Abstract
Antibodies directed against histone posttranslational modifications (PTMs) are critical tools in epigenetics research, particularly in the widely used chromatin immunoprecipitation (ChIP) experiments. However, a lack of quantitative methods for characterizing such antibodies has been a major bottleneck in accurate and reproducible analysis of histone modifications. Here, we report a simple and sensitive method for quantitatively characterizing polyclonal and monoclonal antibodies for histone PTMs in a ChIP-like format. Importantly, it determines the apparent dissociation constants for the interactions of an antibody with peptides harboring cognate or off-target PTMs. Analyses of commercial antibodies revealed large ranges of affinity, specificity and binding capacity as well as substantial lot-to-lot variations, suggesting the importance of quantitatively characterizing each antibody intended to be used in ChIP experiments and optimizing experimental conditions accordingly. Furthermore, using this method we identified additional factors potentially affecting the interpretation of ChIP experiments.
Keywords: epigenetics, histone code, antibody-antigen interaction, flow cytometry
Histone PTMs play important roles in epigenetic regulation.1; 2 Antibodies to histone PTMs are critical tools for epigenetics research, particularly for the widely used ChIP technique. In ChIP experiments, nucleosomes in solution are captured with an antibody immobilized on solid support (Fig. 1a). “ChIP-grade” antibodies for histone PTMs should reliably capture their cognate antigen with high specificity and high efficiency. Thus, considerable efforts have been made to validate anti-histone PTM antibodies. Recent studies have revealed substantial variability in the level of specificity of commercial antibodies sold as ChIP grade.3
Figure 1.
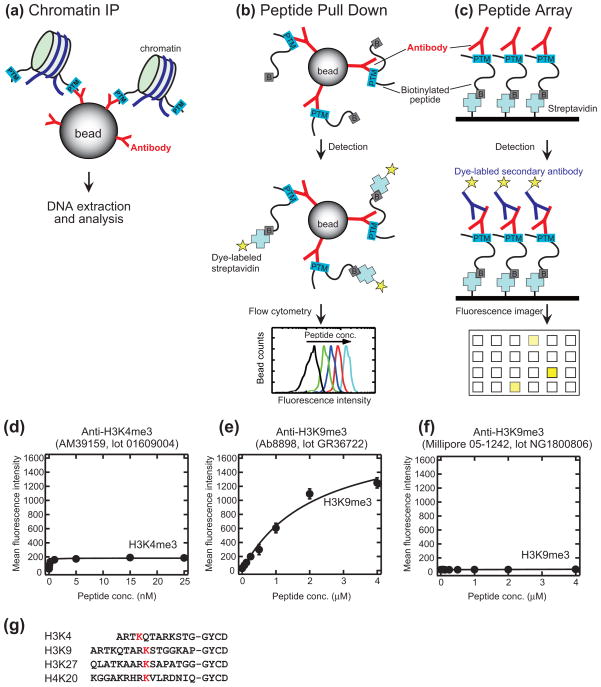
Assay design. (a–c) Schematics comparing the ChIP experiment (a), antibody characterization using the peptide IP assay developed in this work (b) and peptide arrays (c). In the peptide IP assay, the binding of a biotinylated peptide to an antibody immobilized on beads is detected. In peptide arrays, antibody binding to peptides immobilized on solid support through the biotin-streptavidin interaction (or direct coupling) is detected.5 (d–f) Titration curves of three ChIP-grade antibodies to peptides containing their respective cognate PTMs. The identities of the antibodies are given in the figures. Expanded plots of binding data for these antibodies are given in Fig. 2. The lines show the best fit of the 1:1 binding model in (d) and (e). No detectable binding was found in (f), and the line connects the data points. Error bars where not visible are within the size of the symbols. (g) Peptides used in the IP assay. The residues containing PTM are in red. The GYCD tag is for biotinylation and quantification.
A number of methods, from Western blotting and enzyme-linked immunosorbent assay (ELISA) to peptide arrays, have been reported for validating anti-histone antibodies. Typically, specificity of an antibody to its cognate histone is tested using Western blotting against nuclear extracts or cell lysates. Specificity toward a particular histone PTM is then tested using a panel of peptides carrying histone PTMs using ELISA or peptide arrays. Generally, peptides carrying histone PTMs are immobilized on solid support and the binding of an antibody to hundreds of peptides can be examined simultaneously (Fig. 1c).4; 5 These methods exploit the fact that many histone PTMs occur in the flexible tails, and thus synthetic peptides are excellent surrogate antigens. Generally, an antibody is considered specific if it exhibits substantially stronger binding to the peptide representing the cognate PTM over other peptides representing “off targets”.
A major limitation of these assays is that they do not test factors that are most relevant to successful ChIP experiments, i.e. how efficiently and specifically an antibody immunoprecipitates its cognate antigen. Western blotting and peptide arrays test the binding of an antibody in solution to peptides immobilized on solid support. This format is opposite to that of immunoprecipitation, where an antibody immobilized on solid support captures antigens in solution. Therefore, it is not clear how well the common evaluation methods predict the performance of an antibody in ChIP experiment. Indeed, Egelhofer et al. reported perplexing results that more than 20% of antibodies that have been validated to be specific in peptide blots still fail in ChIP experiments, illustrating that the current validation methods overlook an important parameter that defines antibody performance in ChIP experiments.6
Recently, Peach et al. reported a method that characterizes antibodies in immunoprecipitation format.7 In this method, chromatin samples are immunoprecipitated with an antibody of interest and enrichment of different PTMs is quantified using mass spectrometry (referred to as IP-MS here after). Thus, it tests antibodies in a relevant assay format and provides results that can be directly compared with enrichment obtained from ChIP experiments. However, it requires extensive sample preparation and manipulation and access to a high-end mass spectrometer and data analysis expertise.
Another limitation of the current assays is that their results are highly sensitive to experimental conditions. For example, the amount and density of immobilized peptides and the concentration of antibody used can substantially influence results from peptide arrays. Likewise, it is well known that one needs to optimize sample amounts and antibody concentrations to obtain “good” Western blotting data. The enrichment information from the IP-MS method is also influenced by the amounts of different modifications that are present in the starting material. As a consequence, it is difficult to compare validation results performed by different methods and/or at different locations, because it is difficult to describe the performance of antibodies in these assays in numerical terms that are not highly sensitive to experimental conditions.
The dissociation constant (Kd) is a fundamental parameter that defines antibody affinity, and antibody specificity can be quantitatively evaluated using Kd values for different antigens. However, standard biophysical methods for Kd determination are sample-intensive and often require accurate knowledge of the effective antibody concentration, both of which are difficult to attain with current anti-histone PTM antibodies that are mostly polyclonal antibodies in limited quantities.
Results and Discussion
Assay design
We set out to establish an assay that address the two major issues associated with the current assays for anti-histone antibody characterization, i.e., testing antibodies in a format relevant to ChIP and obtaining truly quantitative parameters that are insensitive to experimental variations without consuming large amounts of antibody samples. Our “peptide immunoprecipitation (IP)” assay mimics the IP step of ChIP experiments (Fig. 1a and 1b). An antibody of interest is first immobilized on protein A- or protein G-coated polystyrene beads, as how an antibody is captured in ChIP experiments. A peptide containing a histone PTM and also a biotin moiety in solution is then captured with the beads. After a washing step to eliminate the unbound peptide, the captured peptide is quantified via the attached biotin moiety with fluorescently labeled streptavidin using flow cytometry (Fig. 1b). The fluorescence intensity observed for each bead is proportional to the amount of a biotinylated peptide captured by the immobilized antibody on the bead. By performing the experiment as a function of the peptide concentration, one can obtain a saturation curve from which the apparent Kd value is determined (Fig. 1d–1f and Fig. 2). Our method is similar to that successfully used for protein-protein interactions,8 and to yeast surface display where yeast cells serve as beads.9 The Kd values determined in these bead-based methods were found to be close to those determined with standard biophysical methods.9 Importantly, our method, in the current implementation, consumes only 25 ng of an antibody for each data point. Because we observed excellent signal-to-noise ratios in the flow cytometry data, it is likely that the antibody consumption can be further reduced. Furthermore, it does not require the knowledge of the concentration of the active antibody in a sample, because the antibody concentration is kept substantially lower than the peptide concentration, which allows for the approximation that eliminates the antibody concentration from the equation used in Kd determination (see Methods for details).
Figure 2.
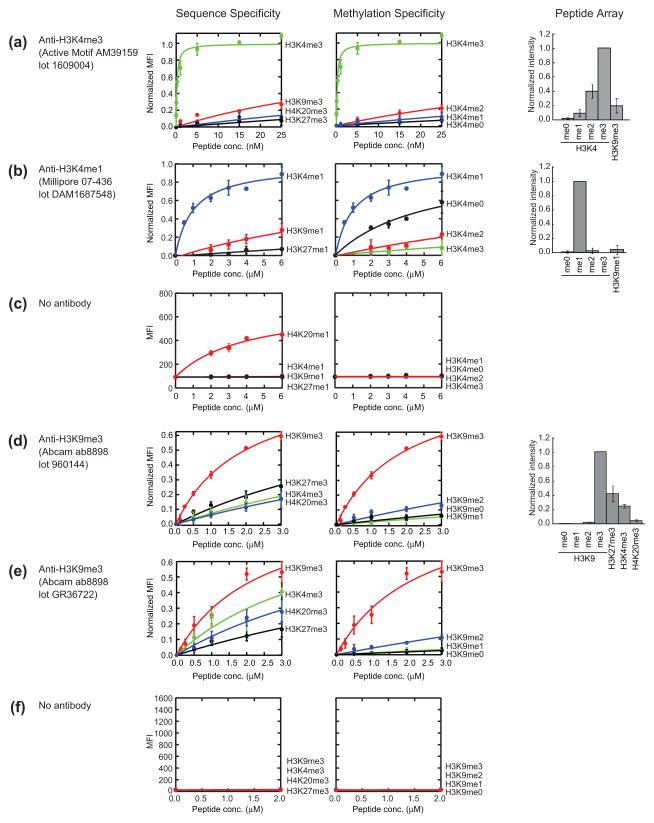
Characterization of anti-histone PTM antibodies. In peptide IP assay (left and center panels), the mean fluorescence intensities (MFI) were normalized to have a range of 0–1 with 1 corresponding to the MFI at saturation of the highest affinity interaction estimated from curve fitting, and they are plotted against peptide concentration. The left panels show binding data to the cognate PTM and to other sequences containing the same methylation state, indicating sequence specificity. The center panels show binding data to the cognate PTM and other methylation states of the same sequence, indicating methylation state specificity. The curves are the best fits of the 1:1 binding model. See Materials and Methods for details of model fitting. In peptide arrays (right panels), spot intensities with s.d. from duplicate experiments are shown for the same PTMs as in peptide IP. Spot intensities are normalized relative to the highest intensity observed among tested peptides. (c and f) Binding of the indicated peptides to the beads without an immobilized antibody. Data in (f) were measured using the same settings as in Figure 1d–f. Data in (c) were measured with higher sensitivity. Thus the MFI values of the two data sets cannot be directly compared.
Large differences in affinity and binding capacity among anti-histone PTM antibodies
Using our new assay, we first characterized three different types of antibodies, a purified polyclonal antibody, an unpurified anti-serum, and a purified monoclonal antibody (Fig. 1d–f). The antibody concentration in the anti-serum was not available. We normalized the amounts of different antibodies in these experiments by saturating the beads with the antibodies and then measuring the same number of beads.
Using these antibody-coated beads, we measured binding of their respective cognate peptides. For two of the three antibodies, we obtained titration curves displaying saturation behavior (Fig. 1d and 1e; also see Fig. 2a and 2e). Interestingly, they had dramatically different titration curves. AM39159, the anti-serum, reached saturation with low nanomolar concentration of its cognate peptide with an apparent Kd value of 0.21 nM (Fig. 1d and 2a). In contrast, Ab8898, the polyclonal antibody, did not reach complete saturation even with micromolar concentrations of its cognate peptide, yielding an apparent Kd value of ~2 μM, i.e. ~10,000 times higher than that for AM39159 (Fig. 1e). These results indicate dramatically different affinity among commercial ChIP-grade antibodies.
The monoclonal antibody, 05-1242, showed no detectable binding, although we tested two different samples for the antibody and also we performed the assays using protein A beads and protein G beads. We were not able to determine whether there was no active antibody in these samples of 05-1242 or if the affinity of the active antibody in these samples was too low to be detected by our method. The datasheet accompanying this antibody (http://www.millipore.com/catalogue/item/05-1242) shows that it binds to a peptide corresponding to trimethylated Lys of histone H3 (K9me3), eliminating the possibility that a short peptide is an inappropriate antigen for this antibody. Regardless of the underlying cause, it is clear that the 05-1242 antibody is not suitable for ChIP experiment, because it captured no detectable amounts of the cognate peptide.
The fluorescence intensities observed at high peptide concentrations were significantly different between AM39159 and Ab8898 (Fig. 1d and 1e). Because the peptides were essentially fully biotinylated, and the experiments were performed with fully saturated beads using the same instrument settings, the fluorescence intensity is proportional to the amount of the peptides captured by the antibodies. Thus, the large difference in the fluorescence intensity, at least 6-fold, observed between the two antibodies suggests a large difference in the “binding capacity” of these antibody samples. The binding capacity is an important parameter for ChIP antibodies, because ideally an antibody should capture all chromatins harboring a histone PTM of interest in a ChIP experiment. Both antibodies are rabbit polyclonal, and thus they are likely to be in the IgG format having two antigen binding sites per molecule. Consequently, the large difference in the binding capacity is probably due to lower effective concentration of an active antibody rather than to differences in antibody format. This interpretation is reasonable because AM39159 is an anti-serum containing additional antibodies that are incapable of binding to the cognate antigen but still compete for protein A on the beads, reducing the concentration of active antibodies.
Differences in the dissociation rate of a bound peptide during washing steps can result in differences in the amount of the retained peptide. The use of streptavidin should quench the dissociation of a bound peptide, because streptavidin is tetrameric and it can greatly increase the effective affinity by forming multivalent interactions between the immobilized antibody and the biotinylated peptide. Usually there is a strong correlation between the dissociation rate and affinity of antibodies, i.e. a high-affinity antibody usually exhibits a slow dissociation rate. However, the observed difference is contrary to this general trend, suggesting that the underlying kinetics is unlikely to be a dominant cause of the observed difference. Together, these results show that our assay can quantify both affinity and binding capacity of an antibody sample.
Quantitative characterization of antibody specificity
We then quantitatively characterized binding specificity of three commercial, ChIP-grade rabbit polyclonal antibodies for methylated lysines using a series of peptides corresponding to different methylation states and different Lys sites (Fig. 2; Supplementary Table 1). The anti-H3K4me3 antibody bound to its cognate target with a Kd of 0.21 nM, and its Kd values to other histone marks were estimated to be >200-fold higher, indicating excellent affinity and specificity (Fig. 2a). In contrast, the affinity of the other antibodies tested was much weaker and they also showed substantial binding to off-target antigens (Fig. 2b, 2d and 2e). We detected binding of a peptide representing H4K20me1 to the beads in the absence of an immobilized antibody at a level similar to the binding of the cognate peptide to the anti-H3K4me1 antibody (Fig. 2c), and thus we excluded this peptide from subsequent analysis. No other peptides showed significant binding to the beads (Fig. 2c and 2f). The anti-H3K4me1 antibody showed significant cross-reactivity to another Lys (H3K9me1) and also to different methylation states (H3K4 without methylation). Two separate lots of the anti-H3K9me3 antibody had similarly low affinity, with micromolar Kd values, for their cognate target, and their Kd values to other methylated peptides were only 2–4 fold higher, indicating both low affinity and low specificity (Fig. 2d and 2e). Furthermore, the specificity profiles, in terms of Kd values to different PTMs, of the two lots were substantially different, suggesting that the two lots would produce substantially different results in ChIP experiments. The Kd values are summarized in Supplementary Table 1. These results illustrate that our assay can quantitatively assess binding specificity of anti-histone PTM antibodies.
Comparison with peptide arrays
We compared results from the peptide IP assay with those from peptide arrays. In the peptide array assay, antibody binding to immobilized peptides is detected using a fluorescently labeled secondary antibody (Fig. 1c). Interestingly, when evaluated with peptide arrays, the anti-H3K4me3 antibody appeared substantially more cross-reactive than in peptide IP (Fig. 2a). This disparity is probably because the effective peptide concentrations in the peptide array were high so that this antibody, with its extremely high affinity, bound strongly even to off-target PTMs. In contrast, the anti-H3K4me1 antibody appeared highly specific in the peptide array, although its specificity was among the worst that we tested using the peptide IP assay. The apparent discrimination of H3K4me1 from the other methylation states at the same site by this antibody is rather puzzling, because it showed similarly weak affinity to these PTMs in the peptide IP assay (Fig. 2b, center panel). We speculate that the very low affinity of this antibody made only its binding to its cognate target detectable in the peptide array and hence exaggerated the small difference in affinity between the cognate and off-target peptides. Among the three antibodies tested, only the H3K9me3 antibodies showed similar rank orders of specificity in both assays. These results suggest the importance of systematically examining conditions for peptide arrays to compensate for differences in antibody affinity.
Effects of antibody and PTM concentrations on IP efficiency
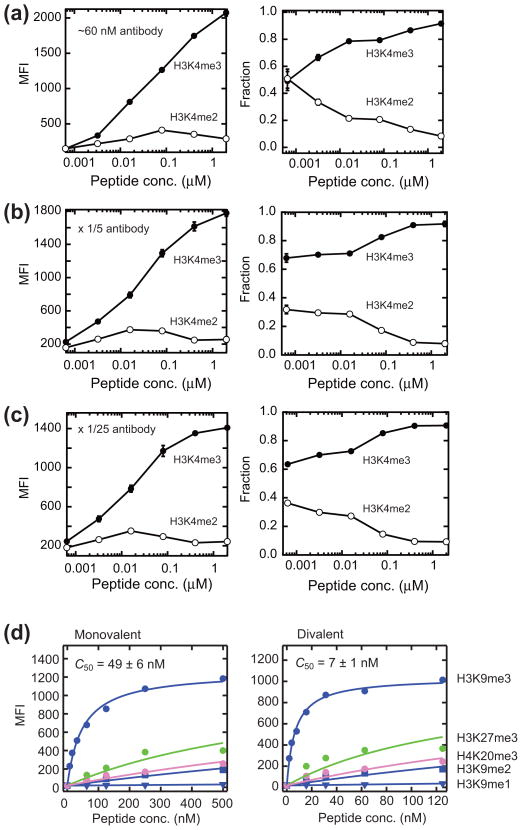
To determine how the concentrations of histone PTMs and antibodies affect the peptide IP performance, we performed “competitive IP” experiments. Here we determined the relative amounts of two PTMs, H3K4me3 and H3K4me2, captured by the anti-H3K4me3 antibody from 1:1 mixtures. This experiment was enabled by the use of monovalent streptavidin.10 The selectivity declined as the peptide concentration was decreased (Fig. 3a–c, right panels), primarily because the amount of the cognate PTM captured by the antibody declined (Fig. 3a–c, left panels). The selectivity was not highly sensitive to antibody concentration, and it slightly improved as the antibody concentration was reduced. These results illustrate the challenge of performing highly selective IP even with an antibody that exhibits very high affinity and high specificity. It should be noted that the Kd value of the antibody to H3K4me3 (0.2 nM) was much lower than the peptide concentrations where the selectivity started to decline (~20 nM) (Fig. 3a–c). These results suggest that, as typical ChIP protocols call for a histone concentration of 200–400 nM,11 an antibody with a micromolar Kd is not likely to achieve high levels of selectivity in actual ChIP experiments (see the next section for additional consideration).
Figure 3.
Effects of peptide and antibody concentrations on the selectivity and effects of antigen valency on peptide IP. (a–c) Selectivity of an antibody assessed with competitive IP experiments. The H3K4me3 peptide bound to fluorescently labeled monovalent streptavidin and H3K4me2 peptides bound to unlabeled monovalent streptavidin from equal mixtures were captured by different concentrations of the anti-H3K4me3-coated beads, and quantified. Binding signals were obtained by measuring fluorescence intensity (MFI) of Alexa 647-labeled streptavidin (left panels). Note that the instrument sensitivity was adjusted for each set of experiments, and thus the MFI for different panels cannot be compared. The fractions of the two peptides were plotted as a function of peptide concentration (right panels). The anti-H3K4me3 antibody was diluted 5- and 25-fold in (b) and (c), respectively, relative to (a). The concentration of the antibody in (a) was estimated to be 60 nM. (d) Binding levels of the indicated peptides coupled to monovalent streptavidin (left panel) and those to divalent streptavidin (right panel) to the anti-H3K9me3 antibody (Ab8898, lot 960144)-coated beads are plotted versus peptide concentration. The apparent half-saturation concentrations, C50, for the H3K9me3 peptide are indicated. The indicated errors are the s.d. of curve fitting.
Effects of PTM valency on IP efficiency
A mononucleosome has two copies of each core histone, and thus contains zero, one or two identical PTM marks. The standard antibody molecule in the form of IgG possesses two antigen-binding sites. Consequently, the number of a PTM within a nucleosome could affect the IP efficiency. To mimic a nucleosome containing one or two identical copies of the modification, respectively, we prepared complexes of streptavidin with one or two copies of the H3K9me3 peptide attached. The affinity of the anti-H3K9me3 antibody to the divalent target was ~7 times higher than to the monovalent target (Fig. 3d), suggesting strong bias in IP efficiency toward the divalent PTM. The specificity profile of this antibody was not substantially affected by the peptide valency (Fig. 3d). This affinity enhancement by a divalent PTM should improve the efficiency of an antibody with low affinity.
Due to technical limitations, we examined only the effect of the valency of an identical PTM. However, our results suggest that, when an antibody has substantial affinity to an off-target PTM, the presence of the cognate PTM and the off-target PTM in the same chromatin should increase the IP efficiency over a chromatin that contains a single instance of the cognate PTM. Again, the knowledge of the Kd values to the cognate and off targets will help estimate the levels of cross-reactivity.
Conclusions
Our peptide IP assay is powerful in that it enables the determination of apparent Kd and binding capacity using small amounts of antibody samples. Our data revealed a large range of affinity, specificity and binding capacity among commercial ChIP-grade antibodies. More than 20% of anti-histone PTM antibodies validated with dot blot still failed in ChIP experiments.6 Because affinity is the primary factor that defines the efficiency of immunoprecipitation, the failures of high-specificity antibodies in ChIP may well be attributed to their low antibody affinity. The large variation of binding capacity among tested samples was rather surprising, and it points to another reason for ChIP failure. Our peptide IP assay complements peptide arrays toward the comprehensive characterization of anti-histone PTM antibodies. Peptide arrays could be first employed to quickly identify off-targets, and then peptide IP would yield Kd values for the cognate and off targets, which will in turn help validate/reject antibodies and optimize ChIP conditions. As low-cost flow cytometers are being introduced to the market, we expect that the peptide IP assay to become an easily accessible method for ChIP antibody characterization. More broadly, our assay is applicable to characterizing many other types of antibody-antigen interactions where a biotinylated antigen can be readily prepared.
MATERIALS AND METHODS
Peptides and antibodies
Histone peptides were purchased from Abgent and Genemed Synthesis. They were biotinylated by incubating 1 mM histone peptides with 3 mM EZ-Link Maleimide-PEG2-Biotin (Thermo Scientific) in PBS (9.57 mM phosphate, 137 mM NaCl, 2.68 mM KCl, pH7.4) for 1 hour on ice. The resulting biotinylated peptides were purified using a RESOURCE S column (GE Healthcare). After purification, biotinylation and peptide identity were confirmed by mass spectrometry using Voyager De-Pro MALDI-TOF (AB Sciex). Peptide concentrations were determined by measuring absorbance at 280 nm using a molar extinction coefficient of 1490 M−1 cm−1. Antibodies were purchased from their respective vendors.
Peptide pull-down assay
To prepare antibody-coated beads, 60 μl of protein A-coated polystyrene beads (Spherotech inc. 4.1 μm diameter, 0.5% w/v) were incubated with 3 μg of an antibody in PBS containing 0.5%w/v bovine serum albumin and 0.1%v/v Tween-20 (PBST/BSA) for 30 min at room temperature. The beads were then washed twice with 500 μl PBST/BSA by centrifugation, and suspended in 60 μl of PBST/BSA. The resultant 0.5 μl of the antibody-coated beads were incubated in 20 μl of PBST/BSA containing various concentrations of biotinylated peptides at 4 °C for 30 min. The bead concentration was set in such a way that it gave a sufficient bead concentration for efficient flow cytometry analysis and also it was sufficiently low to minimize ligand depletion. For the anti-H3K4me3 antibody, which exhibited very high affinity to the cognate histone peptide, the volume of the mixture was increased so that the peptide was in large excess over the antibody and the antibody concentration was much lower than the Kd value in order to minimize ligand depletion. No significant differences were observed when longer incubation periods were used. Then the beads were washed with 300 μl of PBST/BSA three times using a 96-well filter plate (MultiScreenHTS HV, 0.45 μm pore size; Millipore). Then, 20 μl of 10 μg/ml streptavidin-DyLight650 (Thermo Scientific) in PBST/BSA was added to the beads. After incubation at 4 °C for 30 min, the beads were washed with 300 μl of PBST/BSA three times using the filter plate using a vacuum manifold (Millipore). The beads were suspended in 300 μl PBST/BSA and analyzed using a Guava easyCyte 6/L flow cytometer (Millipore). Signals were gated based on the bead size using forward scatter and side scatter intensities so as to reject data from anomalously large beads (< 10% of the total particles). The Kd values for their cognate targets were determined from plots of the mean fluorescence intensity (MFI) for 500 beads versus peptide concentration by nonlinear least-squares fitting in the Igor program (WaveMetrics) with the equation:
Note: This equation does not reproduce in the PDF file for review - please see supplementary material
where MFI(x) is the mean fluorescence intensity observed at the peptide concentration, x; MFI0 is the MFI in the absence of a peptide; ΔMFI is the difference in the MFI in the absence and presence of the saturating concentration of a peptide. The Kd values for off targets were determined based on an assumption that the ΔMFI values are the same for all peptides for a given antibody. We recognize that the ΔMFI values may be different for different peptides, depending on the kinetics of the antibody-peptide interaction. If the dissociation rate is fast, the ΔMFI value would decrease. In such a case, the use of a uniform ΔMFI value would lead to an overestimate of the Kd value to an off target and hence an overestimate of antibody specificity. Notwithstanding this limitation, the assumption is necessary to estimate the Kd values from titration data that do not show clear curvature.
Peptide arrays
Peptide arrays were performed as described in previous report.5
The effect of antigen valency on antibody affinity
In order to control the number of a peptide bound to streptavidin, we prepared monovalent and divalent streptavidin, which contained one and two active streptavidin out of four subunits, respectively. The different streptavidin constructs were synthesized, expressed, and purified following published procedures10 with small modifications. Wild-type streptavidin with N-terminal his-tag and nonfunctional mutant streptavidin (N23A/S27D/S45A) were separately expressed in E. coli BL21(DE3). To generate monovalent and divalent streptavidin, wild-type and mutant streptavidin samples were combined in guanidine hydrochloride, refolded, and then purified using a Chelating Sepharose Fast Flow column (GE Healthcare) charged with Ni2+ ion and a RESOURCE S column (GE Healthcare). They were then conjugated with Alexa Fluor 647 carboxylic acid succinimidyl ester (Invitrogen). For preparing complexes of streptavidin containing one or two copies of histone peptide, monovalent and divalent streptavidin were mixed with a histone peptide with 1:1 or 1:2 molar ratios, respectively. Binding measurements were performed as described above.
Evaluation of antibody selectivity using direct competition
An equal molar mixture of the H3K4me3 peptide bound to fluorescently labeled monovalent streptavidin and the H3K4me2 peptide bound to unlabeled monovalent streptavidin was mixed with anti-H3K4me3-coated beads, and the binding signals were obtained by measuring fluorescence signal of Alexa647. The experiments were repeated in the opposite labeling scheme using H3K4me2 bound to the labeled streptavidin. The fractions of the two peptides were calculated from the binding signals of H3K4me3 and H3K4me2 obtained from the two, complementary experiments. Because the concentration of the antibody in this anti-serum was unknown, we estimated the concentration by determining the amount of the antibody required to saturate the protein A beads and the nominal capacity of the beads provided by the vendor (2.5 μg antibody per mg beads). The highest antibody concentration used was approximately 60 nM.
Supplementary Material
Limited characterization of antibodies is a bottleneck in epigenetics research.
We describe a method that quantitatively determines affinity, specificity and binding capacity.
The method mimics the ChIP format and highly sensitive.
Commercial “ChIP-grade” antibodies show large ranges in their performance.
Acknowledgments
We thank Dr. A. Ruthenburg for discussion; Drs. J. Lavinder and G. Georgiou for assistance in peptide procurement. This work was supported by the NIH grants R21 DA025725 and RC1 DA028779 to SK and R01 GM085394 to BDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs SM, Strahl BD. Antibody recognition of histone post-translational modifications: emerging issues and future prospects. Epigenomics. 2011;3:247–249. doi: 10.2217/epi.11.23. [DOI] [PubMed] [Google Scholar]
- 4.Nady N, Min J, Kareta MS, Chedin F, Arrowsmith CH. A SPOT on the chromatin landscape? Histone peptide arrays as a tool for epigenetic research. Trends Biochem Sci. 2008;33:305–13. doi: 10.1016/j.tibs.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–8. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, Gu T, Kharchenko PV, Kuan S, Latorre I, Linder-Basso D, Luu Y, Ngo Q, Perry M, Rechtsteiner A, Riddle NC, Schwartz YB, Shanower GA, Vielle A, Ahringer J, Elgin SC, Kuroda MI, Pirrotta V, Ren B, Strome S, Park PJ, Karpen GH, Hawkins RD, Lieb JD. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–3. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peach SE, Rudomin EL, Udeshi ND, Carr SA, Jaffe JD. Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Molecular & cellular proteomics : MCP. 2012;11:128–37. doi: 10.1074/mcp.M111.015941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazer LL, Roman DL, Muxlow MR, Neubig RR. Use of flow cytometric methods to quantify protein-protein interactions. Curr Protoc Cytom. 2010;13:11.1–11.15. doi: 10.1002/0471142956.cy1311s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 10.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–45. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3:398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.