 1.0 ST Array System for Human, Mouse and Rat. 2007. [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biology. 2007;8(4):R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4(5):3. [PubMed] [Google Scholar]
- Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. Journal of Biological Chemistry. 1999;274(46):33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- Dho SE, Trejo J, Siderovski DP, McGlade CJ. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: Structural determinants of numb association with the cortical membrane. Molecular Biology of the Cell. 2006;17(9):4142–4155. doi: 10.1091/mbc.E06-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I, Glahn DC, Laird AR, Thelen SM, Bullmore E. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am J Psychiatry. 2008;165(8):1015–1023. doi: 10.1176/appi.ajp.2008.07101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Chandler SD, Bousman CA, Chana G, Lucero GR, Tatro E, May T, Lohr JB, Kremen WS, Everall IP, Tsuang MT. Alternatively Spliced Genes as Biomarkers for Schizophrenia, Bipolar Disorder and Psychosis: A Blood-Based Spliceome-Profiling Exploratory Study. Curr Pharmacogenomics Person Med. 2009;7(3):164–188. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(4):382–392. doi: 10.1002/ajmg.b.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, Han M, Liew CC, Tsuang MT. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goghari VM. Executive functioning-related brain abnormalities associated with the genetic liability for schizophrenia: an activation likelihood estimation meta-analysis. Psychol Med. 2010:1–14. doi: 10.1017/S0033291710001972. [DOI] [PubMed] [Google Scholar]
- Handran S, Pickett S, Verdick D. Key Considerations for Accurate Microarray Scanning and Image Analysis. In: Kamberova G, Shah S, editors. DNA Array Image Analysis: Nuts & Bolts. DNA Press, LLC; Salem, MA: 2002. pp. 83–98. [Google Scholar]
- Heinrich C, Keller C, Boulay A, Vecchi M, Bianchi M, Sack R, Lienhard S, Duss S, Hofsteenge J, Hynes NE. Copine-III interacts with ErbB2 and promotes tumor cell migration. Oncogene. 2010;29(11):1598–1610. doi: 10.1038/onc.2009.456. [DOI] [PubMed] [Google Scholar]
- Horvath S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2011;69(2):157–162. doi: 10.1016/j.biopsych.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kahler AK, Djurovic S, Kulle B, Jonsson EG, Agartz I, Hall H, Opjordsmoen S, Jakobsen KD, Hansen T, Melle I, Werge T, Steen VM, Andreassen OA. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, Law AJ. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A. 2010;107(35):15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Human Molecular Genetics. 2007;16(2):129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nature Reviews Neuroscience. 2007;8(11):819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Mexal S, Berger R, Pearce L, Barton A, Logel J, Adams CE, Ross RG, Freedman R, Leonard S. Regulation of a novel alphaN-catenin splice variant in schizophrenic smokers. American Journal of Medical Genetics B Neuropsychiatric Genetics. 2008;147B(6):759–768. doi: 10.1002/ajmg.b.30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60(2):163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(37):15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Partek Incorporated. Partek Documentation. Partek Incorporated; 2008. [Google Scholar]
- Reugels AM, Boggetti B, Scheer N, Campos-Ortega JA. Asymmetric localization of Numb:EGFP in dividing neuroepithelial cells during neurulation in Danio rerio. Developmental Dynamics. 2006;235(4):934–948. doi: 10.1002/dvdy.20699. [DOI] [PubMed] [Google Scholar]
- Sartorius LJ, Weinberger DR, Hyde TM, Harrison PJ, Kleinman JE, Lipska BK. Expression of a GRM3 splice variant is increased in the dorsolateral prefrontal cortex of individuals carrying a schizophrenia risk SNP. Neuropsychopharmacology. 2008;33(11):2626–2634. doi: 10.1038/sj.npp.1301669. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101(6):671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. American Journal of Medical Genetics B Neuropsychiatric Genetics. 2006;141B(2):142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- Tan W, Wang Y, Gold B, Chen J, Dean M, Harrison PJ, Weinberger DR, Law AJ. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. Journal of Biological Chemistry. 2007;282(33):24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One. 2011;6(1):e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Miranda-Angulo AL, Wong J, Perlman WR, Ward SE, Radhakrishna V, Straub RE, Weinberger DR, Kleinman JE. Variants in the estrogen receptor alpha gene and its mRNA contribute to risk for schizophrenia. Human Molecular Genetics. 2008;17(15):2293–2309. doi: 10.1093/hmg/ddn130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Cheung C, Leung M, Li Q, Chua S, McAlonan G. Are Bipolar Disorder and Schizophrenia Neuroanatomically Distinct? An Anatomical Likelihood Meta-analysis. Front Hum Neurosci. 2010;4:189. doi: 10.3389/fnhum.2010.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends in Biochemical Science. 2006;31(10):581–588. doi: 10.1016/j.tibs.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.