Abstract
Nucleotide excision repair (NER) removes lesions caused by environmental mutagens or UV light from DNA. A hallmark of NER is the extraordinarily wide substrate specificity, raising the question of how one set of proteins is able to recognize structurally diverse lesions. Two key features of good NER substrates are that they are bulky and thermodynamically destabilize DNA duplexes. To understand what the limiting step in damage recognition in NER is, we set out to test the hypothesis that there is a correlation of the degree of thermodynamic destabilization induced by a lesion, binding affinity to the damage recognition protein XPC-RAD23B and overall NER efficiency. We chose to use acetylaminofluorene (AAF) and aminofluorene (AF) adducts at the C8 position of guanine in different positions within the NarI (GGCGCC) sequence, as it is known that the structures of the duplexes depend on the position of the lesion in this context. We found that the efficiency of NER and the binding affinity of the damage recognition factor XPC-RAD23B correlated with the thermodynamic destabilization induced by the lesion. Our study is the first systematic analysis correlating these three parameters and supports the idea that initial damage recognition by XPC-RAD23B is a key rate-limiting step in NER.
Keywords: DNA damage, DNA repair, nucleotide excision repair, acetylaminofluorene, XPC-RAD23B, thermodynamic stability
INTRODUCTION
Nucleotide excision repair (NER) is the main pathway for removing bulky DNA adducts formed by solar UV irradiation, environmental mutagens and chemotherapeutic agents in humans.(1, 2) At least 30 proteins are involved in NER and they assemble at the sites of damage in sequential order with the help of protein-DNA and protein-protein interactions. Although the same set of proteins is responsible for the recognition of all lesions by NER, the efficiencies of the excision of individual lesions can vary greatly.(3) Investigation of the substrate specificity suggested early on that good NER substrates are helix-destabilizing and contain a chemical modification, and that there is a correlation between the degree of helix destabilization and NER efficiency.(4) These observations were framed in the context of the “bipartite substrate discrimination model” suggesting that lesions are processed by NER if they contain a chemical modification and disrupt base pairing.(5) Subsequent studies revealed that local DNA duplex destabilization can include factors such as disrupted base-pairing, bending and flexibility, all in line with the general concept thermodynamic destabilization of the DNA duplex is the main trigger for NER.(6–8)
These observations are in agreement with current models of how NER proteins recognize damaged sites in DNA.(9,10) The initial damage recognition factor XPC-RAD23B employs a lesion-unspecific mechanism for damage recognition, probing for thermodynamic destabilization and binding ssDNA accessible on the non-damaged strand opposite the lesion.(11–13) This binding mode of XPC-RAD23B explains how thermodynamic destabilization rather than lesion structure is the determining factor for damage recognition. As XPC-RAD23B does not appear to directly contact the lesion, it is also prone to binding destabilized sites in DNA that do not contain damage such as mismatches. NER therefore requires a damage verification step that assures the presence of a chemical modification in the substrate.(14) Recent studies strongly suggest that this damage verification step is accomplished by the XPD helicase, a subunit of TFIIH. It is believed that XPD tracks along the DNA until it stalls at a bulky lesion, thereby permitting further assembly of the NER complex.(15–18) In line with this model, certain bulky lesions that do not induce duplex destabilization are refractory to XPC-RAD23B binding and NER.(19, 20)
We set out to probe the model that NER efficiency is directly correlated with the degree of thermodynamic destabilization and XPC-RAD23B binding affinity in a systematic fashion in the context of a specific lesion. We reasoned that major groove adducts of aminofluorene (dG-AF) and acetyl-aminofluorene (dG-AAF), would be particularly well suited for this purpose, as it is known that the structural and thermodynamic features of these adducts on DNA is highly sequence context dependent.(21, 22) As we would not expect any major differences of how AF and AAF adducts would stall the XPD helicase during the damage verification step, we believe that this system is well suited to correlate thermodynamic properties, XPC-RAD23B binding and NER efficiency.
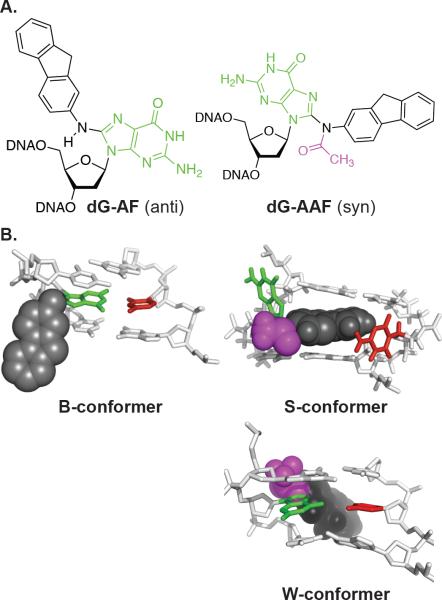
The structural properties of AF and AAF adducts are intriguing and well studied in the context of the NarI sequence (GGCGCC), where dG-AF and dG-AAF can assume dramatically different conformations depending on which G in the sequence bears the adduct.(23, 24) AF and AAF adducts can assume two main conformations:(22, 24–26) If the adduct-containing nucleotide has an anti glycosidic bond conformation, the duplex is fully B-form with all the base pairs intact and the aromatic fluorene ring is located in the major groove (Figure 1, B-conformation). Alternatively, the glycosidic bond can assume a syn conformation, in particular in the presence of the additional bulk of the acetyl group at N8 in AAF. Here, the base is displaced in the major groove and the fluorene intercalates into base stack (Figure 1, S-conformation). Lastly, dG-AAF adducts can also be present in the wedge (W) conformation, where the adducted G also has a syn glycosidic bond and undergoes a Hoogsteen pairing interaction with the C opposite it (Figure 1, W-conformation). Cho and coworkers studied the relative populations and thermodynamic properties in detail and found that dG-AAF and dG-AF adducts with higher propensity for S and W conformations cause higher degrees of duplex destabilization and are more readily processed by bacterial NER.(24) Additional studies have compared the processing of dG-AAF and dG-AF substrates by bacterial and human NER, but have not thoroughly addressed the origins of repair efficiency.(27–32)
Figure 1.
Structures of dG-C8-AF; N-(deoxyguanosin-8-yl)-2-aminofluorene and dG-C8-AAF; N-(deoxyguanosin-8-yl)-2-acetylaminofluorene and anti and syn conformations. dG-C8-AF is believed to have the base mainly in the syn conformation, whereas dG-C8-AAF favorably retains anti conformation.
Yeo et al,
Here we report that there is a strong correlation between the efficiency of human NER, XPC-RAD23B binding affinity and degree of thermodynamic destabilization induced by AF and AAF adducts in various positions of the NarI sequence. Our studies suggest that the efficiency of damage recognition by XPC-RAD23B is a key determinant of the overall NER rate.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and solvents were purchased from Fluka-Sigma-Aldrich. The 1000 Å “Q-columns” were from Biosearch Technologies and all other reagents for DNA synthesis were from Glen Research. DNA syntheses were performed on a PerSeptive Biosystems Expedite 8900 DNA synthesizer, HPLC purifications on a JASCO system equipped with a Phenomenex Clarity Oligo-RP Semi-prep column: C18, 5 μm, 50×10.00 mm. C18-SepPak cartridges were from Waters. T4 PNK, T4 polymerase, and T4 DNA ligase were from New England Biolabs (NEB) and sequenase (T7 DNA polymerase) from USB Corp. [α-32P]-dCTP was from PerkinElmer. Unmodified 12-mer oligonucleotides were purchased from Integrated DNA Technologies (IDT).
Preparation of Oligodeoxynucleotides containing a site specific AAF or AF
The AAF-containing oligonucleotides 12-AAF, 24-AAF and 44-AAF (see Table 1 for sequences) were synthesized using our modified `ultra-mild' DNA synthesis protocol.(27) All DNA syntheses were performed on 1 μM scale using 1000 Å “Q-columns”. The phosphoramidites for T, Ac-dC, Pac-dA, iPrPac-dG were dissolved to 0.1M in CH3CN, that for iPrPac-dG-AAF in CH2Cl2 to 0.1 M. The 5'-DMTr protective group was retained for all the syntheses (`DMT-on' synthesis). After completion of oligonucleotide synthesis, the solid support was dried and incubated overnight at 55°C with a solution containing 10% (V/V) of diisopropylamine (iPr2NH) and 0.25 M of β-mercaptoethanol in MeOH. The supernatant containing the oligonucleotide was decanted and concentrated. Oligonucleotides were then dissolved in 1 ml of 1 M triethylamoniumacetate (TEAA, pH 7) and filtered through 0.45 μm filter and purified by reverse-phase HPLC purification. The peak of `DMTr-ON' oligonucleotide was collected, concentrated, and treated with 80% acetic acid solution for 40 min at room temperature to remove the DMTr. Oligonucleotides containing dG-AF were prepared by treatment of the dG-AAF-containing oligonucleotides with 1M NaOH as described.(33) After purification, all oligonucleotides were desalted by Sep-Pak (Waters), eluted with H2O and stored in TE (pH 8). The identity of 12-AAF/AF and 24-AAF/AF-containing oligonucleotides was confirmed by ESI-MS, that of the 44-AAF/AF-containing oligonucleotides by MALDI-MS.
Table 1.
Oligonucleotides used in this study*
| 12–dG1–AF/AAF | 5'–TACCXGCGCCAC |
| 12–dG2–AF/AAF | 5'–TACCGXCGCCAC |
| 12–dG3–AF/AAF | 5'–TACCGGCXCCAC |
| 24–dG1–AF/AAF | 5'–TAGCTATTACCXGCGCCACATGTC |
| 24–dG2–AF/AAF | 5'–GCTATTACCGXCGCCACATGTCAG |
| 24–dG3–AF/AAF | 5'–CTATTACCGGCXCCACATGTCAGC |
| 44–dG1–AF/AAF | 5'–CCCTAGCTAGAGCTACGTAGCTATTACCXGCGCCACATGTCAGC |
| 44–dG2–AF/AAF | 5'–CCCTAGCTAGAGCTACGTAGCTATTACCGXCGCCACATGTCAGC |
| 44–dG2–AF/AAF | 5'–CCCTAGCTAGAGCTACGTAGCTATTACCGGCXCCACATGTCAGC |
X denotes dG–AAF, dG–AF or dG residues; the NarI sequence is underlined
XPC RAD23B expression and purification and cell extract preparation
Polyhistidine-tagged RAD23B was expressed in E. coli BL21(DE3)LysS using the expression vector pET-24d and purified on nickel beads (Qiagen) as described.(34) Polyhistidine and MBP-tagged XPC was expressed in Sf9 cells. The cells were lysed as described and S3 was combined with partially purified RAD23B.(14) The correctly folded heterodimer was further purified through nickel beads (Qiagen), gel filtration (Pharmacia) and heparin (Amersham) columns. HeLa whole-cell extracts were prepared as described.(35)
Generation of plasmids and in vitro NER assay
Single-stranded circular DNA was generated from p98 (with a NarI site engineered into pBlusscript II SK+) as described.(27, 36) The oligonucleotides 24-AAF or 24-AF (100 pmol) were 5'-phosphorylated by incubation with 20 units of T4 PNK and 2 mM of ATP at 37 °C for 2h. After annealing with 30pmol of single-stranded p98, the mixture was incubated with dNTPs (800 μM), T4 DNA polymerase (90 units) and T4 DNA ligase (100 units) to generate the covalently closed circular DNA containing a single dG-AAF or dG-AF adduct. The closed circular DNA was purified by cesium chloride/ethidium bromide density gradient centrifugation, followed by consecutive butanol extractions to remove the ethidium bromide and finally concentrated on a Centricon YM-30 (Millipore). The collected covalently closed circular DNA was further purified by sucrose gradient centrifugation to remove traces of ethidium bromide and nicked DNA. Plasmids containing dG-AAF and dG-AF were aliquoted and stored at −80 °C.
The in vitro NER assay was performed as described.(27,36) HeLa cell extract (2μl of 21mg/ml), 2 μl of 5× repair buffer (200 mM Hepes-KOH, 25 mM MgCl2, 110 mM phosphocreatine (di-Tris salt, Sigma), 10 mM ATP, 2.5 mM DTT and 1.8 mg/ml BSA, adjusted to pH 7.8), 0.2 μl of creatine phosphokinase (2.5 mg/ml, rabbit muscle CPK, Sigma), and 0.5 μL 0.4 M NaCl (final NaCl concentration was 70 mM) in a total volume of 10 μl were pre-incubated at 30°C for 10 min. Covalently-closed circular DNA plasmid (1μl of 50 ng/μl) containing either the AAF or the AF adduct was added and the mixture incubated at 30 °C for 45 min. After placing the samples on ice, 0.5 μl of 1 μM of a 3'-phophorylated oligonucleotide: d(GGGGCATGTGGCGCCGGTAATAGCTACGTAGCTC) was added and the mixture heated at 95°C for 5 min. The samples were allowed to cool to room temperature for 15 min. Sequenase/[α-32P]-dCTP mix (1 μl of 0.25 units of Sequenase and 2.5 μCi of [α-32P]-dCTP per reaction) was added before incubating at 37 °C for 3 min, followed by addition of 1.2 μl of dNTP mix (100 μM of each dATP, dTTP, dGTP; 50 μM dCTP) and incubation for another 12 min. The reactions were stopped by adding 12 μl of loading dye (80% formamide/10 mM EDTA) and heating at 95 °C for 5 min. The samples were run on a 14% sequencing gel (0.5× TBE) at 45 W for 2.5 hrs. The reactions products were visualized using a PhosphorImager (Typhoon 9400, Amersham Biosciences). Each NER was performed at least three times with each substrate for quantification. Relative repair efficiency of each substrate was normalized to a non-specific band and quantified by the Image Quant TL program from Amersham Biosciences.
Electrophoretic Mobility Shift Assay (EMSA)
AAF- or AF-modified 44mer oligonucleotides (4 nM) were annealed to a complementary strand labeled 5' with Cy5 (1.3 nM) in 10 mM Tris-HCl (pH 8), 6.6 mM NaCl and 0.66 mM MgCl. The annealed oligo was incubated with XPC-RAD23B (0–150 nM) in a 15 μL mixture containing 25 mM Tris-HCl (pH 7.5), 40 mM NaCl, 0.1 mg/mL BSA, 10% glycerol, 4 nM non-modified 44mer duplex (competitor), at 25 °C for 30 minutes. The reaction mixture was loaded onto a native 5% polyacrylamide gel pre-equilibrated with 0.5 × TBE buffer and run at 4 °C for 50 min at 20 mA. Gels were scanned using a Typhoon 9400 imager. The band intensities of free 44mer oligonucleotides and XPC-RAD23B-bound were determined using the Image Quant TL program from Amersham Biosciences. EMSA with each substrate was performed three times to give average binding data. The sigmoid curve was drawn by the Sigmaplot software V10.0 program using the Hill equation, f=y+a*x^b(c^b+x^b), (where Y is the % of protein-bound substrate, x is the concentration (nM) of XPC-RAD23B, and c is the Kd value) and Kd values were obtained from the half-maximal binding point.
Measuring melting temperatures and thermodynamic parameters
UV-absorption thermal denaturation experiments were carried out as described using the `thermal' program of a CARY 100 Bio UV-VIS spectrophotometer equipped with a multicell block temperature regulation unit and a fluid conduction thermal regulation enhancement attachment (Varian, Inc.).(37) Duplexes (at concentrations between 2 and 11 μM) were dissolved in 1 mL of 25 mM sodium phosphate buffer solution, pH 6.8, containing 100 mM NaCl and 0.5 mM EDTA. After temperature equilibration, samples were heated from 15°C or 80°C or cooled from 80°C to 15°C at a rate of 0.2°C/min. Eight different sample concentrations and three independent melting profiles at each concentration composed a set of 24 independent determinations performed on each duplex. The enthalpy (ΔH°) and entropy (ΔS°) were calculated using a published protocol:(38,39) van't Hoff plots of Tm−1 versus ln(Ct) to fit to Tm−1=R/ΔH° lnCt/4 + ΔS°/ΔH°, in which Tm is a melting transition point in K, Ct is a total duplex concentration, and R is the universal gas constant (1.987cal/Kmol); ΔG°= ΔH°-TΔS°. Reported enthalpy and entropy values are the average of individual determinations, and error estimates represent the standard deviation of the data. The Gibbs free energy (ΔG°) of duplex formation was then calculated from these values.
RESULTS
The NER efficiency of dG-AAF and dG-AF adducts varies greatly within the NarI site
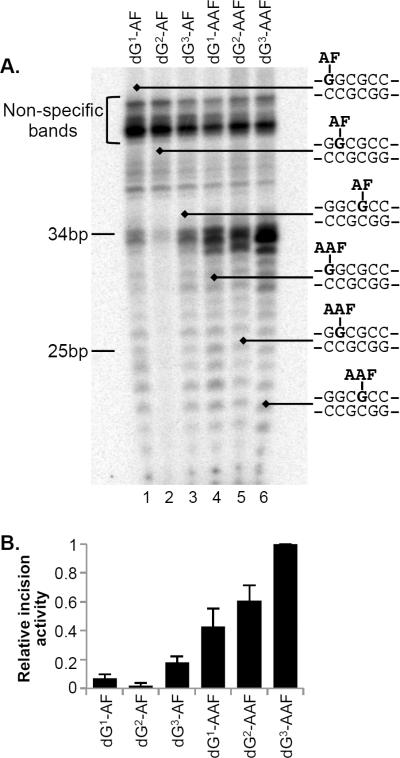
We incorporated dG-AAF and dG-AF residues at each of the three guanine residues in the NarI sequence (Table 1) in 24mer oligonucleotides by solid-phase synthesis using our established protocol.(27,40) These 24-mers were annealed to a single-stranded plasmid and subjected to a primer extension, ligation and purification protocol to generate plasmids containing site-specific dG-AF or dG-AAF adducts.(27,36) The plasmids were incubated with NER-proficient HeLa whole cell extracts, leading to the formation of excision products of a characteristic length of around 26–31 nucleotides. These reaction products were detected by annealing to a complementary oligonucleotide with a 4G overhang and a fill-in reaction with polymerase and α-[32P]-dCTP. Under identical conditions in three independent experiments, we observed the different levels of products for the six substrates (Figure 2A). The relative efficiencies of incision were quantified and we assigned the value for the best substrate dG3-AAF as 100%. The relative intensities of repair averaged from three independent experiments were 100% for dG3-AAF, 60% for dG2-AAF and 40% for dG1-AAF, while the relative intensities for the AF substrates was 20% (dG3-AF), 2% (dG2-AF) and 7% (dG1-AF) (Figure 2B). Therefore, the efficiency of NER of dG-AAF and dG-AF varies up to 50-fold within context of the NarI sequence.
Figure 2. Relative NER efficiencies of dG AF and dG AAF adducts in the NarI sequence.
A. Plasmids containing site-specific dG-AF (lane 1–3) or dG-AAF (lane 4–6) adducts were incubated with HeLa whole cell extracts. The excision products were detected by annealing to complementary oligonucleotides with a 4G overhang, which served as a template for end-labeling with [α-32P] dCTP with sequenase. The reaction products were resolved on a 14% denaturing polyacrylamide gel. The position of 25nt and 34nt markers is indicated on the left of the gel. B. Quantification of the relative NER efficiencies from at least three independent experiments.
Yeo et al.,
Binding affinities of XPC-RAD23B to dG-AAF and dG-AF substrates correlate with NER efficiency
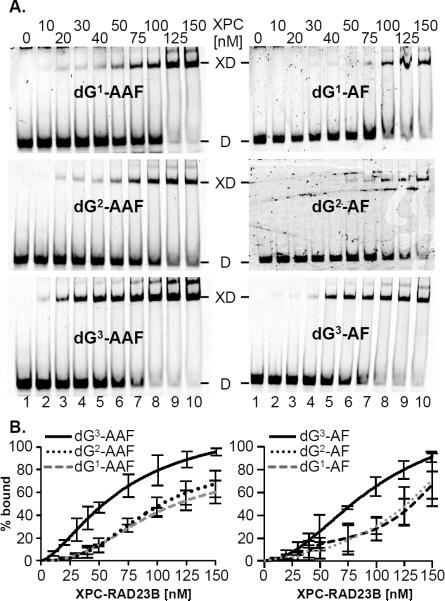
As XPC-RAD23B is the initial damage recognition factor in NER,(14) we wished to determine to what extent the differences in NER efficiency of the various dG-AF and dG-AAF adducts correlated with XPC-RAD23B binding affinities. Therefore we conducted electrophoretic mobility shift assays (EMSA) to obtain the binding constants between XPC-RAD23B and these substrates. 44mer oligonucleotides containing a central dG-AF or dG-AAF lesion at the three G positions in the NarI sequence (Table 1) were annealed to a fluorescently labeled complementary strand, incubated with increasing amounts of XPC-RAD23B and three equivalents of non-damaged competitor DNA and equilibrated. Bound and unbound fractions were separated on 5% native polyacrylamide gels (Figure 3A). Comparison of the binding patterns revealed that the dG3-AAF-containing oligonucleotides bound XPC-RAD23B more tightly than those containing dG2-AAF or dG1-AAF, while all of the dG-AF oligonucleotides had lower affinity (Figure 3A). To calculate the dissociation constants, the binding data were fit to a sigmoidal curve (see Experimental procedures). For the dG-AAF oligonucleotides, the dissociation constants qualitatively correlated with the NER efficiencies with dG3-AAF having a significantly lower Kd value than dG1-AAF and dG2-AAF (Table 3).
Figure 3. Binding affinities of XPC RAD23B bound to dG AF and dG AAF substrates correlates with NER efficiency.
A. 44mer oligonucleotides containing a dG-AF or dG-AAF were annealed to complementary oligonucleotides containing a fluorescent label (5'-Cy5). The substrates were incubated with the indicated concentrations of XPC-RAD23B and the mixtures analyzed on to a 5% native polyacrylamide gel. The positions of the unbound DNA (D) and XPC-RAD23B-bound DNA (XD) are indicated. B. The binding data of three independent experiments were quantified by fitting to a sigmoidal curve and Kd values calculated using the Hill equation.
Yeo et al.,
Table 3.
Correlation of NER efficiency, XPC-RAD23B binding affinity and Gibbs free energy
| Substrate | relative NER efficiency | XPC-RAD23B Kd (nM) | ΔG (kcal/mol) |
|---|---|---|---|
| dG1-AF | 7% | >120 | −13.9 |
| dG2-AF | 2% | >120 | −13.8 |
| dG3-AF | 20% | 79 ± 5 | −13.2 |
| dG1-AAF | 40% | 117 ± 4 | −12.2 |
| dG2-AAF | 60% | 103 ± 6 | −12.2 |
| dG3-AAF | 100% | 54 ± 7 | −9.8 |
Of the AF series, dG3-AF, had the lowest Kd value and we were unable to accurately determine the Kd value for dG1-AF or dG2-AF as binding failed to reach saturation under our experimental conditions. We estimate the Kd for these two 44-mers to clearly higher than 120 M, based on the concentration at which 50% of maximal binding is reached. The Kd value for dG3-AF (79 nM) was lower than that of dG2-AAF and dG3-AAF sequences, despite it being less efficiently processed by NER. However, the binding affinity of XPC-RAD23B to the various adducts correlated well with the NER efficiency within the dG-AAF and dG-AF series.
Better NER substrates have lower melting temperatures and thermodynamic stabilities
As our studies revealed a strong correlation between NER efficiency and XPC-RAD23B binding affinity, we investigated whether there was an additional correlation with the thermodynamic properties of the corresponding substrates. We therefore synthesized duplexes containing the 12 central residues of the NER/binding substrates (Table 1) and measured their melting temperatures. All of the dG-AAF and dG-AF duplexes showed lower melting temperatures compared to a control oligonucleotide containing a dG at the position of the lesion. Consistent with earlier studies, the presence of either AF or AAF decreased thermal and thermodynamic stability of DNA duplex (Table 2).(24) The melting temperature of the control oligonucleotide was 10 to 18 °C higher than that of dG-AF or dG-AAF-containing DNA duplexes. Van't Hoff plots were derived by measuring the Tm over a concentration range of 2–11μM and plotting ln(Ct/4) versus 1/Tm to calculate thermodynamic parameters (Figure 4).(39) dG3-AAF had the lowest melting temperature (48°C) and the highest Gibbs free energy (ΔG, −9.8 kcal/mol), indicating that the presence of the AAF adduct at this position affected duplex stability most severely, consistent with it being the best NER substrate. The melting temperature and in particular the Gibbs free energies correlated well with the NER efficiencies, and the order of increasing stability (ΔG) was as follows: dG3-AAF > dG2-AAF ≈ dG1-AAF > dG3-AF ≈ dG1-AF > dG2-AF > dG (Table 2).
Table 2.
Tm values and thermodynamic properties of dG-AF and dG-AAF containing duplexes.
| Substrate | Tmb (°C) | ΔH (kcal/mol) | ΔS (cal/mol•k) | ΔG (kcal/mol)c |
|---|---|---|---|---|
| dG (control) | 66 ± 1 | −102 ± 1 | −278 ± 1 | −15.8 ± 0.4 |
| dG1-AF | 55 ± 0 | −110 ± 0 | −310 ± 0 | −13.9 ± 0.1 |
| dG2-AF | 56 ± 0 | −105 ± 0 | −294 ± 1 | −13.8 ± 0.1 |
| dG3-AF | 51 ± 1 | −123 ± 1 | −354 ± 1 | −13.2 ± 0.4 |
| dG1-AAF | 54 ± 1 | −91 ± 1 | −254 ± 1 | −12.2 ± 0.3 |
| dG2-AAF | 50 ± 1 | −113 ± 1 | −325 ± 1 | −12.2 ± 0.3 |
| dGb-AAF | 48 ± 1 | −65 ± 2 | −178 ± 1 | −9.8 ± 0.2 |
Thermodynamic parameters for oligonucleotides containing modifications were acquired from absorbance versus temperature melting curves.
Melting temperatures were measured at an oligonucleotide concentration of 5.8 μM.
Gibbs free energy calculated at 37°C. Reported errors are standard deviations rounded to the next integral.
Figure 4.
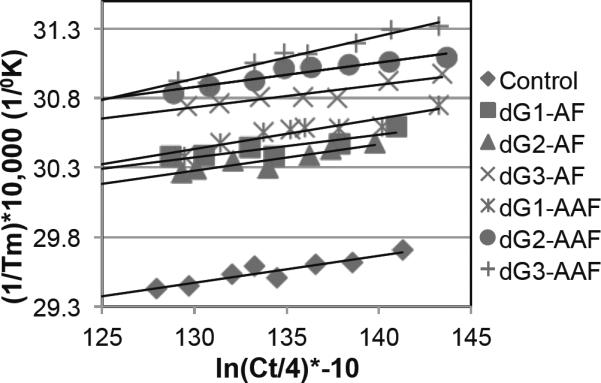
Van't Hoff plots of Tm−1 versus ln(Ct/4) derived from the melting curves for the control, dG-AF and dG-AAF 12mer duplexes (at concentrations between 2 and 11μM). Ct represents the total oligonucleotide concentration
Yeo et al.,
DISCUSSION
Human NER employs a remarkable strategy for the recognition of structurally diverse lesions that fulfill two criteria:(9) 1) they induce a local thermodynamic destabilization to permit binding by the damage recognition factor XPC-RAD23B and 2) they are bulky and have the ability to stall the XPD helicase in the damage verification step. Here we tested one prediction of this model, that for structurally similar lesions the degree of thermodynamic helix destabilization determines the binding affinity for XPC-RAD23B and overall NER rate. In our studies using a series of dG-AF and dG-AAF lesions in the context of the NarI sequence, we observe this predicted correlation, although our data suggest that the steps following XPC-RAD23B can also influence to the overall reaction rate (Table 3).
Structural features of dG-AF and dG-AAF adducts determine repair outcomes
Duplexes containing dG-AF and dG-AAF have been subject to extensive structural studies and the structure of these adducts is strongly dependent on the sequence context.(22,24) In general, dG-AF is present in higher proportion in the base-paired conformation with the fluorene in the major groove. dG-AF lesions are therefore less distorting than dG-AAF lesions and are less well repaired in all sequence contexts both in bacterial and human systems.(25,27–29,31) The differences within the dG-AF and dG-AAF series are less pronounced, but we note that our data of repair efficiencies are in good agreement with the degree of thermodynamic destabilization (this study) and the fraction of adduct present in the S/W versus B conformation.(25) It is however likely that more subtle changes within these conformations could also contribute to repair efficiencies, as been shown for example for the repair of various benz[a]pyrene adducts.(6,41,42) Such differences may also result from more distant neighbor contexts or DNA conformation (eg. supercoiled versus linear). They might also explain modest differences in relative repair rates of dG-AAF adducts between our studies and those of Mu et al., who also investigated the NER efficiencies of the repair of dG-AF and dG-AAF adducts in the NarI sequences.(43) While the overall results and conclusions of these studies are in good agreement, a minor difference in the relative NER efficiencies was observed for the dG2-AAF and dG3-AAF substrates.(30.43) We believe that this may be due to the use of linear versus circular substrates in the two studies. Additionally, Mu et al used molecular dynamics simulations to explore the structures of dG-AF and dG-AAF in these sequences.(43) They found sequence-dependent differences in the base-stacking pattern of the S-conformers, which may influence repair efficiency. Thus it is likely that the sequence-specific NER rates are not only determined by the fraction of adducts present in the B- versus S-/W-conformers, but also by subtle structural differences within those conformers.
Moderate differences in XPC binding affinity determine hierarchy of repair
XPC-RAD23B is widely regarded as the key damage recognition factor in NER, and structural and biochemical studies of the human and yeast proteins have shown that it binds specifically to the non-damaged strand opposite the lesion.(11–13) Several studies have been concerned with the binding affinity and specificity of XPC-RAD23B binding to DNA lesions and two important features are emerging:(14, 20, 44, 45) First, the difference in binding affinity of XPC-RAD23B for the best substrates (such as a 6–4 photoproduct or dG-AAF) and non-specific DNA is only about 2–10 fold. Although absolute values of the Kd are the are in the range of 1–100nM, depending on the protein preparations and technique used for Kd determination, there appears to be only a moderate difference in binding affinity, raising the question how this might be sufficient for specifically recognizing lesions in the context of genomic DNA. Second, XPC-RAD23B binding to NER substrates is very resistant to inhibition by competitors, and the extent of resistance to inhibition correlates well with the suitability as an NER substrate.(14,44) These observations suggest that XPC-RAD23B has a very slow koff rate on good NER substrates, implying a long residence time of the protein on the damage, allowing for the recruitment of TFIIH/XPD and other downstream factors to continue progression through the NER reaction. In this context, a relatively small increase in binding affinity may well be very significant in increasing damage recognition and the rate of NER. XPC as well as the bacterial NER protein UvrB insert a beta-hairpin into the protein at the lesion site and it is possible that this process alters the structure of the XPC-DNA complex in a way to facilitate the recruitment of the subsequent factors.(3) One apparent discrepancy in our results is that the dG3-AF is bound with lower affinity that the dG2-AAF and dG1-AAF adducts, although the two dG-AAF adducts are processed more efficiently by NER. It is possible that this may due to structural features of the XPC-lesion complex that might differ for dG-AF and dG-AAF. Such structural alterations may influence recruitment and binding of TFIIH and also possibly translocation of the XPD helicase.
Conclusion
In this study, we have shown that there is a good correlation of the extent of thermodynamic destabilization, binding to the damage recognition protein XPC-RAD23B and overall NER rate for DNA lesions, supporting and extending current models of damage recognition in NER. It will be interesting to determine, in quantitative terms, how subtle differences in the structures of DNA lesion influence damage recognition and how steps downstream of XPC-RAD23B in NER, such as damage verification by XPD/TFIIH and preincision complex assembly (involving XPA, RPA, XPG and ERCC1-XPF) amplify damage discrimination in this multi-step repair pathway.
ACKNOWLEDGEMENTS
We are grateful to Robert Rieger and the Proteomic Center of Stony Brook University (supported by grant NIH/NCRR1 S10 RR023680-1) and Ilaria Zanardi, Bela Ruzsicska at the Institute of Chemical Biology and Drug Discovery of Stony Brook University for MS spectral analyses. We thank Nicolas Geacintov and Suse Broyde (New York University) for communication of data prior to publication and Tanya Zalyznyak and Carlos de los Santos (Stony Brook University) for help with the Tm measurements and helpful discussions.
FUNDING SOURCES This work was supported by National Institutes of Health grants ES004068 and GM080454.
Abbreviations
- AAF
acetylaminofluorene
- AF
aminofluorene
- Ct
total DNA concentration
- EMSA
electrophoretic mobility shift assay
- NER
nucleotide excision repair
REFERENCES
- (1).Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edition ed. ASM Press; Washington DC: 2005. [Google Scholar]
- (2).Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- (3).Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- (5).Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. U S A. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids. 2010;2010:174252. doi: 10.4061/2010/174252. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- (8).Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst) 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- (9).Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- (10).Schärer OD. Achieving broad substrate specificity in damage recognition by binding accessible nondamaged DNA. Mol. Cell. 2007;28:184–186. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- (11).Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- (12).Maillard O, Solyom S, Naegeli H. An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS Biol. 2007;5:e79. doi: 10.1371/journal.pbio.0050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- (14).Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mathieu N, Kaczmarek N, Naegeli H. Strand- and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc. Natl. Acad. Sci. USA. 2010;107:17545–17550. doi: 10.1073/pnas.1004339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- (17).Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pugh RA, Wu CG, Spies M. Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J. 2012;31:503–514. doi: 10.1038/emboj.2011.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lukin M, Zaliznyak T, Johnson F, de los Santos C. Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts. Nucleic Acids Res. 2012;40:2759–2770. doi: 10.1093/nar/gkr1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Schärer OD, Grollman AP. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactamy-DNA adducts. Nucleic Acids Res. 2012;40:2494–2505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair (Amst) 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- (22).Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- (23).Belguise-Valladier P, Fuchs RP. N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J. Mol. Biol. 1995;249:903–913. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- (24).Jain V, Hilton B, Patnaik S, Zou Y, Chiarelli MP, Cho BP. Conformational and thermodynamic properties modulate the nucleotide excision repair of 2-aminofluorene and 2-acetylaminofluorene dG adducts in the NarI sequence. Nucleic Acids Res. 2012;40:3939–3951. doi: 10.1093/nar/gkr1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jain N, Meneni S, Jain V, Cho BP. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 2009;37:1628–1637. doi: 10.1093/nar/gkn1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang L, Broyde S. A new anti conformation for N-(deoxyguanosin-8-yl)-2- acetylaminofluorene (AAF-dG) allows Watson-Crick pairing in the Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) Nucleic Acids Res. 2006;34:785–795. doi: 10.1093/nar/gkj479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gillet LC, Alzeer J, Schärer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified `ultra-mild' DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mekhovich O, Tang M, Romano LJ. Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein-DNA complex formation. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- (29).Meneni SR, Shell SM, Gao L, Jurecka P, Lee W, Sponer J, Zou Y, Chiarelli MP, Cho BP. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. 2007;46:11263–11278. doi: 10.1021/bi700858s. [DOI] [PubMed] [Google Scholar]
- (30).Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Seeberg E, Fuchs RP. Acetylaminofluorene bound to different guanines of the sequence -GGCGCC- is excised with different efficiencies by the UvrABC excision nuclease in a pattern not correlated to the potency of mutation induction. Proc. Natl. Acad. Sci. USA. 1990;87:191–194. doi: 10.1073/pnas.87.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shibutani S, Gentles R, Johnson F, Grollman AP. Isolation and characterization of oligodeoxynucleotides containing dG-N2-AAF and oxidation products of dG-C8-AF. Carcinogenesis. 1991;12:813–818. doi: 10.1093/carcin/12.5.813. [DOI] [PubMed] [Google Scholar]
- (34).Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek PJ, Bootsma D, Hoeijmakers JH, Hanaoka F. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 1996;16:4852–4861. doi: 10.1128/mcb.16.9.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Biggerstaff M, Wood RD. Assay for nucleotide excision repair protein activity using fractionated cell extracts and UV-damaged plasmid DNA. Methods Mol. Biol. 1999;113:357–372. doi: 10.1385/1-59259-675-4:357. [DOI] [PubMed] [Google Scholar]
- (36).Shivji MK, Moggs JG, Kuraoka I, Wood RD. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 1999;113:373–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- (37).Bohon J, de los Santos CR. Effect of 6-thioguanine on the stability of duplex DNA. Nucleic Acids Res. 2005;33:2880–2886. doi: 10.1093/nar/gki572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Allawi HT, SantaLucia J., Jr. Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- (39).Zaliznyak T, Bonala R, Johnson F, de Los Santos C. Structure and stability of duplex DNA containing the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG(N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem. Res. Toxicol. 2006;19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- (40).Gillet LC, Schärer OD. Preparation of C8-amine and acetylamine adducts of 2'-deoxyguanosine suitably protected for DNA synthesis. Org. Lett. 2002;4:4205–4208. doi: 10.1021/ol026474f. [DOI] [PubMed] [Google Scholar]
- (41).Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J. Mol. Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. Embo J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S. Nucleotide excision repair of 2-acetylaminofluoreneand 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks788. published on August 16, 2012 as doi:10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- (45).Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]