Abstract
It is generally accepted that the presentation of multiple ligands on a nanoparticle surface can improve cell targeting; however, little work has been done to determine whether an optimal ligand density exists. We have recently developed a site-specific bioconjugation strategy that allows for distinct control of ligand density on a nanoparticle through the combined utilization of expressed protein ligation (EPL) and copper-free click chemistry. This EPL-Click conjugation strategy was applied to create superparamagnetic iron oxide (SPIO) nanoparticles labeled with HER2/neu targeting affibodies at differing ligand densities. It was discovered that an intermediate ligand density provided statistically significant improvements in cell binding compared with higher and lower ligand densities. This intermediate optimal ligand density was conserved across nanoparticles with differing hydrodynamic diameters, different HER2/neu targeting ligands and also to cells with lower receptor densities. Additionally, an intermediate optimal ligand density was also evident when nanoparticles were labeled with folic acid.
Keywords: Ligand Density, SPIO, Targeting, Click Chemistry, nanoparticle
Introduction
Continuing advancements in nanoparticle technologies has led to their use in a myriad of biological applications extending from cell tracking to advances in drug delivery. Further, with the limitless ability to modulate nanoparticle syntheses and modifications to fit specific areas of need, it is envisioned that their utility will only continue to grow. In applications where nanoparticles are used to improve the differential diagnosis or therapeutic treatment of disease, it is becoming increasingly popular to modify nanoparticle surface characteristics in an effort to increase their accumulation at sites of interest and decrease their non-specific uptake in other areas of the body. One of the most common modifications involves decorating the nanoparticle surface with multiple affinity ligands in order to actively ‘target’ nanoparticles to pathology-associated biomarkers of interest. The introduction of multiple targeting ligands onto nanoparticle surfaces (multivalency) has been shown to increase binding avidity, increase the rate of internalization and ultimately improve therapeutic efficacy and/or image contrast.1–5
One particular class of nanoparticles that has become increasingly dependent on targeted delivery is superparamagnetic iron oxide (SPIO) nanoparticles (NPs). SPIO NPs are an attractive magnetic resonance (MR) contrast agent, providing T2*-weighted contrast enhancement in MR imaging applications. Due to their strong contrast-enhancing capabilities, biocompatibility and ability to provide functional data concomitant with anatomic information, SPIO have recently been evaluated as molecular imaging agents, whereby they are used to report the expression level of target cell-surface receptors to improve the specificity of disease detection. However, while SPIO NPs have seen extensive biological applications, their full transition to the clinic as molecular imaging agents has been slow developing due to the high concentrations of accumulated SPIO NPs needed to generate detectable MR contrast in an area of interest.
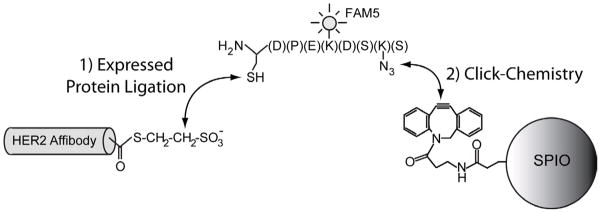
To date, affinity ligands have been used to deliver SPIO NPs to a range of different pathologies, including tumor cells6,7, tumor vasculature8,9, atherosclerosis10–12 and several other biological phenomena13–18; however, in all cases, little work has been done to determine optimal ligand densities for receptor targeting and cell binding.19–21 Given the sensitivity drawbacks associated with contrast-enhanced MRI, optimizing the cell binding capabilities of targeted SPIO is critical and could vastly enhance their applicability in several areas of clinical need. One reason for the lack of studies pertaining to the optimization of ligand density stems from the inability to tightly control the efficiency of nanoparticle labeling and the inability to control the orientation of the targeting ligands on the nanoparticle surface, using conventional bioconjugation techniques.22 Recently, we have reported on the development of a novel bioconjugation strategy that combines expressed protein ligation (EPL) and click chemistry to produce a highly efficient and site-specific bioconjugation reaction that ensures that all targeting ligands have the same orientation.5 Using EPL-Click bioconjugations, we have been able to gain more precise control over nanoparticle ligand density and study the implications that ligand density has on cell receptor targeting. In our approach, bacterially expressed HER2/neu-targeted Affibodies (HER2-Affibodies) were ligated to an azido-fluorescent peptide (AzFP) via EPL (Figure 1). The product of this reaction, which we refer to as HER2-AzFP, was then clicked to aza-dibenzocyclooctyne (ADIBO)-labeled SPIO NPs at different concentrations to create nanoparticles with differing HER2/neu targeting ligand densities. ADIBO is a dibenzocyclooctyne derivative capable of copper-free click reactions with azides.23, 24 The resulting HER2-SPIO conjugates were then assessed on the basis of their cell targeting capabilities. To determine the generalizibility of our findings, the effect of ligand density on cell targeting was evaluated under diverse experimental conditions, including with SPIO NPs that possess differing hydrodynamic diameters, cells with diverse receptor densities, and SPIO NPs with different targeting ligands. In all, three different targeting ligands were assessed, HER2-Affibodies,25 a homology derived HER2/neu targeting peptide (AHNP)26 and folic acid, which was used to target the folate receptor.
Figure 1.
Schematic of EPL-Click conjugation strategy. Expressed protein ligation between a HER2-Affibody containing a C-terminal thioester and an azido-fluorescent peptide (AzFP) containing an N-terminal cysteine results in the chemoselective attachment of a “clickable” azido-group onto the affibody (HER2-AzFP). Subsequent Copper-free click chemistry between ADIBO modified SPIO-NPs and HER2-AzFP results in the site-specific attachment of the HER2-Affibody onto the SPIO NPs.
Methods
Results
Copper-Free Click Conjugation
An effective analysis of ligand density requires a bioconjugation method that provides a high degree of control and consistency when functionalizing nanoparticles with targeting ligands. Previously, we reported that the combination of expressed protein ligation and copper-catalyzed click chemistry can provide these necessary requirements. Herein we have evolved the conjugation scheme to utilize aza-dibenzocyclooctyne (ADIBO), a new functional group that allowed the click chemistry reactions to proceed without the need for copper catalysts. Copper is known to cause transmetalation and even small amounts of the ions can be highly toxic,27,28 thus the evolution of our bioconjugation to utilize copper-free click chemistries may have a higher degree of clinical applicability as compared to copper-dependent techniques. To demonstrate the consistency and controllability of the copper-free conjugation strategy, a fixed concentration of ADIBO-labeled SPIO NPs was labeled with increasing concentrations of HER2-AzFP. Notably, since there is a 1:1 stoichiometric ratio between the HER2-affibody and the fluorescent dye on the AzFP, the extent of labeling can generally be assessed by measuring the absorbance of the attached dye. The relationship between the fluorescence intensity of fluorescein (FAM5) on HER2-SPIO and the number of HER2 affibodies per SPIO is shown in Figure S1. The fluorescence intensity of HER2-SPIO increased linearly with ligand density, with perhaps some self-quenching evident at the highest labeling ratios examined.
HER2-SPIO Characterization
Cell binding of actively targeted nanoparticles is dependent on many factors, several of which can be attributed directly to the nanoparticle itself. Specifically, shape, size, and surface charge have been shown to have significant effects on the non-specific binding of nanoparticles.29,30 In order to accurately compare SPIO conjugates, it was first necessary to establish which, if any, nanoparticle physico-chemical parameters were altered by increasing ligand surface density. Following surface functionalization with the HER2-AzFP, measurements were taken to assess the hydrodynamic diameter, surface zeta potential and magnetic relaxivity of each conjugate (Table 1). Additionally, unlabeled ADIBO-SPIO NPs were also analyzed. As indicated in Table 1, the addition of HER2-AzFP did not significantly alter any of the basic nanoparticle characteristics. All nanoparticles had a hydrodynamic diameter of roughly ~26 nm, and also a negative surface zeta potential (~ -15 mV). A representative size distribution for a 26 nm NP can be found in Figure S2. Nanoparticle surface zeta potentials were kept at a negative charge as we have found that positive surface zeta potentials for nanoparticles can induce non-specific cellular associations and uptake (See Figure S3).29, 31 Surface labeling with HER2-AzFP also had little effect on the magnetic properties of the nanoparticles, as each SPIO conjugate and the unlabeled SPIO had R2 relaxivities of ~133 mM−1s−1 and R1 relaxivities of ~13 mM−1s−1.
Table 1.
Comparison of HER2-SPIO Physico-Chemical Parameters
| Ligand/SPIO | Hydrodynamic Diameter (nm) | R2 (mM−1*s−1) | R1 (mM−1*s−1) | R2/R1 | Surface Zeta (mV) |
|---|---|---|---|---|---|
|
| |||||
| 0 | 26 | 133.4 | 13.0 | 10.28 | −17.9 ± 1.0 |
| 11.5 | 28 | 131.6 | 12.8 | 10.27 | −15.0 ± 1.0 |
| 23 | 24 | 136.1 | 14.8 | 9.222 | −14.8 ± 0.8 |
| 30.2 | 28 | 138.3 | 13.2 | 10.45 | −15.0 ± 1.4 |
| 35.8 | 25 | 131.5 | 13.0 | 10.08 | −14.4 ± 1.1 |
Effect of Ligand Density on Receptor Targeting
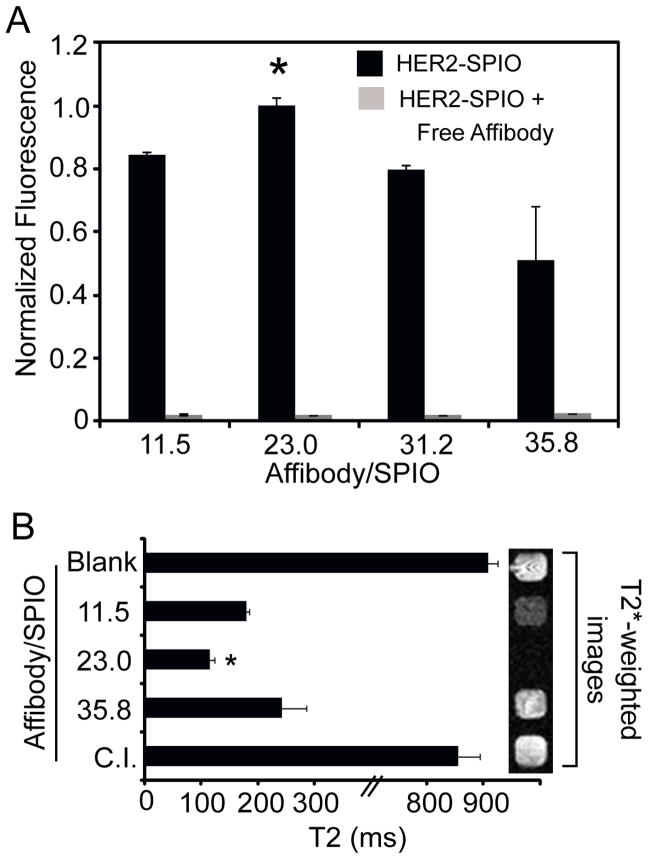
The cell targeting capabilities of HER2-SPIO conjugates with differing ligand densities were assessed by conducting cell-labeling assays with HER2/neu-positive (T6-17) cells. Flow cytometric analysis of targeted cells revealed strong cell labeling for each HER2-SPIO conjugate, with each conjugate displaying a high degree of specificity for the HER2/neu receptor as determined by competitive inhibition analysis (Figure 2A). All flow cytometry measurements were adjusted to account for differences in the fluorescence per NP that existed between SPIO NPs with different ligand densities (Figure S1). Unexpectedly, the highest degree of cell labeling was noted for the SPIO-conjugates that had an intermediate number of ligands per nanoparticle. Specifically the SPIO conjugates with 23 ligands/SPIO exhibited a statistically significant higher degree of cell labeling (p <0.05) compared to conjugates with fewer (11.5) and higher (31.2 and 35.8) numbers of ligands/SPIO. Increases in ligand density were expected to result in SPIO NPs with higher avidities; however, our empirical evidence indicates that there exists an optimal intermediate ligand density for HER2/neu receptor targeting. Representative raw flow cytometry data (unadjusted and unnormalized) is provided as supporting information (Figure S4A). Interestingly, even when the flow cytometry data was not adjusted for differences in NP fluorescence, cells labeled with HER2-SPIO with an intermediate ligand density exhibited a higher mean fluorescent intensity (MFI) than cells labeled with HER2-SPIO with a higher ligand density. This was despite the fact that the HER2-SPIO with an intermediate ligand density had less fluorescence per NP than HER2-SPIO with a higher ligand density (Figure S1). Notably, flow cytometry measurements were highly reproducible, with minimal variability between repetitive experiments (Figure S4B).
Figure 2.
Flow cytometric and MR analysis of T6-17 cells incubated with HER2-SPIO conjugates. T6-17 cells were incubated with 26nm HER2-SPIO conjugates, with ligand densities ranging from 11.5 to 35.8 affibodies per SPIO, for 30 minutes at 37°C. Analogous competitive inhibition (C.I.) studies were performed in the presence of excess free HER2-affibody. (A) Cell samples were then evaluated by flow cytometry. Each measurement represents the normalized mean fluorescence intensity (MFI). (B) SPIO-labeled cells and unlabeled cells (Blank) were also evaluated by acquiring T2 relaxation times and T2*-weighted MR images of each sample. Statistically significant (p<0.05) improvements in cell binding were found to occur for targeted SPIO NPs with an intermediate ligand density (23.0 Affibody/SPIO).
Implications of ligand density on MR Imaging
To underscore the importance of optimizing ligand density for cell targeting studies and to eliminate concerns that fluorescent artifacts (e.g. self-quenching, inaccurate adjustments when accounting for differences in fluorescence per NP) were responsible for the findings reported in Figure 2A, T6-17 cell cells were targeted with 26 nm HER2-SPIO conjugates (11.5, 23.0 35.8 Affibody/SPIO) and were analyzed and compared based on T2-relaxation times of the cell suspensions. T2-relaxation times were measured using a Bruker mq60 MR relaxometer operating at 1.41 T (60 MHz). All six conjugates showed a marked decrease in T2-relaxation times as compared to non-targeted cells and competitive inhibition assays (Figure 2B). Quantitative analysis within the targeted conjugate groups revealed that the 23.0 Affibody/SPIO conjugate produced a statistically significant decrease in T2-relaxation time when compared to the other conjugates (p < 0.05), which is consistent with flow cytometry measurements. However, it is important to note that while relative changes in T2-relaxation time, as a function of ligand density, were in close agreement with the flow cytometry studies, T2 relaxation times are inversely and non-linearly dependent on iron (i.e. SPIO) concentration and are influenced by other factors such as magnetic field strength and pulse sequence. Therefore, the total effect size will differ from fluorescence measurements, which generally increase linearly with the amount of contrast agent.
T2*-weighted MR images collected for each cell pellet using a 9.4T, 31 cm horizontal bore MR spectrometer highlighted the importance of optimizing cell targeting (Figure 2B, right panel). Despite the significant decreases in T2-relaxation times acquired for each sample using the benchtop relaxometer, only two of the targeted cell pellets elicited observable negative contrast upon MR-imaging, with the conjugates possessing 23.0 Affibody/SPIO producing the greatest loss in signal. Notably, the observed discrepancies between T2-relaxation times and negative contrast in the MR images can likely be attributed to the specific pulse sequences that were applied and the intrinsic sensitivities of the systems.
Effect of Nanoparticle Hydrodynamic Diameter on Receptor Targeting
To investigate whether the hydrodynamic diameter of targeted nanoparticles alters the sensitivity of cell labeling to variations in ligand density, ~50 nm SPIO NPs were synthesized and surface functionalized with the ADIBO click agent. A fixed concentration of ADIBO-SPIO was then ‘clicked’ with increasing concentrations HER2-AzFP ligands (Figure S5). In total, six distinct HER2-SPIO conjugates with differing ligand densities were used for cell labeling studies with HER2/neu positive T6-17 cells. A representative size distribution for a 50 nm NP can be found in Figure S6.
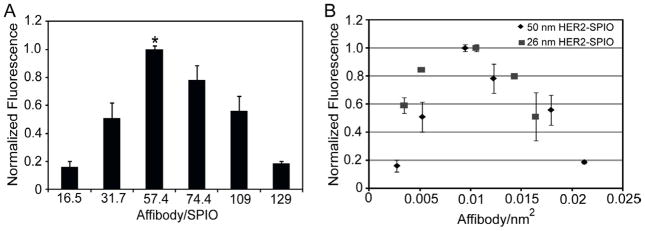
Flow cytometric analysis of targeted cells revealed that the 50 nm HER2-SPIO conjugates elicited a similar ligand density effect as the smaller 26 nm HER2-SPIO conjugates (Figure 3A). Specifically, optimal cell targeting was achieved with at an intermediate ligand density (57.4 ligands/SPIO), with a statistically significant increase in cell labeling compared to SPIO NPs with both higher and lower ligand densities (p<0.05). Representative raw flow cytometry data (unadjusted and unnormalized) is provided in Figure S7.
Figure 3.
Flow cytometric analysis of T6-17 cells incubated with 50nm HER2-SPIO conjugates. (A) T6-17 cells were incubated with 50nm HER2-SPIO conjugates, with ligand densities ranging from 16.5 to 129 affibodies per SPIO, for 30 minutes at 37°C. Cell samples were then evaluated by flow cytometry. Each measurement represents the normalized mean fluorescence intensity (MFI). Statistically significant (p<0.05) improvements in cell binding were found to occur for targeted SPIO NPs with an intermediate ligand density (57.4 Affibody/SPIO). (B) The effect of differing ligand densities (HER2-AzFP per surface area) on cell targeting was compared for 26nm and 50nm SPIO. Despite the differences in nanoparticle hydrodynamic diameter, optimized cell targeting occurs at near identical ligand surface densities.
Comparison of the 26 nm and 50 nm HER2-SPIO conjugates on the basis of the number of ligands per nanoparticle surface area revealed that for the two different sizes, there exists a nearly identical optimal ligand density for cell targeting (Figure 3B). Specifically, HER2-SPIO conjugates with both hydrodynamic diameters experienced the highest degree of cell labeling at ~0.01 ligands/nm2.
Effect of Receptor Density on Receptor Targeting
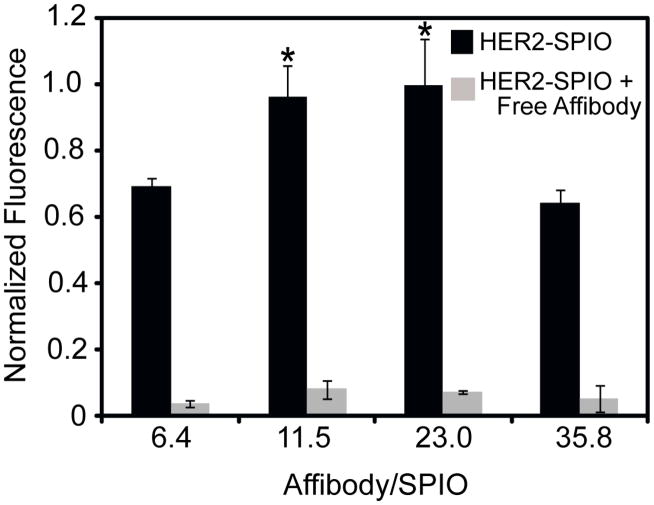
The density of cell surface receptors plays an integral role in nanoparticle targeting studies. An ideal target for diagnostic and therapeutic applications should exist at high concentrations with a high specificity for the pathology of interest, yet not all diseases of interest are associated with a highly over-expressed biomarker. To this end, we investigated the extent to which levels of HER2/neu receptor expression affect the cell targeting capabilities of HER2-SPIO conjugates with differing ligand densities. For these studies, the BT-20 cell line was selected which is known to express the HER2/neu receptor at low concentrations32 in contrast to the T6-17 cells described above which are genetically engineered to express high levels of HER2/neu receptor expression. Four 26 nm HER2-SPIO conjugates with differing ligand densities (6, 12, 23, 36 ligands/SPIO) were targeted to BT-20 cells, and the extent of cell labeling was determined by flow cytometric analysis (Figure 4). Despite the low receptor density, the effect of nanoparticle ligand density on cell labeling was conserved. Specifically, the highest cell labeling was noted for HER2-SPIO conjugates with intermediate ligand densities (11 and 23 ligands/SPIO), while cells targeted with conjugates possessing 6 and 36 ligands/SPIO displayed a statistically significant lower level of cell labeling (p < 0.05). Conservation of the ligand density effect at low receptor concentrations could have significant implications on active targeting studies. Notably, it is suspected that no statistical difference was observed between BT-20 cells labeled with NPs at ligand densities of 11 and 23 ligands/SPIO because of the much narrower dynamic range between maximally labeled and unlabeled cells, compared with T6-17 cells Figure S8).
Figure 4.
Flow cytometric analysis of BT-20 cells labeled with various HER2-SPIO conjugates. BT-20 cells, which express low levels of HER2/neu receptor, were incubated with 26 nm HER2-SPIO conjugates, with ligand densities ranging from 6.4 to 35.8 affibodies per SPIO, for 30 minutes at 37°C. Analogous competitive inhibition studies were performed in the presence of excess free HER2-affibody. Cell samples were then evaluated by flow cytometry. Each measurement represents the normalized mean fluorescence intensity (MFI). SPIO NPs with low and intermediate ligand densities (11.5 and 23.0 Affibodies/SPIO, respectively) exhibited statistically significant (p<0.05) improvements in cell binding as compared to lower and higher ligand densities.
Extension of Ligand Density Effect to Other Targeting Ligands
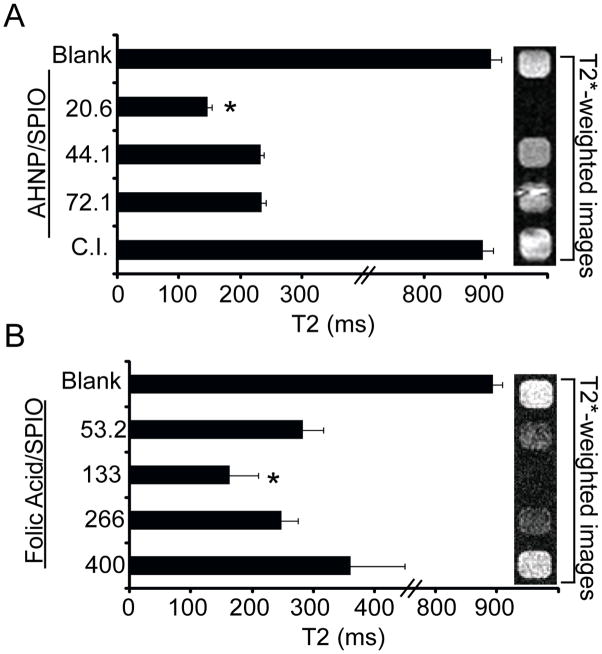
While the effect of ligand density was established for targeted HER-SPIO nanoparticles on the basis of nanoparticle size and receptor density, it remained unclear whether these effects extended to other targeting ligands. First, it was examined whether SPIO NPs (26 nm) conjugated to a different HER2/neu-targeting ligand would also have an intermediate optimal ligand density for cell labeling experiments. Specifically, a small, homology derived anti-HER2/neu peptide (AHNP) was selected for investigation.26 Several AHNP-SPIO conjugates, with a range of ligand densities, were prepared and incubated with T6-17 cells. The degree of cell binding was investigated by measuring the T2-relaxation times of cell suspensions and by collecting T2*-weighted MR images of analogous cell pellets (Figure 5A). As was observed with the HER2-SPIO conjugates, T6-17 cells targeted with AHNP-SPIO conjugates all showed a marked decrease in T2-relaxation times, with the most significant decreases coming from AHNP-SPIO labeled with the lowest ligand density tested (p<0.01). It is not surprising that different HER2/neu-targeting ligands would exhibit different trends in cell binding; however, for both ligands the optimal targeting of the HER2/neu receptor occurred for ligand densities well below saturating levels, underscoring the importance of assessing and controlling ligand densities for targeted nanoparticle studies.
Figure 5.
MR analysis of T6-17 cells labeled with AHNP-SPIO and KB cells labeled with folic acid (FA)-SPIO. (A) T6-17 cells were targeted with AHNP-SPIO, with ligand densities ranging from 20.6 to 72.1 ligands per SPIO, for 30 minutes at 37°C. Analogous competitive inhibition studies (C.I.) were performed in the presence of excess free AHNP. SPIO-labeled cells and unlabeled cells (Blank) were evaluated by acquiring T2 relaxation times and T2*-weighted MR images of each sample. (B) KB cells were targeted with FA-SPIO for 30 minutes at 37°C. Note, the folic acid/SPIO reflects the reaction conditions not the final labeling ratio, which could not be precisely determined. NPs reacted with 20.6 AHNP per SPIO and 133 FA per SPIO exhibited statistically significant (p<0.05) improvements in MR contrast as compared to SPIO with lower and higher ligand densities.
To examine whether the observed differences in cell binding that result from varying the ligand density could be extended to other popular biomarkers, the 26 nm SPIO NPs were functionalized with folic acid (FA), a ligand for the folate receptor. Specifically, aminated SPIO NPs were reacted with increasing concentrations of NHS-FA to produce FA-SPIO with increasing FA ligand densities. FA-SPIO conjugates were incubated with KB cells, which are known to overexpress the folate receptor. Cells were washed and analyzed for cell binding by measuring their T2 relaxation times (Figure 5B). Similar to HER2/neu-targeted SPIO, FA-SPIO showed the greatest degree of cell binding at an intermediate FA density, with marked decreases in cell binding noted for higher and lower ligand densities of FA. Moreover, when T2*-weighted images were collected for FA-SPIO targeted KB cells, a drastic improvement in negative contrast generation was noted for the optimal ligand density as compared to higher and lower variants (Figure 5B, right panel). Extending the observed ligand density effect to a new receptor class emphasizes the importance of optimizing ligand densities for targeted nanoparticle studies as it is now evident that these results are not just applicable to HER2/neu targeting, but likely many systems.
Discussion
The ability to optimize targeted nanoparticle platforms by controlling ligand density could have significant downstream effects on generated contrast and dose delivered for diagnostic and therapeutic studies. Herein, we have shown that for at least some nanoparticle platforms and targeting ligands there exist an optimal ligand density below nanoparticle surface saturation levels and that selecting this density for targeted cell studies results in significant improvements in cell binding and contrast. Additionally, we have shown that this optimal ligand density effect persists across nanoparticles with different hydrodynamic diameters and different targeting ligands, as well as across cells with different receptor densities and different targeted receptors. Importantly, when evaluating the effect of ligand density on nanoparticle systems, it is important to take into account that there is an inherent degree of variability and limited precision with all measurements. For example, most NP systems, particularly those above 20 nm are not monodisperse, including SPIO NPs. Further, in most cases only the average number of ligands per NP can be varied not the exact number of ligands on every NP. Therefore, there will be a distribution in the actual number of ligands per NP, with some NPs having fewer ligands and some more. The width of this distribution is largely dependent on the number of ligand grafting sites and the conjugation efficiency.33 In order to facilitate narrow ligand distribution, a large number of grafting sites (e.g. ~50–100 per NP or more) and efficient bioconjugation reactions are required. Notably, the 26 nm SPIO-NP that was primarily used in this study was labeled with as many as 72 ligands (AHNP) and the 50 nm SPIO-NP was labeled with over 100 HER2-affibodies. Moreover, click conjugation reactions are known to have reaction efficiencies nearing 100%.34 Therefore, although it cannot be assessed directly, it is expected that the distribution of functional ligands in this study is relatively narrow. This is further supported by the clear differences in cell labeling that we observed with SPIO-NPs with different ligand densities. One of the complications associated with a broad distribution of functional ligands is that there is a large overlap between NP formulations with different average ligand densities. This high degree of overlap makes it difficult to identify differences in cell binding between two NP formulations, because both formulations would be largely composed of NPs with very similar ligand densities.
Several factors may contribute to the observed ligand density effect, including but not limited to steric interference stemming from ligand size, packing densities, and ligand orientation, receptor mobility and receptor internalization. Steric interference between ligands on the nanoparticle surface may occur due to their tight packing at high densities. The relatively small size of the HER2-Affibody, AHNP peptide and folate molecule theoretically allow for high packing densities on the nanoparticle surface. Increasing the average number of ligands per nanoparticle will greatly increase the propensity of two ligands existing in close vicinity. This can create competition between multiple ligands for a single receptor and also has the potential to limit the access that ligands have to receptors. For example, over crowding could prevent ligands from obtaining the correct orientation necessary for binding. This effect would presumably be amplified when all of the targeting ligands possess the same orientation, as is the case in this study and in most studies where peptides and small molecules are used. Such an effect would not be expected to occur for larger targeting ligands and would likely be less pronounced when the surface orientation of the targeting ligands is random, i.e. when bioconjugation is not site-specific. Indeed, nanoparticle ligand density studies that have been conducted with randomly oriented antibodies, single chain antibody fragments (ScFv’s), and large proteins, have shown that cell binding generally increases with ligand density.20,33 In several of these reports it was further shown that once a certain threshold in the ligand density is reached, there is no further improvement in cell labeling.35, 36 Whether this saturation phenomena occurs seems to be dependent on the affinity of the targeting ligand. For example, it was found that only scFv’s with affinities above 15 nM exhibited this behavior. Binding affinities for the Affibody (22 pM)25, AHNP peptide (300 nM)26 and folate receptor (10–1000 pM)37 fall on either side of the 15 nM cut-off, yet optimal non-saturating ligand densities were noted for all ligands. There do not seem to be any prior reports that suggest that nanoparticles labeled with antibodies or other large targeting ligands have an optimal intermediate ligand density. However, several recent publications have cited optimal intermediate ligand densities for folate-labeled nanoparticles, which is consistent with our findings.21, 38–40 This seems to suggest that ligand size and orientation may be two key factors that dictate whether an optimal intermediate ligand density exists or whether cell labeling varies more asymptotically with ligand density.
In addition to steric hindrances, another possible explanation of the observed ligand density effect can be garnered by examining the state in which the HER2/neu receptor resides in the cellular membrane. It has been reported that the HER2/neu receptors exist as ‘receptor clusters’ in cellular membranes, with an average cluster size of 67 nm stemming from an average of 9 proteins.41 Targeting these ‘clusters’ with very low ligand densities may not provide a high enough nanoparticle avidity to result in stable cell binding. Intermediate ligand densities may be optimal, allowing multiple nanoparticles to bind to each cluster. In contrast, nanoparticles with a high ligand density may consume too many receptors in each cluster and thus hinder other nanoparticles from binding to the same cluster. It is important to note that “clustering” effects have been noted for many popular biomarkers in either their active or inactive states, including the folate receptor42, ICAM-119, αvβ3 integrin43 and the transferrin receptor44.
Interestingly, in silico analysis of ligand-directed nanoparticles towards diffusible, membrane bound receptors has predicted optimal non-saturating ligand densities for cellular uptake.45 Specifically, thermodynamic modeling revealed that adhesion strength between nanoparticles and cells governs cellular uptake, and the enthalpic and entropic energies governing this process are inherently linked to nanoparticle size and ligand density. When ligand density is increased beyond optimal values, decreases in cellular uptake were predicted for a range of nanoparticle sizes. At high ligand densities, these decreases were partially attributed to a diminishing availability of accessible receptors for nanoparticle binding due to high ratios of receptors bound per nanoparticle. Therefore, over-recruitment and diminishing receptor availability is a possible explanation for the observed decrease in cell binding at high ligand densities. With a limited number of receptors available for binding, over-recruitment via high ligand density nanoparticles could decrease overall cell binding. It should be noted, however, that these predictive models make assumptions that ignore some variability that intrinsically exists with experimental targeting studies. For example, the thermodynamic models assume monodisperse NPs with even distributions of ligands over the NP surface. The NPs used in this study, although relatively uniform in size, were not monodisperse. Further, while the applied conjugation strategy can control for ligand orientation and is expected to produce relatively narrow distributions in ligand density, as discussed above, there will certainly be some variability in ligand density, which is not accounted for in the model.
The presence of an intermediate optimal ligand density does not seem to be limited to the NP platform presented here (i.e. SPIO). An intermediate optimal density of folic acid has been reported on folate receptor-targeted liposomes21, 40 and micellar nanoparticles19, 38, and simulated for general spherical particles.45 Thus, it is our opinion that the ligand density effect is not constrained to specific nanoparticle platforms, but rather is dependent on the surface packing density of functional ligands, the size of the ligands, and the orientation of the surface bound ligand. For example, linking a ligand to a nanoparticle via a bioconjugation technique that lacks chemoselectivity and cannot control for ligand orientation can result in surface ligands with reduced or eliminated functionality, thus altering the perceived ‘functional’ density of ligands. Additionally, use of inefficient bioconjugation techniques46, 47 or construction of nanoparticles with limited conjugation sites may hinder targeted nanoparticles from achieving optimal ligand densities. Also, as was previously mentioned, ligand densities responsible for decreases in nanoparticle cell binding may not be achievable with larger targeting proteins (i.e. scFvs, diabodies, antibodies).
Notably, this study utilized a short, PEG linker (dPEG4) to connect the nanoparticle surface and targeting ligand. Change in linker lengths and flexibility could increase the availability of targeting ligands and alleviate potential steric hindrances. Computational analysis of the effect of spacer arm length and flexibility for liposomal particles determined that longer linkers increase the ‘area of influence’ for a nanoparticle thereby decreasing negative steric effects.39 It should be noted, however, that utilizing long spacer arms for ligand attachment could result in an increase in NP size and affect pharmacokinetics. As was noted in Table 1 for this study, no major differences were noted in SPIO nanoparticle hydrodynamic diameters following addition of ligands.
Ultimately, the extension of these findings to in vivo targeting applications must be considered. Surface functionalization of nanoparticles can effect known pharmacokinetic properties of passively targeted nanoparticle precursors.48 The presence of different targeting ligand surface densities may alter serum protein opsonization patterns and accelerate systemic nanoparticle clearance49. In fact, one study has shown that while in vitro cell binding studies were unable to distinguish between targeted nanoparticles with different ligand densities, in vivo studies revealed that nanoparticles with ligand densities below the highest conditions tested resulted in an increase in tumoral localization.4 While this result presumably stems from improved nanoparticle pharmacokinetics, it does underscore the multifaceted role that ligand density plays in vivo. Moreover, for popular targeted cancer receptors that are known to also be expressed in healthy tissues (e.g. transferrin receptor, folate receptor, EGFR), maximizing cell binding through ligand density could manifest through off-site nanoparticle accumulation.50 Therefore, the optimal ligand densities for in vivo use may be those which confer the greatest selectivity for the target tissue as oppose to the highest avidity.
Actively targeted nanocarriers have emerged as a new tool with the potential to expand the applicability of diagnostic imaging, and improve the selectivity of drug delivery systems. Herein, we have shown that controlling the ligand density on a nanoparticles surface can have a significant impact on target cell binding. Specifically, intermediate ligand densities were shown to provide optimal SPIO NP cell targeting regardless of nanoparticle size, receptor density or the selected targeting ligand utilized. Such implications merit consideration in targeted nanoparticle design, as optimized targeting could provide significant benefits both in vitro and in vivo.
Supplementary Material
Acknowledgments
Sources of Support: This work was supported by the National Institute of Health R01-EB012065 (NIBIB), R01-CA157766 (NCI), R01-HL087036 (NHLBI), and R21-EB013226(NIBIB).
Footnotes
The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 2.Chiu GN, Edwards LA, Kapanen AI, Malinen MM, Dragowska WH, Warburton C, et al. Modulation of cancer cell survival pathways using multivalent liposomal therapeutic antibody constructs. Mol Cancer Ther. 2007;6:844–55. doi: 10.1158/1535-7163.MCT-06-0159. [DOI] [PubMed] [Google Scholar]
- 3.Gindy ME, Ji S, Hoye TR, Panagiotopoulos AZ, Prud’homme RK. Preparation of poly(ethylene glycol) protected nanoparticles with variable bioconjugate ligand density. Biomacromolecules. 2008;9:2705–11. doi: 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- 4.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci U S A. 2008;105:2586–91. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elias DR, Cheng Z, Tsourkas A. An intein-mediated site-specific click conjugation strategy for improved tumor targeting of nanoparticle systems. Small. 2010;6:2460–8. doi: 10.1002/smll.201001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan C, Gao W, Chen Z, Fan H, Li M, Deng F. Tumor selectivity of stealth multi-functionalized superparamagnetic iron oxide nanoparticles. Int J Pharm. 2011;404:180–90. doi: 10.1016/j.ijpharm.2010.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang L, et al. Biomimetic amplification of nanoparticle homing to tumors. Proc Natl Acad Sci U S A. 2007;104:932–6. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiessling F, Huppert J, Zhang C, Jayapaul J, Zwick S, Woenne EC, et al. RGD-labeled USPIO inhibits adhesion and endocytotic activity of alpha v beta3-integrin-expressing glioma cells and only accumulates in the vascular tumor compartment. Radiology. 2009;253:462–9. doi: 10.1148/radiol.2532081815. [DOI] [PubMed] [Google Scholar]
- 9.Fang C, Veiseh O, Kievit F, Bhattarai N, Wang F, Stephen Z, et al. Functionalization of iron oxide magnetic nanoparticles with targeting ligands: their physicochemical properties and in vivo behavior. Nanomedicine (Lond) 2010;5:1357–69. doi: 10.2217/nnm.10.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morishige K, Kacher DF, Libby P, Josephson L, Ganz P, Weissleder R, et al. High-resolution magnetic resonance imaging enhanced with superparamagnetic nanoparticles measures macrophage burden in atherosclerosis. Circulation. 2010;122:1707–15. doi: 10.1161/CIRCULATIONAHA.109.891804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsourkas A, Shinde-Patil VR, Kelly KA, Patel P, Wolley A, Allport JR, et al. In vivo imaging of activated endothelium using an anti-VCAM-1 magnetooptical probe. Bioconjug Chem. 2005;16:576–81. doi: 10.1021/bc050002e. [DOI] [PubMed] [Google Scholar]
- 12.Kelly KA, Allport JR, Tsourkas A, Shinde-Patil VR, Josephson L, Weissleder R. Detection of vascular adhesion molecule-1 expression using a novel multimodal nanoparticle. Circ Res. 2005;96:327–36. doi: 10.1161/01.RES.0000155722.17881.dd. [DOI] [PubMed] [Google Scholar]
- 13.Thorek DL, Tsao PY, Arora V, Zhou L, Eisenberg RA, Tsourkas A. In vivo, multimodal imaging of B cell distribution and response to antibody immunotherapy in mice. PLoS One. 2010;5:e10655. doi: 10.1371/journal.pone.0010655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, et al. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18:410–4. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 15.Zhao M, Beauregard DA, Loizou L, Davletov B, Brindle KM. Non-invasive detection of apoptosis using magnetic resonance imaging and a targeted contrast agent. Nat Med. 2001;7:1241–4. doi: 10.1038/nm1101-1241. [DOI] [PubMed] [Google Scholar]
- 16.Mouli SK, Zhao LC, Omary RA, Thaxton CS. Lymphotropic nanoparticle enhanced MRI for the staging of genitourinary tumors. Nat Rev Urol. 2010;7:84–93. doi: 10.1038/nrurol.2009.254. [DOI] [PubMed] [Google Scholar]
- 17.Branca RT, Cleveland ZI, Fubara B, Kumar CS, Maronpot RR, Leuschner C, et al. Molecular MRI for sensitive and specific detection of lung metastases. Proc Natl Acad Sci U S A. 2010;107:3693–7. doi: 10.1073/pnas.1000386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman R, Uchida N, Bliss TM, He D, Christopherson KK, Stellwagen D, et al. Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc Natl Acad Sci U S A. 2007;104:10211–6. doi: 10.1073/pnas.0608519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fakhari A, Baoum A, Siahaan TJ, Le KB, Berkland C. Controlling ligand surface density optimizes nanoparticle binding to ICAM-1. J Pharm Sci. 2011;100:1045–56. doi: 10.1002/jps.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Tian S, Petros RA, Napier ME, Desimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J Am Chem Soc. 2010;132:11306–13. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawano K, Maitani Y. Effects of polyethylene glycol spacer length and ligand density on folate receptor targeting of liposomal Doxorubicin in vitro. J Drug Deliv. 2011;2011:160967. doi: 10.1155/2011/160967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorek DL, Elias DR, Tsourkas A. Comparative analysis of nanoparticle-antibody conjugations: carbodiimide versus click chemistry. Mol Imaging. 2009;8:221–9. [PubMed] [Google Scholar]
- 23.Cheng Z, Elias DR, Kamat NP, Johnston ED, Poloukhtine A, Popik V, et al. Improved Tumor Targeting of Polymer-based Nanovesicles Using Polymer-Lipid Blends. 2011. In Submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orski SV, Poloukhtine AA, Arumugam S, Mao L, Popik VV, Locklin J. High density orthogonal surface immobilization via photoactivated copper-free click chemistry. J Am Chem Soc. 2010;132:11024–6. doi: 10.1021/ja105066t. [DOI] [PubMed] [Google Scholar]
- 25.Orlova A, Magnusson M, Eriksson TL, Nilsson M, Larsson B, Hoiden-Guthenberg I, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66:4339–48. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 26.Park BW, Zhang HT, Wu C, Berezov A, Zhang X, Dua R, et al. Rationally designed anti- HER2/neu peptide mimetic disables P185HER2/neu tyrosine kinases in vitro and in vivo. Nat Biotechnol. 2000;18:194–8. doi: 10.1038/72651. [DOI] [PubMed] [Google Scholar]
- 27.Bleackley MR, Macgillivray RT. Transition metal homeostasis: from yeast to human disease. Biometals. 2011 doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- 28.Apostoli P, Catalani S. Metal ions affecting reproduction and development. Met Ions Life Sci. 2011;8:263–303. [PubMed] [Google Scholar]
- 29.Thorek DL, Tsourkas A. Size, charge and concentration dependent uptake of iron oxide particles by non-phagocytic cells. Biomaterials. 2008;29:3583–90. doi: 10.1016/j.biomaterials.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, Fang H, Chen Z, Taylor JS, Wooley KL. Shape effects of nanoparticles conjugated with cell-penetrating peptides (HIV Tat PTD) on CHO cell uptake. Bioconjug Chem. 2008;19:1880–7. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crayton SH, Elias DR, Al Zaki A, Cheng Z, Tsourkas A. ICP-MS analysis of lanthanide-doped nanoparticles as a non-radiative, multiplex approach to quantify biodistribution and blood clearance. Biomaterials. 2012;33:1509–19. doi: 10.1016/j.biomaterials.2011.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes A. Developing a cell line standard for HER2/neu. Cancer Biomark. 2005;1:229–32. doi: 10.3233/cbm-2005-14-505. [DOI] [PubMed] [Google Scholar]
- 33.Hakem IF, Leech AM, Johnson JD, Donahue SJ, Walker JP, Bockstaller MR. Understanding ligand distributions in modified particle and particlelike systems. J Am Chem Soc. 2010;132:16593–8. doi: 10.1021/ja107139c. [DOI] [PubMed] [Google Scholar]
- 34.Hein CD, Liu XM, Wang D. Click chemistry, a powerful tool for pharmaceutical sciences. Pharm Res. 2008;25:2216–30. doi: 10.1007/s11095-008-9616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Weller GE, Zern B, Ayyaswamy PS, Eckmann DM, Muzykantov VR, et al. Computational model for nanocarrier binding to endothelium validated using in vivo, in vitro, and atomic force microscopy experiments. Proc Natl Acad Sci U S A. 2010;107:16530–5. doi: 10.1073/pnas.1006611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, et al. Impact of single-chain Fv antibody fragment affinity on nanoparticle targeting of epidermal growth factor receptor-expressing tumor cells. J Mol Biol. 2007;371:934–47. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon KH, Mulligan T, Chung KN, Elwood PC, Cowan KH. Effects of folate receptor expression following stable transfection into wild type and methotrexate transport-deficient ZR-75–1 human breast cancer cells. J Biol Chem. 1992;267:24140–7. [PubMed] [Google Scholar]
- 38.Poon Z, Chen S, Engler AC, Lee HI, Atas E, von Maltzahn G, et al. Ligand-clustered “patchy” nanoparticles for modulated cellular uptake and in vivo tumor targeting. Angew Chem Int Ed Engl. 2010;49:7266–70. doi: 10.1002/anie.201003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghaghada KB, Saul J, Natarajan JV, Bellamkonda RV, Annapragada AV. Folate targeting of drug carriers: a mathematical model. J Control Release. 2005;104:113–28. doi: 10.1016/j.jconrel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Yamada A, Taniguchi Y, Kawano K, Honda T, Hattori Y, Maitani Y. Design of folate-linked liposomal doxorubicin to its antitumor effect in mice. Clin Cancer Res. 2008;14:8161–8. doi: 10.1158/1078-0432.CCR-08-0159. [DOI] [PubMed] [Google Scholar]
- 41.Kaufmann R, Muller P, Hildenbrand G, Hausmann M, Cremer C. Analysis of Her2/neu membrane protein clusters in different types of breast cancer cells using localization microscopy. J Microsc. 2011;242:46–54. doi: 10.1111/j.1365-2818.2010.03436.x. [DOI] [PubMed] [Google Scholar]
- 42.Smart EJ, Mineo C, Anderson RG. Clustered folate receptors deliver 5-methyltetrahydrofolate to cytoplasm of MA104 cells. J Cell Biol. 1996;134:1169–77. doi: 10.1083/jcb.134.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cluzel C, Saltel F, Lussi J, Paulhe F, Imhof BA, Wehrle-Haller B. The mechanisms and dynamics of (alpha)v(beta)3 integrin clustering in living cells. J Cell Biol. 2005;171:383–92. doi: 10.1083/jcb.200503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu AP, Aguet F, Danuser G, Schmid SL. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J Cell Biol. 2010;191:1381–93. doi: 10.1083/jcb.201008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan H, Li J, Bao G, Zhang S. Variable nanoparticle-cell adhesion strength regulates cellular uptake. Phys Rev Lett. 2010;105:138101. doi: 10.1103/PhysRevLett.105.138101. [DOI] [PubMed] [Google Scholar]
- 46.Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J Control Release. 2007;120:18–26. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Natarajan A, Gruettner C, Ivkov R, DeNardo GL, Mirick G, Yuan A, et al. NanoFerrite particle based radioimmunonanoparticles: binding affinity and in vivo pharmacokinetics. Bioconjug Chem. 2008;19:1211–8. doi: 10.1021/bc800015n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huynh NT, Roger E, Lautram N, Benoit JP, Passirani C. The rise and rise of stealth nanocarriers for cancer therapy: passive versus active targeting. Nanomedicine (Lond) 2010;5:1415–33. doi: 10.2217/nnm.10.113. [DOI] [PubMed] [Google Scholar]
- 49.Hansen CB, Kao GY, Moase EH, Zalipsky S, Allen TM. Attachment of antibodies to sterically stabilized liposomes: evaluation, comparison and optimization of coupling procedures. Biochim Biophys Acta. 1995;1239:133–44. doi: 10.1016/0005-2736(95)00138-s. [DOI] [PubMed] [Google Scholar]
- 50.Haun JB, Hammer DA. Quantifying nanoparticle adhesion mediated by specific molecular interactions. Langmuir. 2008;24:8821–32. doi: 10.1021/la8005844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.