Abstract
MORF (monocytic leukemia zinc-finger protein (MOZ)-related factor) and MOZ are catalytic subunits of histone acetyltransferase (HAT) complexes essential in hematopoiesis, neurogenesis, skeletogenesis and other developmental programs and implicated in human leukemias. The canonical HAT domain of MORF/MOZ is preceded by a tandem of plant homeodomain (PHD) fingers whose biological roles and requirements for MORF/MOZ activity are unknown. Here we demonstrate that the tandem PHD1/2 fingers of MORF recognize the N-terminal tail of histone H3. Acetylation of Lys9 (H3K9ac) or Lys14 (H3K14ac) enhances binding of MORF PHD1/2 to unmodified H3 peptides two to three fold. The selectivity for acetylated H3 tail is conserved in the double PHD1/2 fingers of MOZ. This interaction requires the intact N-terminus of histone H3 and is inhibited by trimethylation of Lys4. Biochemical analysis using NMR, fluorescence spectroscopy and mutagenesis identified key amino acids of MORF PHD1/2 necessary for the interaction with histones. Fluorescence microscopy and immunoprecipitation experiments reveal that both PHD fingers are required for binding to H3K14ac in vivo and localization to chromatin. The HAT assays indicate that the interaction with H3K14ac may promote enzymatic activity in trans. Together, our data suggest that the PHD1/2 fingers play a role in MOZ/MORF HATs association with the chromatic regions enriched in acetylated marks.
Keywords: plant homeodomain finger, MORF, histone, binding, NMR
INTRODUCTION
MORF (monocytic leukemic zinc-finger protein (MOZ)-related factor) and MOZ are catalytic subunits of the major histone acetyltransferase (HAT) complexes (reviewed in 1; 2; 3). The HAT activity of MORF/MOZ is required for normal developmental programs, including hematopoiesis and skeletogeneis and for transcriptional regulation of various genes, particularly the Hox family 2; 3; 4. Dysregulation of the MOZ/MORF HAT function caused by chromosomal translocations is associated with aggressive forms of leukemia, myelodysplastic syndromes, leiomyomata, and other human malignancies 2; 3. Both MORF and MOZ fuse with multiple genes including CBP and p300 HATs 5; 6; 7; 8. The resulting transformed proteins possess two catalytic HAT domains, one from the MOZ/MORF fragment and another from the CBP/p300 fragment. Formation of such super-HAT chimeras may trigger aberrant histone acetylation and rapid transcriptional activation and overexpression of oncogenes 2; 9; 10.
Human MORF and MOZ together with HBO1, MOF and Tip60 comprise one of the three families of acetyltransferase enzymes, the MYST (Moz, Ybf2/Sas3, Sas2, Tip60) family. The MOZ/MORF HATs are conserved throughout the eukaryotic kingdom, widely expressed and are responsible for acetylation of a substantial portion of histone H3 1. Both MORF and MOZ form stable complexes with the adaptor proteins, such as BRPF (bromodomain PHD finger) proteins 1, 2 or 3, the tumor suppressor ING5 (inhibitor of growth 5), and EAF6 (homolog of Esa1-associated factor 6). BRPFs appear to mediate the assembly of the complexes at specific genomic regions 11; 12. Although the role of the EAF6 component remains unclear, ING5 has been shown to bind histone H3 trimethylated at Lys4 (H3K4me3) via its C-terminal PHD finger and stabilize the complex at promoters of actively transcribed genes 13.
MORF, also known as MYST4 or KAT6B (lysine acetyltransferase 6B), is a large, 1781-residue multimodular protein 14. It contains an NEMM (N-terminal part of Enok, MOZ or MORF) domain, two PHD fingers, and the MYST module followed by the ED (glutamate/aspartate-rich) and SM (serine/methionine-rich) regions (Fig. 1a). The ED and SM regions likely play a role in transcriptional activity of MORF 15. The MYST domain, composed of a zinc finger and an acetyl-CoA binding motif, catalyzes transfer of the acetyl group from acetyl-CoA to a lysine residue of histone substrate. A cysteine-rich sequence preceding the catalytic MYST domain encompasses a tandem of PHD fingers the biological function of which is not well understood.
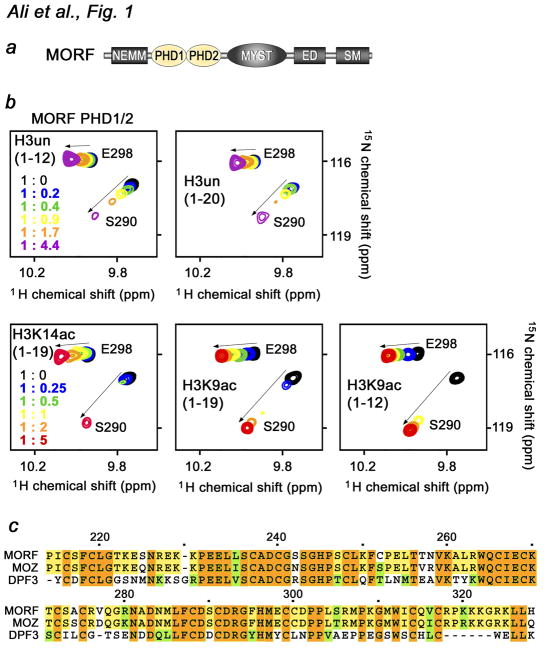
Fig. 1.
The MORF PHD1/2 fingers bind histone H3. (a) Architecture of MORF. The tandem PHD1/2 fingers are colored yellow. (b) Superimposed 1H,15N HSQC spectra of 0.1 mM MORF PHD1/2, collected as indicated H3 peptides were separately titrated in. The spectra are color-coded according to the protein-peptide ratio (inset). (c) Alignment of the PHD1/2 sequences: absolutely, moderately and weakly conserved residues are colored brown, yellow and green, respectively. Each tenth residue of MORF PHD1/2 is marked by a dot and labeled.
The PHD fingers comprise a large family of protein modules found in various chromatin regulatory complexes (reviewed in 16; 17). They are structurally related to the E3 ubiquitin ligase RING fingers 18 and the phosphoinositide-binding FYVE domains 19, however are distinguished because of their biological activities. According to specificity toward posttranslationally modified or unmodified histones, the PHD finger family can be divided into several subsets. One of the major subsets binds strongly and specifically to H3K4me3, whereas another subset recognizes unmodified histone H3 (H3un) tail 20; 21; 22; 23; 24; 25. A few PHD fingers are capable of ‘reading’ two epigenetic marks at the same time 26 or concomitantly interacting with modified histones and non-histone ligands 27; 28; 29; 30. The double PHD1/2 fingers of DPF3 associate with histone H3 acetylated at Lys14 (H3K14ac) 31; 32, and more recently, the double PHD1/2 fingers of MOZ have been shown to bind H3K14ac 33. In the last few years a wealth of information on PHD fingers activities was generated, however it remains challenging to predict a physiological ligand of a PHD finger based on the primary sequence as its specificity or selectivity is defined by the motifs outside of zinc-coordinating Cys/His residues.
In these studies, we demonstrate that the double PHD1/2 fingers of MORF, the catalytic subunit of the MORF/MOZ HAT complexes, exhibit selectivity toward acetylated histone H3 tail. The interaction with histone peptides induces large chemical shift changes particularly in the second PHD finger (PHD2) and is inhibited by methylation of Lys4 of histone H3 or by mutating the binding site residues in PHD2. We show that the preference for acetylated H3 is conserved in the MOZ PHD1/2 fingers. Our findings provide new insight into the biological role of MORF which can function both as a writer and a reader of the same epigenetic marks.
RESULTS AND DISCUSSION
The MORF PHD1/2 fingers bind histone H3 tail
The MORF HAT subunit contains a unique assembly of closely linked PHD fingers the biological role of which remains unclear. As single PHD modules have been shown to associate with histone H3 peptides, we tested whether the tandem MORF PHD1/2 fingers are capable of binding to histones. 1H,15N HSQC spectra of uniformly 15N-labeled PHD1/2 were collected while a peptide corresponding to amino acids 1–12 of unmodified histone H3 (H3un) was added stepwise (Fig. 1b). Substantial chemical shift changes observed in the spectra demonstrated that the MORF PHD1/2 fingers recognize H3un. Addition of a longer H3 peptide (amino acids 1–20) induced similar resonance perturbations, suggesting that the twelve N-terminal residues of H3 are sufficient for the interaction. The intermediate exchange regime on the NMR time scale indicated strong binding.
Acetylation of Lys9 or Lys14 enhances binding of MORF PHD1/2 to H3
Histone binding of PHD fingers can be influenced by post-translational modifications (PTMs) in H3 tail 16; 17. Furthermore, alignment of the amino acid sequences of MORF PHD1/2 and PHD modules with known functions reveals a high degree similarity to the double PHD fingers of DPF3, which have been found to prefer acetylated histone peptides, particularly H3K14ac 31; 32 (Fig. 1c). To examine the effect of acetylation, we investigated interaction of the MORF PHD1/2 fingers with histone H3 peptides acetylated at Lys9 (H3K9ac) and Lys14 (H3K14ac) by NMR (Fig. 1b, second row). Analysis of resonance perturbations induced in MORF PHD1/2 by long (amino acids 1–19) and short (amino acids 1–12) H3K9ac revealed that these peptides are bound to the same extent and slightly better in comparison with the H3un peptides. To determine if position of the acetylation mark on the histone tail plays a role, H3K14ac peptide (1–19) was titrated in. As shown in Figure 1b, interaction of the MORF PHD1/2 fingers with H3K14ac triggered comparable chemical shift changes, implying that acetylation of either lysine residue enhances binding.
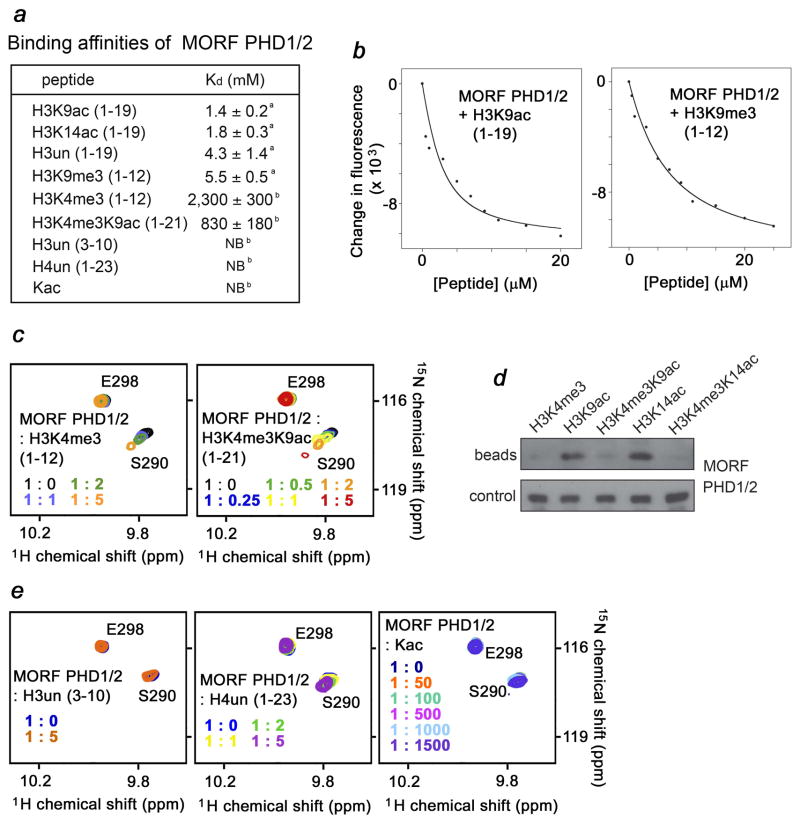
To assess the magnitude of enhancement, we measured binding affinity of the MORF PHD1/2 fingers for the unmodified and acetylated H3 peptides using tryptophan fluorescence (Fig. 2a, b). The dissociation constants (Kds) for the interactions with H3K9ac and H3K14ac were found to be 1.4 ± 0.2 μM and 1.8 ± 0.3 μM, respectively. However binding to the H3un peptide was two to three fold weaker (Kd of 4.3 ± 1.4 μM). Altogether, NMR and fluorescence data demonstrate that acetylation of either Lys9 or Lys14 increases binding of MORF PHD1/2 to the histone H3 tail. Similar, four-fold augmentation of binding affinity upon acetylation of Lys14 of histone H3 has been reported for the double PHD fingers of DPF3 (Kds of 2 μM for H3un and 0.5 μM for H3K14ac) 31. We also note that the low μM-range affinities are exhibited by single PHD fingers, chromodomains, Tudor and other histone-binding modules 34, inferring that the association of MORF PHD1/2 with acetylated or unmodified histone H3 is physiologically relevant.
Fig. 2.
Selectivity of the MORF PHD1/2 fingers. (a) Binding affinities of the MORF PHD1/2 fingers for indicated peptides were measured by tryptophan fluorescence (a) and NMR (b). NB- no binding. (b) Representative binding curves used to determine Kds of the PHD1/2-H3K9ac and PHD1/2-H3K9me3 interactions. (c, e) Superimposed 1H,15N HSQC spectra of MORF PHD1/2, collected as indicated H3 peptides were added stepwise. (d) Western blot of bound MORF PHD1/2 (beads) following in-solution pulldowns with the indicated peptides. Control shows detection of unbound protein following incubation with peptide-bead conjugate.
Methylation of Lys4 inhibits binding of MORF PHD1/2
Methylation of lysine residues is known to differentially affect binding of PHD fingers to histone H3. We assessed the effect of trimethylation of Lys4 and Lys9 on the interaction of MORF PHD1/2. As shown in Fig 2c (first panel), titration of the H3K4me3 peptide induced small changes in the NMR spectra of PHD1/2. Plotting these changes over peptide concentration yielded a ~ 2 mM binding affinity, which is three orders of magnitude lower than affinity of this protein toward H3un or H3K9ac. Thus, methylation of Lys4 disrupts the association of MORF PHD1/2 with histone H3, and concurring acetylation of Lys9 only slightly improves binding. The MORF PHD1/2 fingers associate with the doubly modified H3K4me3K9ac peptide ~four fold stronger than they associate with H3K4me3 but still three orders of magnitude weaker than with H3K9ac, underscoring the robust inhibitory effect of Lys4 methylation (Fig. 2a and c, second panel). The inhibitory effect of Lys4 methylation on H3 binding in the presence of K9ac and K14ac was also evident in peptide pulldown assays (Fig. 2d). In contrast, methylation of Lys9 did not alter this interaction. Binding affinity of the MORF PHD1/2 fingers for H3K9me3, measured by tryptophan fluorescence, remained unchanged as compared to the affinity for H3un (Fig. 2a). This result suggests that coordination of the acetyl group rather than an increase in hydrophobicity enhances interaction of MORF PHD1/2 with H3K9ac.
The intact N-terminus of H3 is required for binding of MORF PHD1/2
To define the role of the N-terminal residues of H3 in the association with MORF PHD1/2, a peptide comprising amino acids Thr3-Ser10 but missing Ala1 and Arg2 [H3 (3–10)], was tested in NMR titration experiments. Addition of a 5-fold excess of H3 (3–10) caused no changes in the NMR spectra of PHD1/2, implying that this module does not bind H3 deficient in the first two N-terminal residues and that the Ala1-Arg2 sequence is required for the interaction (Fig. 2c, third panel). In agreement, H4 peptide (residues 1–23) was unable to trigger chemical shift changes in MORF PHD1/2 (Fig. 2c, fourth panel). Thus, the Ser1-Gly2-Arg2 sequence of H4 cannot substitute for the Ala1-Arg2 sequence of H3. In the complexes of PHD fingers with H3un, the N-amino group of Ala1 of the peptide forms hydrogen bonds with two or three backbone carbonyl groups of the PxGxW motif, whereas the guanidinium moiety of Arg2 is usually involved in hydrogen bonding and ionic interactions 16; 24. The PxGxW motif is also present in the MORF PHD2 sequence (Fig. 1b), suggesting a similar mechanism for coordinating the N-terminus of H3. A lack of any significant resonance perturbations upon titrating acetylated lysine amino acid alone indicated that the acetylated group is bound by MORF PHD1/2 only in the context of the intact histone tail (Fig. 2c, fifth panel).
Selectivity for acetylated H3 is conserved in MOZ PHD1/2
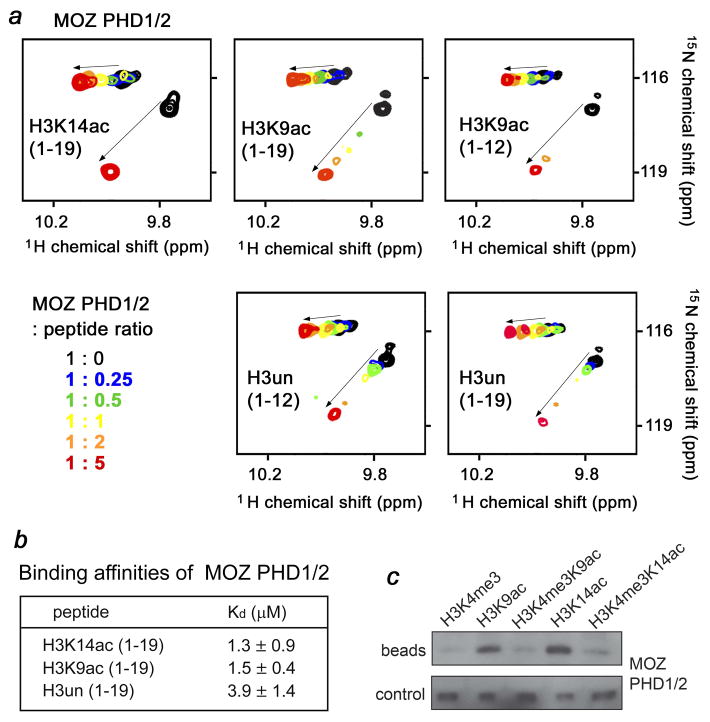
A similar assembly of PHD fingers exists in a paralog of MORF, the MOZ HAT protein. We examined whether the MOZ PHD1/2 fingers are able to bind acetylated histone H3. Titration of either H3K14ac or H3K9ac peptide into the MOZ PHD1/2 NMR sample induced chemical shift changes almost identical in magnitude, implying that the peptides are bound to the same degree (Fig. 3a). In agreement, binding affinities of MOZ PHD1/2 for these peptides measured by tryptophan fluorescence were 1.3 ± 0.9 μM and 1.5 ± 0.4 μM, respectively (Fig. 3b). The H3un peptide was bound two to three fold weaker (Kd of 3.9 ± 1.4 μM) as seen in NMR and fluorescence assays. Similarly to MORF PHD1/2, peptide pulldown assays revealed that methylation of Lys4 disrupts binding of MOZ PHD1/2 to the histone H3 tail acetylated at Lys9 or Lys14 (Fig. 3c). Collectively, these data demonstrate that the preference for the acetylated histone H3 tail is a general feature, which is conserved in the double PHD1/2 fingers of MORF, MOZ and DPF3, and are also in line with the recent report demonstrating binding of the MOZ PHD1/2 fingers to H3K14ac 33.
Fig. 3.
The selectivity for acetylated H3 is conserved. (a) Superimposed 1H,15N HSQC spectra of MOZ PHD1/2, recorded during addition of the indicated H3 peptides. The spectra are color-coded according to the protein-peptide ratio. (b) Binding affinities of the MOZ PHD1/2 fingers for indicated H3 peptides were measured by tryptophan fluorescence. (c) Western blot of bound MOZ PHD1/2 (beads) following in-solution pulldowns with the indicated peptides. Control shows detection of unbound protein following incubation with peptide-bead conjugate.
The molecular basis for the interaction of MORF PHD1/2 with H3K9ac
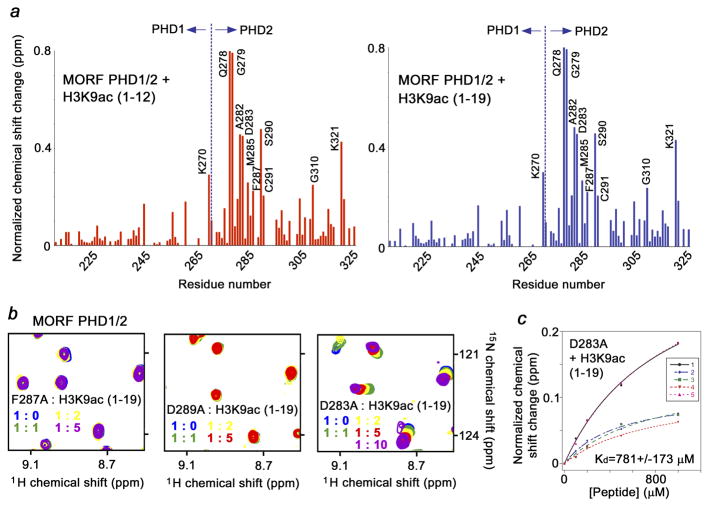
To identify the active site residues of the MORF PHD1/2 fingers, 1H, 13C and 15N resonances of the protein were assigned using a set of triple resonance NMR experiments. The differences in chemical shifts for backbone amides of MORF PHD1/2 in the apo-state and bound to the long or short H3K9ac peptides are shown as the histogram plots in Figure 4a. A similar pattern of resonance perturbations suggested that these peptides occupy the same binding pocket. Notably, chemical shift changes were detected primarily in the PHD2 finger, whereas resonances of PHD1 were less perturbed, with the exception of K270 at the far C-terminus of PHD1. The Q278-C291 sequence of PHD2, particularly residues Q278, G279, A282, D283, M285, F287, S290 and C291, as well as G310 and K321 exhibited largest chemical shift changes upon binding to the histone peptides, inferring that these residues are directly or indirectly involved in the interaction. The corresponding residues of DPF PHD1/2 comprise an extended site running across PHD2, which accommodates the N-terminal Ala1-Lys9 residues of the H3K14ac tail, while the C-terminal residues of the H3K14ac peptide, particularly those surrounding K14ac, are bound by the PHD1 finger 31. The structure of the MOZ PHD1/2-H3K14ac complex, where the electron density is seen for the first seven N-terminal residues of the H3K14ac peptide, also reveals that the N-terminus of H3 is bound by the PHD2 finger 33.
Fig. 4.
Identification of the histone H3K9ac binding site of the MORF PHD1/2 fingers. (a) Histograms show normalized 1H,15N chemical shift changes in backbone amides of 15N-labeled MORF PHD1/2 upon addition of short and long H3K9ac peptides at a protein:peptide ratio of 1:5. The most perturbed residues are labeled. (b) Mutations of the binding site residues abolish interaction of the MORF PHD1/2 fingers with H3K9ac. Superimposed 1H,15N HSQC spectra of MORF PHD1/2 mutants, collected as H3K9ac peptide was titrated in. (c) Binding affinity of D285A MORF PHD1/2 was estimated by NMR.
The importance of the active site residues was confirmed by mutagenesis. The D283, F287 and D289 residues of MORF PHD1/2 were replaced with alanine and binding of the mutant proteins to H3K9ac was examined by NMR (Fig. 4b). According to the position of these residues in known PHD-H3un complexes, D283 and D289 may form hydrogen bonds to Lys4 and Arg2 of the peptide, whereas F287 may separate the binding pockets for Lys4 and Arg2. As anticipated, substitution of F287 and D289 completely abolished the interaction of MORF PHD1/2 with H3K9ac as no chemical shift changes were observed in NMR spectra (Fig. 4b). Mutation of D283 reduced binding affinity of MORF PHD1/2 by two orders of magnitude (Fig. 4c).
Interaction of the MORF PHD1/2 fingers with histone H3 is required for the in vivo localization to chromatin
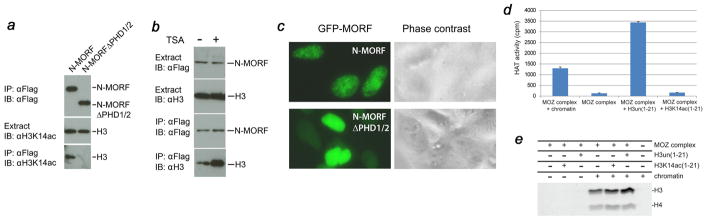
The functional importance of the histone H3 recognition by MORF PHD1/2 was investigated in HEK293 cells using co-immunoprecipitation and fluorescence microscopy. The MORF constructs containing the entire N-terminal region of MORF (N-MORF, amino acids 1–352) and N-MORFΔPHD1/2 (amino acids 1–215), in which the PHD1/2 fingers are deleted, were generated as Flag- and GFP-tag fusions. The Flag-MORF constructs were individually expressed in HEK293 cells for immunoprecipitation on anti-Flag agarose, and acetylated at Lys14 histone H3 was detected by immunoblotting with an anti-H3K14ac antibody. As shown in Figure 5a, N-MORF co-immunoprecipitated endogenous H3K14ac. In contrast, N-MORFΔPHD1/2 was unable to pull down acetylated histone H3, indicating that the PHD1/2 fingers are necessary for the interaction with H3K14ac in vivo. Furthermore, treatment of the HEK293 cells with the HDAC inhibitor trichostatin A (TSA) enhanced the association of N-MORF with histone H3, implying that hyperacetylation promotes binding (Fig. 5b). The significance of the PHD1/2 fingers for subcellular localization of MORF was examined through monitoring the distribution of GFP-tagged N-MORF and N-MORFΔPHD1/2 constructs in HEK293 cells using green fluorescence microscopy. The GFP-N-MORF protein clearly associated with chromatin forming nuclear speckles, whereas GFP-N-MORFΔPHD1/2 was evenly dispersed in the nucleus (Fig. 5c). Similar speckled localization has been observed for the PHD1/2 fingers of DPF3 and MOZ 31; 33. Taken together, these data demonstrate that the PHD1/2 fingers are required for in vivo interaction with acetylated histone H3 and the recruitment to specific chromatin regions.
Fig. 5.
The PHD1/2 fingers of MORF are required for the interaction with histone H3 in cells. (a) IPs of the Flag-MORF [N-terminal region of MORF (N-MORF, amino acids 1–352) and N-MORFΔPHD1/2 (amino acids 1–215), in which the PHD1/2 fingers are deleted] from HEK293 cells and immunoblotting with indicated antibodies. (b) IPs of the Flag-N-MORF from HEK293 cells +/− TSA and immunoblotting with indicated antibodies. (c) HEK293 cells were transfected with indicated GFP-MORF constructs, and subcellular localization of the GFP signal was detected by live green fluorescence microscopy. (d, e) HAT assays with indicated substrates and MOZ-BRPF1-ING5-hEaf6 complex purified from co-transfected cells were quantified by liquid scintillation (d) and gel fluorography (e).
Association of the MOZ PHD1/2 fingers with H3K14ac may provide a mechanism for propagation of HAT activity
Others and we have previously shown that the catalytic activity of the MORF/MORF complexes is fine-tuned by histone PTMs 11; 12; 13. To assess whether binding of the MOZ PHD1/2 fingers to acetylated histone H3 tail affects the HAT function, MOZ-BRPF1-ING5-hEaf6 complex was purified from co-transfected HEK293 cells and its enzymatic activity was tested on synthetic H3un and H3K14ac peptides and purified chromatin (Fig. 5d and e). As expected, the purified MOZ complex can efficiently acetylate native chromatin or H3un peptide. However when H3K14ac peptide was used as a substrate, no significant acetylation was detected, indicating that the MOZ complex is unable to acetylate other lysine residues on the same H3K14ac peptide, including Lys4, Lys9 and Lys18. This suggests that once H3K14ac is established and bound by MORF/MOZ PHD1/2, the HAT domain 11; 12; 13 targets Lys14 in a distinct H3 tail of the same or adjacent nucleosome or acts in trans. In agreement with such a model, pre-incubation of MOZ with the H3K14ac peptide led to a slight decrease in HAT activity of the MOZ complex on chromatin in comparison with the activity seen upon pre-incubation with the H3un peptide (Fig. 5e).
Conclusions
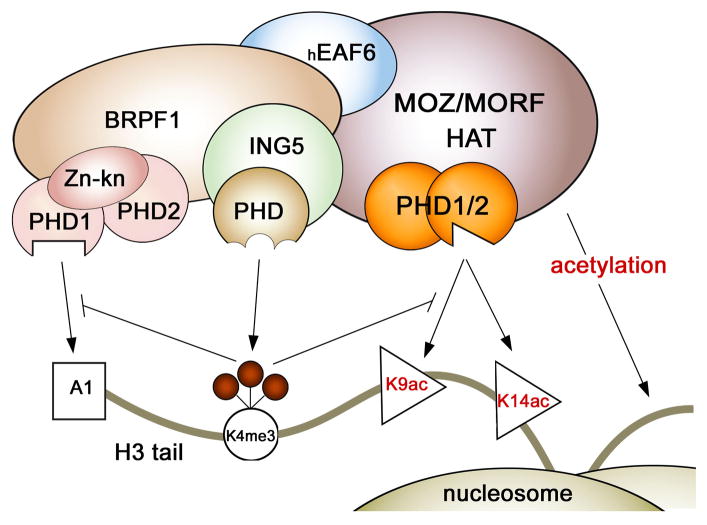
In summary, in this work we demonstrate that the tandem PHD1/2 fingers of MORF and MOZ recognize the amino-terminus of histone H3 and exhibit selectivity for acetylated Lys9 and Lys14. Acetylation of the histone tail augments binding affinity of PHD1/2 two to three fold. This interaction is abolished by methylation of Lys4 but is unaffected by methylation of Lys9. The preference for acetylated H3 is conserved and seen for other double PHD fingers, including DPF3 31. Our results indicate that the PHD1/2 fingers of MORF/MOZ may be involved in stabilization of the MOZ/MORF HAT complexes at chromatin through binding to acetylated H3 tails. We note that the PHD-like domains are present in three of the four subunits of the MORF/MOZ complexes, however these domains differ considerably (Fig. 6). While the MORF/MOZ PHD1/2 fingers are closely coupled, two PHD fingers in the BRPF1 subunit are separated by a zinc knuckle, and a single PHD finger is present in ING5. It has been found that the PHD1 finger of BRPF2 12 and BRPF1 (Lalonde et al., manuscript in preparation) recognizes unmodified H3 tail and methylation of Lys4 abolishes this interaction. In contrast, ING5 PHD finger is specific for H3K4me3 and does not associate with H3un 13. Furthermore, our findings suggest that binding of the PHD1/2 fingers of MOZ to H3K14ac may provide a mechanism for promoting acetylation of nearby histone tails. This mechanism could be critical for creating acetylated chromatin domains through propagation of the same histone mark from one H3 tail to another and from one nucleosome to another, a concept proposed to be at the heart of epigenetic marking and memory 35. Thus, the MOZ/MORF complexes represent an intriguing example of a chromatin binding assembly whose recruitment to a particular genomic site and activity can be carefully regulated through PTMs on histone tails. Future studies should decipher the significance of multiple interactions of the MOZ/MORF subunits with chromatin via crosstalk of the PHD fingers as well as the interplay of adjacent histone readers.
Fig. 6.
Crosstalk between the PHD fingers present in the MORF/MOZ, BRPF1 and ING5 subunits of the MOZ/MORF HAT complexes. The ING5 PHD finger recognizes H3K4me3, an epigenetic mark that inhibits binding of the PHD fingers of either MORF/MOZ or BRPF1.
MATERIALS AND METHODS
Cloning and Protein Purification
The double PHD1/2 fingers of MORF (aa 211–329) and MOZ (aa 207–318) were cloned into a pCool (modified pGEX2T) expression vector with Ampicillin resistance. The proteins were expressed in E. coli Rosetta-2 (DE3) pLysS cells grown in LB or 15NH4Cl minimal media supplemented with 60 μM ZnCl2. After induction with IPTG (0.5 mM) for 16 hrs at 18 °C, cells were harvested by centrifugation at 6,500 rpm and lysed by sonication. The unlabeled and 15N-labeled GST-fusion proteins were purified on glutatahione Sepharose 4B beads (GE Healthcare). The GST tag was cleaved with Thrombin protease (GE Healthcare). The proteins were concentrated into PBS buffer pH 7.4, supplemented with 100 mM NaCl and 5 mM dithiothreitol.
PCR mutagenesis
Point mutants were prepared using the Stratagene QuickChange XL Site Directed Mutagenesis kit according to the manufacturer’s instructions.
NMR Spectroscopy and sequence specific assignments
NMR experiments were performed at 298K on a Varian INOVA 800 MHz spectrometer using pulse field gradients to suppress artifacts and eliminate water signal. The NMR samples contained 0.5 mM uniformly 15N/13C-labeled MORF PHD1/2 fingers in PBS buffer pH 6.5 and 10 % D2O. The 1H,15N heteronuclear single quantum coherence (HSQC), HNCACB 36, CBCA(CO)NH 37, HNCO 37, C(CO)NH 38, and HC(CO)NH 38 were recorded and analyzed for sequential and spin system assignments. The NMR data were processed with nmrPipe 39 and analyzed with CCPNMR Analysis v1.6 40, and nmrDraw.
NMR titrations of histone peptides
The 1H,15N HSQC spectra of 0.1 mM uniformly 15N-labeled wild type or mutant MORF and MOZ PHD1/2 were collected on a Varian INOVA 600 MHz spectrometer. The spectra were recorded at 298K using 1024 × 160 increments, and a spectral width of 8804.8 × 1944.3 Hz in the 1H and 15N dimensions, respectively. The binding was characterized by monitoring chemical shift changes in 1H,15N HSQC spectra of MORF and MOZ PHD1/2 as differently modified histone tail peptides (synthesized by the University of Colorado Denver Peptide Core Facility) were added stepwise. The dissociation constants (Kds) were determined by a nonlinear least-squares analysis in Kaleidagraph using the equation:
where [L] is concentration of the peptide, [P] is concentration of the protein, Δδ is the observed chemical shift change, and Δδmax is the normalized chemical shift change at saturation. Normalized 41 chemical shift changes were calculated using the equation , where Δδ is the change in chemical shift in parts per million (ppm).
Fluorescence Spectroscopy
Spectra were recorded at 25 °C on a Fluoromax-3 spectrofluorometer (HORIBA). The samples containing 2 μM MORF or MOZ PHD1/2 and progressively increasing concentrations of the histone peptide were excited at 295 nm. Emission spectra were recorded over a range of wavelengths between 305 and 405 nm with a 0.5 nm step size and a 1 s integration time and averaged over 3 scans. The Kd values were determined using a nonlinear least-squares analysis and the equation:
where [L] is the concentration of the histone peptide, [P] is the concentration of PHD1/2, ΔI is the observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states of the PHD1/2. The Kd value was averaged over four, three or two separate experiments, with error calculated as the standard deviation between the runs.
In-solution peptide pulldown assays
Biotinylated peptide pulldown assays were performed essentially as described 42. MORF PHD1/2 and MOZ PHD1/2 were detected with an anti-GST antibody (Sigma).
Co-immunoprecipitation (IP)
Co-IP was performed as previously described 11. Briefly, HEK293 cells were seeded (300,000 cells per 6 cm dish) and transfected with expression plasmids encoding for the Flag-tagged MORF constructs. Lipofactamine 2000 (Invitrogen) was used as the transaction reagent. About 48 h post-transfection, cells were lysed in buffer B containing 150 mM KCl and used for immunoprecipitation on anti-Flag M2 agarose (Sigma). After extensive washing, specifically bound proteins were eluted in 50 mM phosphoric acid for immunoblotting with anti-Flag (Sigma), anti-H3K14ac (Abcam) or anti-H3 (Abcam) antibodies. Where indicated, TSA (3 μM) was added to the cell culture medium 6 h prior to harvesting.
Live green fluorescence microscopy
HEK293 cells (40,000 per well on a 12-well plate) were transfected with expression plasmids for GFP-tagged MORF constructs. 18 h post-transfection, subcellular localization of GFP-fusion proteins in live cells were examined under a fluorescence microscope. Selective green fluorescence and phase contrast images were taken for further processing via Adobe Photoshop.
Histone acetyltransferase assay
MOZ complex was purified by Flag-IP from 293T cells transfected with 3xFlag-BRPF1, HA-hEaf6, HA-ING5 and Flag-MOZ expression plasmids. Pre-incubation of 300 ng of biotinylated peptides corresponding to amino acids 1–21 of human histone H3, either unmodified or acetylated at Lys14 and/or 500 ng of native short oligonucleosomes (purified from HeLa cells) was done at 4°C for 15 min. HAT reaction was performed in 15 μl final volume of 50 mM KCl, 50 mM Tris (pH 8), 1 mM DTT, 5 % glycerol, 10 mM Na-Butyrate and 0.1 mM EDTA with 1.25 μL of [3H] acetyl-CoA (0.1 μCi/μL, 4.9 Ci/mmol) for 60 min at 30°C. Each reaction was spotted onto p81 filters, which were then washed three times with 50 mM Na Carbonate (pH 9.2). The amount of incorporated [3H] acetyl was determined using a scintillation counter. Alternatively, reaction were loaded on a 18% SDS-PAGE, stained and processed for fluorography.
We found that the tandem PHD1/2 fingers of MORF and MOZ recognize the N-terminal tail of histone H3.
Acetylation of Lys9 or Lys14 of histone H3 enhances this interaction, whereas methylation of Lys4 inhibits it.
We identified key residues of PHD1/2 necessary for the interaction with histones.
The PHD1/2 fingers are required for binding to H3K14ac in vivo and localization to chromatin.
Acknowledgments
We thank N. Pelletier for help with functional experiments. This research is supported by grants from the NIH, GM096863 and CA113472 (T.G.K.), GM085394 (B.D.S.), CA09156 (S.B.R.), operating grants from the Canadian Institutes of Health Research (MOP-64289/J.C. and X.J.Y.), and operating grants from the Canadian Cancer Society (X.J.Y.). M.-E.L. holds a National Science and Engineering Research Council doctoral studentship and J.C. is a Canada Research Chair. T.G.K. is an Independent NARSAD Investigator.
Abbreviations used
- MOZ
monocytic leukemic zinc-finger protein
- MORF
MOZ-related factor
- HAT
histone acetyltransferase
- PHD
plant homeodomain
- H3K9ac
histone H3 acetylated at Lys9
- H3K14ac
histone H3 acetylated at Lys14
- HSQC
heteronuclear single quantum coherence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sapountzi V, Cote J. MYST-family histone acetyltransferases: beyond chromatin. Cellular and molecular life sciences : CMLS. 2011;68:1147–56. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang XJ, Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–19. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]
- 3.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113:4866–74. doi: 10.1182/blood-2008-04-152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;16)(p11; p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 6.Panagopoulos I, Fioretos T, Isaksson M, Samuelsson U, Billstrom R, Strombeck B, Mitelman F, Johansson B. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22; p13) Hum Mol Genet. 2001;10:395–404. doi: 10.1093/hmg/10.4.395. [DOI] [PubMed] [Google Scholar]
- 7.Terui K, Sato T, Sasaki S, Kudo K, Kamio T, Ito E. Two novel variants of MOZ-CBP fusion transcripts in spontaneously remitted infant leukemia with t(1;16;8)(p13; p13; p11), a new variant of t(8;16)(p11; p13) Haematologica. 2008;93:1591–3. doi: 10.3324/haematol.13020. [DOI] [PubMed] [Google Scholar]
- 8.Boyd EM, Bench AJ, Vaghela KJ, Campbell GN, Chowdhury FB, Gudgin EJ, Scott MA, Erber WN. Therapy-related acute myeloid leukaemia with t(8;16)(p11; p13); MOZ-CBP and polymorphisms in detoxifying and DNA repair genes. Leukemia. 2009;23:1164–7. doi: 10.1038/leu.2008.383. [DOI] [PubMed] [Google Scholar]
- 9.Champagne N, Pelletier N, Yang XJ. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20:404–9. doi: 10.1038/sj.onc.1204114. [DOI] [PubMed] [Google Scholar]
- 10.Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedeu J, Montserrat E, Campo E. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11; p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–54. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 11.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, Champagne N, Cote J, Yang XJ. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol. 2008;28:6828–43. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin S, Jin L, Zhang J, Liu L, Ji P, Wu M, Wu J, Shi Y. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF) The Journal of biological chemistry. 2011;286:36944–55. doi: 10.1074/jbc.M111.244400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champagne KS, Saksouk N, Pena PV, Johnson K, Ullah M, Yang XJ, Cote J, Kutateladze TG. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins. 2008;72:1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y, Heng HH, Yang XJ. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–36. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier N, Champagne N, Stifani S, Yang XJ. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene. 2002;21:2729–40. doi: 10.1038/sj.onc.1205367. [DOI] [PubMed] [Google Scholar]
- 16.Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–23. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic acids research. 2011;39:9061–71. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends in biochemical sciences. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta. 2006;1761:868–77. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 24.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–8. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, Gozani O, Kutateladze TG, Cleary ML. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400:137–44. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hom RA, Chang PY, Roy S, Musselman CA, Glass KC, Selezneva AI, Gozani O, Ismagilov RF, Cleary ML, Kutateladze TG. Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain. J Mol Biol. 2010;400:145–54. doi: 10.1016/j.jmb.2010.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, Osmers U, Raman G, Schwantes RH, Diaz MO, Bushweller JH. The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression. Biochemistry. 2010;49:6576–86. doi: 10.1021/bi1009387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro Isomerization in MLL1 PHD3-Bromo Cassette Connects H3K4me Readout to CyP33 and HDAC-Mediated Repression. Cell. 2010;141:1183–94. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–62. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–84. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, Wu J, Shi Y. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes & development. 2012;26:1376–91. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature reviews Genetics. 2010;11:285–96. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittekind M, Mueller L. HNCACB, A high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J Magn Reson. 1993;101:201–5. [Google Scholar]
- 37.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J Magn Reson. 1992;96:432–40. [Google Scholar]
- 38.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson. 1993;101:114–9. [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 41.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–61. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 42.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2391. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]