Abstract
Next-generation sequencing has been used in numerous investigations to characterize and quantify the genetic diversity of a virus sample through the mapping of polymorphisms and measurement of mutation frequencies. Next-generation sequencing has also been employed to identify recombination events occurring within the genomes of higher organisms, for example, detecting alternative RNA splicing events and oncogenic chromosomal rearrangements. Here, we combine these two approaches to profile RNA recombination within the encapsidated genome of a eukaryotic RNA virus, flock house virus. We detect hundreds of thousands of recombination events, with single-nucleotide resolution, which result in diversity in the encapsidated genome rivaling that due to mismatch mutation. We detect previously identified defective RNAs as well as many other abundant and novel defective RNAs. Our approach is exceptionally sensitive and unbiased and requires no prior knowledge beyond the virus genome sequence. RNA recombination is a powerful driving force behind the evolution and adaptation of RNA viruses. The strategy implemented here is widely applicable and provides a highly detailed description of the complex mutational landscape of the transmissible viral genome.
Abbreviations: FHV, flock house virus; RdRp, RNA-dependent RNA polymerase; DI-RNA, defective-interfering RNA; VLP, virus-like particle; NGS, next-generation sequencing; ORF, open reading frame
Keywords: flock house virus, defective RNAs, deep sequencing, virus-like particles
Graphical Abstract
Highlights
► Diversity in RNA packaged by flock house virus was assessed by deep sequencing. ► RNA recombination was quantitatively profiled with single-nucleotide resolution. ► RNA recombination is prolific, with a frequency comparable to mismatch mutation. ► Previously known, as well as novel, defective RNAs are identified. ► Our strategy may be applied to detect recombination in a wide range of viruses.
Introduction
Recombination within RNA viral genomes is a powerful driving force behind the evolution of RNA viruses and has been widely documented in bacterial, plant, and animal viruses (for a review, see Ref. 1). There are two possible models for RNA recombination: non-replicative or breakage rejoining[2], [3] and copy-choice recombination.[4], [5] Breakage rejoining mechanisms are similar to that occurring during mRNA splicing and involve a catalytic ligation of two RNA molecules. During copy-choice recombination, RNA replication is halted mid-flow and the template RNA dissociates. The nascent RNA remains associated with its polymerase, however, and is able to re-prime replication either at a new position on the original template or on a new template strand. Copy-choice recombination is thought to be the most common mechanism for RNA recombination in many RNA viruses and is thought to primarily occur during the synthesis of the negative-sense RNA strand from the positive-sense RNA genome.[6], [7]
RNA recombination may be advantageous to viruses through a variety of proposed mechanisms including the generation of viral escape mutants, the removal of deleterious mutations, the mating of viral genes and genomes, or the capturing of other viral or host genes. As a consequence, RNA recombination contributes to the extraordinary and rapid adaptability of RNA viruses and has been attributed to be the source of a number of recent outbreaks of, for example, dengue virus and echovirus[8], [9], [10] as well as the emergence of entirely new viruses, including severe acute respiratory syndrome-associated coronavirus.11
Conversely, viral RNA recombination may also facilitate the stepwise generation of defective RNAs.12 Defective RNAs have lost their ability to independently encode functional viral proteins but maintain the sequence motifs required for RNA replication by the viral RNA-dependent RNA polymerase (RdRp) and so are able to proliferate parasitically. Additionally, defective RNAs are often packaged into viral particles and so may contain the necessary motifs required for encapsidation. If a defective RNA is able to attenuate the replication of the full-length RNA genome through competition for access to the RdRp, then they are known as defective-interfering RNAs (DI-RNAs).13 DI-RNAs arise during both persistent and acute infections most commonly in cell culture[14], [15] but also occur during wild infections.[16], [17]
Flock house virus (FHV) is an icosahedral T = 3 non-enveloped virus containing a positive-sense, single-stranded RNA genome. It is the prototypic member of the Nodaviridae family and provides an important model for the study of many pathogenic human viruses including poliovirus, human rhinovirus, and hepatitis C virus (reviewed in Refs. 18 and 19). FHV naturally infects insects including Drosophila melanogaster as well as medically important insects such as mosquitoes (including Anopheles gambiae) and the tsetse fly.20 The FHV genome consists of two positive-sense, single-stranded RNAs. RNA1 (3.1 kb) encodes the RdRp, which can autonomously replicate the FHV genome. RNA2 (1.4 kb) encodes the capsid precursor protein, α, 180 copies of which form the viral capsid. FHV packages one copy each of RNA1 and RNA2, but on average, nearly 2% of the packaged RNA is derived from transcripts of the host genome.21 Additionally, virus-like particles (VLPs) of FHV can be made in cell culture by expressing the single capsid protein from a baculoviral vector in Sf21 cells.22 VLPs spontaneously assemble into particles that are closely similar to native FHV, except that they package ribosomal RNAs, transcripts derived from the baculoviral expression vector and other cellular RNAs.21
Next-generation sequencing (NGS) has been used in multiple investigations to assess the polymorphism and mutation frequency within a viral genome, for example, in human immunodeficiency virus,23 human rhinovirus,24 and foot-and-mouth disease virus.25 With NGS technology, it is possible to detect fusion events that may occur during RNA/DNA recombination and has been used to characterize mRNA splicing events26 and to identify oncogenic chromosomal rearrangements.[27], [28], [29] Here, we use NGS to investigate the sequence variation as a result of RNA recombination within the genome encapsidated by FHV. We profile with single-nucleotide resolution hundreds of thousands of recombination events that have occurred during FHV genome replication and which have subsequently been encapsidated into virus particles.
Results and Discussion
Data acquisition
FHV virions and VLPs were generated in cell culture and purified with multiple sucrose gradients as is well established for structural and biochemical studies.30 We thus obtained highly purified viral RNA for RNAseq analysis by virtue of its encapsidation inside FHV virions and VLPs. We did not employ any additional RNA-selection steps, such as PCR or poly-A capture, which would yield a non-random population of RNA molecules. Moreover, the RNA encapsidated in FHV virions is directly relevant to virus infection as it is what may be delivered to another host cell during an infection.
The purified RNAs were fragmented and prepared for sequencing analysis using Illumina protocols for obtaining millions of single short reads. We thus obtained two raw data sets corresponding to the RNA encapsidated by FHV virions and VLPs—hereafter termed the FHV-RNAseq and VLP-RNAseq data sets. We applied a stringent quality filter to our data sets, as described in Materials and Methods, obtaining a total of 28.9 millions and 19.6 millions reads all exactly 95 nt in length for the FHV-RNAseq and VLP-RNAseq data sets, respectively.
Mapping of RNAseq reads to known encapsidated RNAs
To establish the identity of the RNA reads in the data sets, we mapped single reads using the Bowtie aligner31 to genes already known to be present in either the FHV virions or VLPs. We have previously thoroughly characterized the contents of FHV virions and VLPs and so used these sequences as references here.21 Reads were mapped in an end-to-end and un-gapped manner. The number of mapped reads for both data sets is shown in Table 1 . FHV virions packaged roughly equal stoichiometric amounts of FHV RNA1 and RNA2. Out of the 24.6 millions reads that aligned to the FHV genome, only 4 of these mapped to the negative-sense RNA transcript. In addition to the FHV genome, we also detected some encapsidated host RNA, as we have recently reported.21 VLPs primarily packaged host RNA, most of which consisted of ribosomal RNA. VLPs also packaged transcripts from the baculoviral expression vector, which included the FHV RNA2 gene encoding the FHV capsid protein. We also detected a small amount of FHV RNA1 in our VLP-RNAseq (1103 reads), indicating that there was a low-level persistent infection with FHV in our Sf21 cells, as has also been reported for a number other cell lines.[21], [32], [33], [34] Consequently, we would also expect approximately 300 of the FHV RNA2 reads from the VLP-RNAseq data set to be present as a result of FHV replication (judging from the stoichiometric ratios of FHV RNA1 and RNA2 seen in the FHV-RNAseq data set), which is well below the total number observed in the VLP-RNAseq data set (1.46 millions). Note that the FHV RdRp would not be able to replicate the FHV RNA2 derived from the baculovirus vector as this is cloned without the appropriate 5′ and 3′ untranslated regions required for viral replication.6
Table 1.
Mapping of RNAseq reads
| FHV-RNAseq | % | VLP-RNAseq | % | |
|---|---|---|---|---|
| Totals reads | 28,939,991 | 100 | 19,604,376 | 100 |
| FHV RNA1 | 17,888,032 | 61.8 | 1103 | < 0.1 |
| FHV RNA2 | 6,744,282 | 23.3 | 1,462,744 | 7.5 |
| Host genome | 1,081,696 | 3.7 | 12,510,924 | 63.8 |
| AcMNPVa | — | 1,718,522 | 8.8 | |
| FHV RNA1–RNA1 | 1,167,929 | 4.0 | 82 | < 0.1 |
| Insertions | 19,494 | 0 | ||
| Deletions | 1,106,870 | 0 | ||
| MicroInDels | 41,565 | 82 | ||
| FHV RNA1–RNA2 | 11,068 | < 0.1 | 0 | |
| FHV RNA2–RNA1 | 22,288 | < 0.1 | 0 | |
| FHV RNA2–RNA2 | 33,897 | 0.1 | 2168 | < 0.1 |
| Insertions | 10,872 | 226 | ||
| Deletions | 16,086 | 130 | ||
| MicroInDels | 6939 | 1812 | ||
| Other junctions | 19,044 | 0.1 | ||
| Recombinations | 1220 | |||
| MicroInDels | 17,824 | |||
| Unaligned reads | 1,990,779 | 6.9 | 3,889,789 | 19.8 |
AcMNPV, Autographa californica multiple nuclear polyhedrosis virus (baculovirus expression vector).
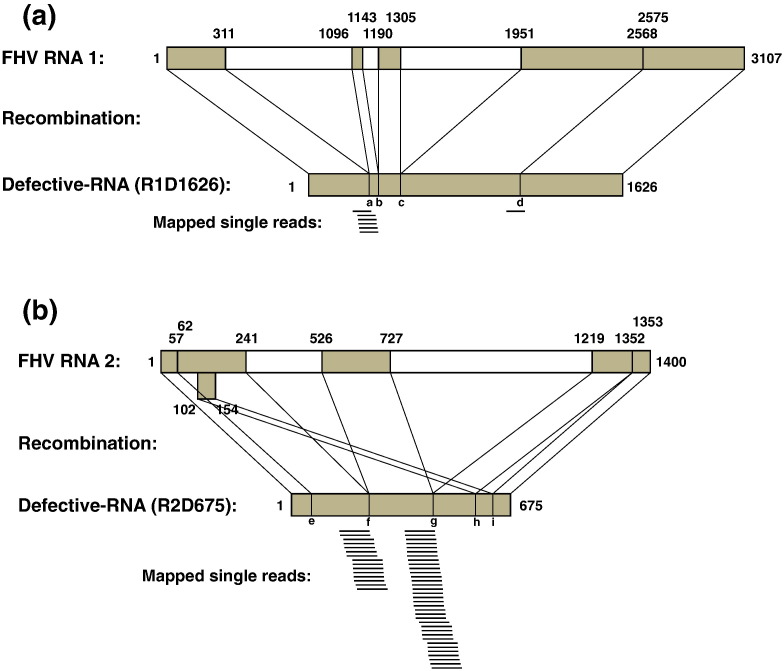
To illustrate the detection of recombination, we mapped the reads to two specific defective RNAs (R2D675 and R1D1626) that are known to arise during passaging of FHV in cell culture.[14], [15] These defective RNAs are formed by recombination events that occur between different portions of the FHV genome as illustrated in Fig. 1 . Many of these result in large deletions of the RNA genome while others result in insertions or duplications (e.g., in R2D675, nucleotides 102–154 are duplicated and reinserted near the 3′ terminus). When these recombination events occur, a junction is generated with a novel nucleic acid sequence. From the FHV-RNAseq data set, 15 single reads mapped to R1D1626, and 101 mapped to R2D675, all of which mapped to junction sites that characterize the respective defective RNA. The locations of the mapped reads are illustrated in Fig. 1. As expected, no reads aligned to the portions of the defective RNAs that maintain a wild-type sequence as these reads were already aligned to the fully intact FHV genome. However, not all junction events were detected, indicating that the precise character of the defective RNAs generated in our sample was different to that previously identified. The mapping of reads over junctions present in known defective RNAs is important to illustrate how individual RNA recombination events within the FHV genome can be detected.
Fig. 1.
Defective RNAs are detected by virtue of their unique recombination junctions. Defective RNAs that have been previously characterized in FHV infections[14], [15] are formed by recombination within. (a) FHV RNA1 to form R1D1626 and (b) FHV RNA2 to form R2D675. These recombination events generate unique junction sequences, labeled a–i for reference in Fig. 2. Reads from the FHV-RNAseq data set were mapped to these sequences and are indicated underneath the schematic for each defective RNA to illustrate their mapping over the recombination junction sites.
A reference pseudo-library containing sequences to all possible recombination events in FHV
We extended our analysis to detect novel recombination events occurring within the FHV genome. To achieve this, we generated a reference pseudo-library containing millions of short reference sequences (≤ 150 nt in length) each corresponding to potential recombination events within the FHV genome. The pseudo-library was generated using a Python script whereby every 75-base sequence from the FHV genome was appended to every other 75-base sequence. We also allowed shorter sequences to be generated for reference sequences corresponding to recombination events occurring near the edges (≥ 20 bp) of the FHV genes. The reference pseudo-library contained 19,965,801 sequences describing junctions within FHV RNA1 and FHV RNA2 as well as between these two. For each reference, the first 75 nt on the 5′ side of the junction site is defined as the 5′ strand, while the 3′ nucleotides form the 3′ strand. Thus, by mapping 95-nt reads to reference sequences 150 nt in length where the recombination junction is placed between bases 75 and 76, a minimum of 20 nt from the single read must align to both the 5′ and 3′ strands. Junctions appearing in either 20-bp extremity of the single reads would not be detected. Consequently, there are 56 of a possible 94 ‘cutting’ sites in the 95-bp single reads where junctions may be detected. While this will result in an underreporting of all possible recombination events, this prevents the possibility of mapping reads with too few nucleotides on one side of a junction to unambiguously assign its identity.
Alignment of the data sets to the pseudo-library for recombination profiling
The single reads from both the FHV-RNAseq and VLP-RNAseq data sets that did not align to the wild-type full-length reference sequences were next mapped to the junction reference library (Table 1). Every read that mapped to the junction reference library mapped to the positive-sense strand. In the FHV-RNAseq data set, we detected a wide variety of junctions within each of the FHV genes. The majority of these were found within RNA1, accounting for approximately 4% of the single reads from the FHV-RNAseq data set and primarily corresponded to large deletions. A smaller number (~ 0.1%) were found in RNA2 and, again, primarily corresponded to deletion events. Junctions that result in effective insertions or duplications of the FHV genes (where the 5′ strand is downstream of the 3′ strand) were observed. Junctions were also detected between FHV RNA1 and RNA2, indicating that RNA recombination between non-homologous templates has occurred, although these events are less frequent than those within RNA1 or RNA2 despite there being a greater number of possible recombination events that can be mapped.
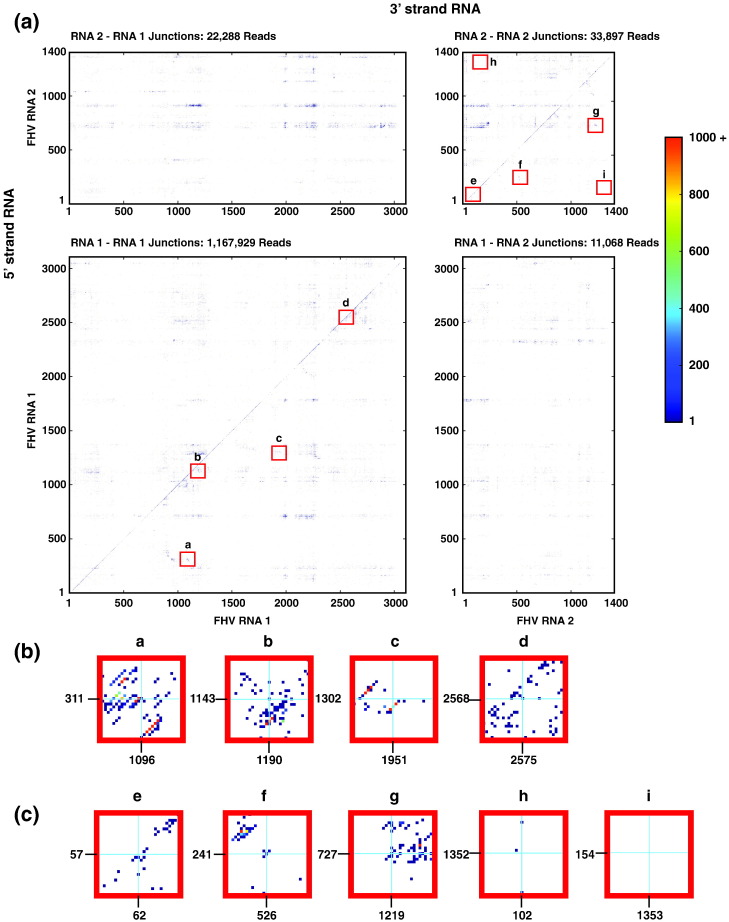
These data are represented as heat maps in Fig. 2 . Here, the y-axis describes the last nucleotide of the 5′ strand and the x-axis describes the first nucleotide of the 3′ strand. The number of reads that map to each particular junction is indicated with a color bar. The heat maps illustrate the wide array of junctions detected, reflecting the diversity of the packaged RNA as a result of recombination.
Fig. 2.
Recombination events are widely detected throughout the FHV genome as illustrated using heat maps. (a) Heat maps show the location of junctions within RNA1, within RNA2, or between RNA1 and RNA2. The y-axis corresponds to the last nucleotide of the mapped 5′ strand and the x-axis corresponds to the first nucleotide of the 3′ strand. The number of reads mapping to each event is indicated with a color bar. The red boxes labeled a–i indicate the positions of the junctions in the defective RNAs illustrated in Fig. 1 and are enlarged in (b) for RNA1 and in (c) for RNA2 to show the mapping in these locations. Blue cross-hairs indicate the precise position of the expected junctions.
Prominent horizontal and vertical striations are visible in the heat maps in Fig. 2. Interestingly, the locations of the striations are maintained regardless of the identity of the recombination partner; for example, the horizontal striation occurring between nucleotides 710 and 730 in RNA2 is present regardless of whether RNA2 or RNA1 provides the 3′ strands. These striations may therefore indicate a sequence-dependent preference in the selection of recombination sites. It is interesting to note that some of these striations pass through locations of previously characterized defective RNAs[14], [15] as well as locations of high-frequency events detected here. The positions of the horizontal and vertical striations do not correlate (Pearson coefficients are − 0.0035 and − 0.0019 for RNA1 and RNA2 recombinations, respectively). This indicates that the sequence-dependent selection of 5′ strands differs from that of 3′ strands. However, no apparent nucleotide preference at either strand could be detected, other than a weak underrepresentation of guanines at the junction sites (Fig. S1). Additionally, there was no average sequence identity detected between nucleotides upstream of the junction site in the 5′ strand and nucleotides upstream of the 3′ strand, as has been reported for defective RNAs, for example, in dengue virus.16 The selection of recombination sites may therefore have a more complex origin, for example, in the character of the local RNA secondary structure.6
The positions in the heat map that correspond to mapping to the defective RNA illustrated in Fig. 1 are shown in Fig. 2b and c. The precise sites of the junctions corresponding to the previously identified defective RNAs are marked with blue cross-hairs and are relatively poorly represented (the reads indicated under the cross-hairs in the heat map are the same reads illustrated in Fig. 1). However, we detected two high-frequency junctions nearby that join nucleotides 249–517 and nucleotides 730–1229 (8544 and 1080 mapped reads, respectively—these can be seen in Fig. 2c insets f and g, respectively) and are two highest-frequency events detected within RNA2. This indicates that a different yet similar population of defective RNAs was favored in our virus sample.
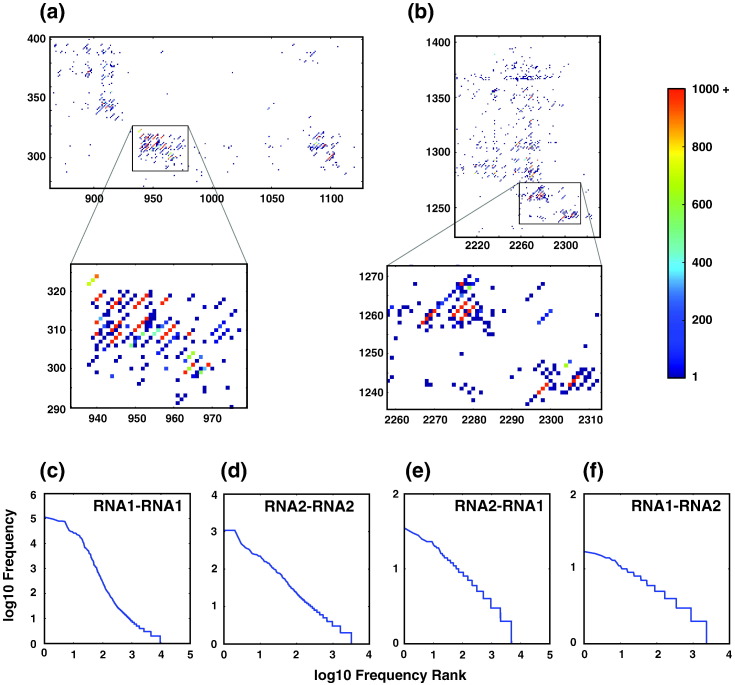
We detected many high-frequency events in FHV RNA1 (58 unique junctions with > 1000 mapped reads). Only one of these resulted in an effective insertion event (from nucleotide 1605 back to nucleotide 1087). The remainder resulted in deletions ranging from 13 to 1063 nt in length. Figure 3 shows two regions of RNA1 to RNA1 recombination that are enriched with high-frequency events (349,194 reads in Fig. 3a and 224,347 reads in Fig. 3b). Interestingly, almost the entirety of these events resulted in deletions exactly 3n nucleotides in length (99% of the reads shown). As a result, the open reading frame (ORF) was maintained in each of these cases, suggesting that the RNA arising from these recombination events has been selected by virtue of their ability to produce a viable protein product. This may be either because RNA encapsidation is coupled to translation (as has been demonstrated for FHV35) or because a functional, yet truncated, form of the viral RdRp is being selected.
Fig. 3.
High-resolution portions of the heat maps demonstrate regions containing multiple high-frequency recombination events. (a) and (b) show two regions of RNA1 that are highly enriched in recombination events. This reveals clusters of high-frequency events that potentially represent defective RNAs. Log–log plots of the ranked frequencies of unique recombination events between (c) FHV RNA1 and FHV RNA1, (d) FHV RNA2 and FHV RNA2, (e) FHV RNA2 and FHV RNA1, and (f) FHV RNA1 and FHV RNA2 indicate that their distribution is heavy tailed.
The high frequency of these recombination events suggests that they are present due to successive rounds of replication rather than to independent instances of RNA recombination. This is supported by the fact that the distribution of frequencies with which individual recombination events were detected is heavy tailed, reminiscent of a power-law distribution (Fig. 3c–f). This indicates that there is an overrepresentation of a small number of unique recombination events. Heavy-tailed distributions can arise when events are initially randomly generated, but some selected events are favorably duplicated,36 as has been observed for a number of replicable components of eukaryotic and prokaryotic genomes.37 Such a scenario is what we would expect during the generation of defective RNAs, which are initially generated by stochastic RNA recombination but are subsequently highly (sometimes competitively) replicated by viral RdRps.12
Detection of sequence reads spanning two recombination events
Many of the recombination events that we detected above occurred within close proximity to one another and it is likely that many of these were present on the same original defective RNA molecule. Consequently, single reads would be present in our data set that will span two recombination events and thus would not be able to map to our pseudo-library containing reference sequence with only single recombination events. To address this, we designed a second pseudo-library of reference sequences that contained combinations of two previously detected recombination events that occurred within close proximity. As before, we designed this library to enforce the single reads to map with at least 20 nt on the 5′ and 3′ sides of the recombination events. Consequently, the maximum distance allowed between junction sites was 55 nt (95 nt from a single read minus two 20-nt ‘seeds’) and we allowed a minimum of 5 nt. This second pseudo-library contained 19,379,090 reference sequences ranging from 95 to 145 nt in length. From the 1,990,779 reads that remained unaligned (Table 1), we mapped an additional 80,374 reads to RNA1 recombinations and 58 reads to RNA2 recombinations. As each reference contained two recombination junctions, this corresponds to an extra 160,748 and 116 junction sites detected in RNA1 and RNA2 respectively.
VLP-RNAseq control demonstrates a low level of artifactual recombination in non-replicated RNAs
We also evaluated the VLP-RNAseq data set for the presence of recombination events. The VLPs were generated by expressing the capsid protein (FHV RNA2) from a baculoviral expression vector. In the absence of viral RNA replication, VLPs package host RNA and RNA transcripts from the baculoviral expression vector (Table 1), which are transcribed by the host DNA-dependent RNA polymerases. We would not therefore expect any junctions to be present in the VLP-RNAseq data set as a result of recombination during viral RNA replication. The VLP-RNAseq data set was generated using the same procedure as for the FHV-RNAseq data set. Consequently, by searching for junctions within the VLP-RNAseq data set, we can estimate the amount of recombination that has occurred during PCR steps in the cDNA library preparation used for RNAseq, as is a recognized artifact.38
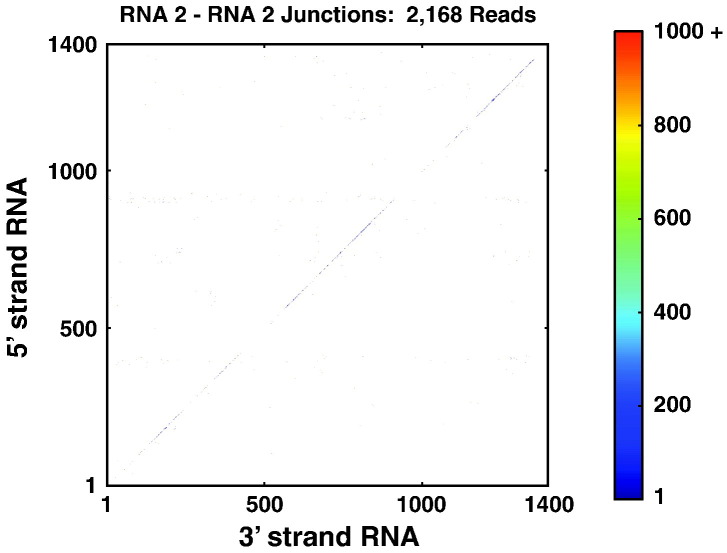
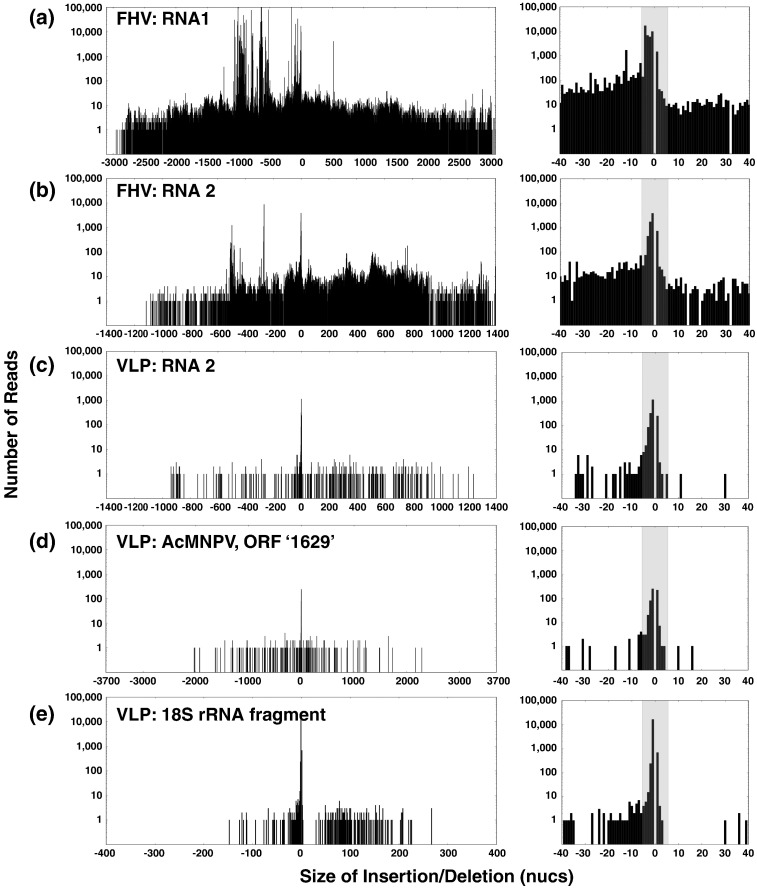
From the VLP-RNAseq data set, 82 reads mapped to RNA1–RNA1 junctions (note that RNA1 was present in very small quantities due to a potential low-level persistent infection of Sf21 cells with FHV) and 2168 reads mapped to RNA2–RNA2 junctions (Fig. 4 and Table 1). A prominent diagonal striation is visible, spanning the heat map of RNA2–RNA2 junctions. These events correspond to very short insertions and deletions. A histogram of the lengths of insertions and deletions that occurs due to each recombination event (Fig. 5 ) shows that insertions and deletions 5 nt or shorter (known as MicroInDels) are abundant in the VLP-RNAseq data set as well as in the FHV-RNAseq data set. This indicates that their formation was not unique to the FHV viral polymerase but was most likely to have arisen during the amplification of the cDNA library used for sequencing. DNA polymerases are known to accrue MicroInDels[39], [40] and have been reported to be abundant in other NGS data sets.[41], [42] Consequently, we exclude all MicroInDels when counting and comparing recombination events between the FHV-RNAseq and VLP-RNAseq data sets, as indicated in Table 1. Excluding these MicroInDels, only 356 reads from the VLP-RNAseq data set mapped to recombination events in FHV RNA2 (none were detected in FHV RNA1). In contrast, we detected 26,958 junctions in FHV RNA2 within the FHV-RNAseq data set.
Fig. 4.
Few recombination events are detected in the control VLP-RNAseq data set. A heat map similar to those illustrated in Fig. 2 of the junctions detected in the VLP-RNAseq data set demonstrate that the majority of events correspond to MicroInDels (N = 1812), evident as the strong diagonal striation. Other artifactual junctions were also detected but with low frequency (N = 356).
Fig. 5.
Frequencies of insertions and deletions of a defined length. Histograms of the sizes of insertions or deletions formed by recombination events are shown. Recombinations are (a) within FHV RNA1 for the FHV-RNAseq data set, (b) within FHV RNA2 for the FHV-RNAseq data set, (c) within FHV RNA2 for the VLP-RNAseq data set, (d) within AcMNPV ORF ‘1629’ for the VLP-RNAseq data set, and (e) within a portion of the 18S rRNA for the VLP-RNAseq data set. Frequencies of insertions and deletions are indicated in the y-axis and their size is indicated on the x-axis (negatives are deletions). Insets on the right show blown-up regions of insertions and deletions ≤ 40 nt in length. The gray-shaded areas illustrate events corresponding to MicroInDels.
In addition to the FHV genome, we also made reference libraries for junctions occurring within two other highly abundant genes that were present in the VLP-RNAseq data set: ORF ‘1629’ from the baculovirus expression vector and a portion of the 18S rRNA. Together with FHV RNA2, these transcripts made up a similar proportion of the VLP-RNAseq data set (4.19M of 19.6M reads = 21.3%) as FHV RNA2 alone did in the FHV-RNAseq data set (6.75M of 28.9M reads = 23.4%). They thus provide a suitable control against which to compare the FHV-RNAseq data set. We detected 1220 reads that mapped to recombination events occurring within these genes and between these genes (Table S1), thus giving a total of 1576 artifactual recombination events per 4.19M mapped reads. This would be equivalent to 2539 reads per 6.75M mapped reads. Excluding the double recombinations, we detected 26,958 recombination events within FHV RNA2 (6.75M mapped reads) and 1,167,929 reads within FHV RNA1 (17.9M mapped reads) and a total of 33,356 between these two, which is clearly in great excess of the artifactual recombination detected in the VLP-RNAseq data set. We can therefore be confident that the background recombination noise in our FHV-RNAseq data set is low and that the recombination events observed in the FHV-RNAseq data set primarily reflect the activity of the viral RdRp.
Mutational frequency in the FHV genome
The coverage of reads across the wild-type FHV genome was not constant (Fig. S2), as is common in RNAseq studies owing to PCR-mediated bias. Consequently, the number of detected junctions over FHV RNA1 and RNA2 were normalized by dividing the number of 5′ strand recombination events or 3′ strand recombination events by the number of wild-type reads that mapped at each nucleotide position to obtain the frequencies of recombination events. From this, we can make an estimate of the average recombination frequency across the FHV genome (Table 2 ). This will reflect both the frequency of individually generated recombination events and the replication of junctions that are found in the defective RNAs. However, the observed frequencies provided a metric with which to evaluate the amount of genetic variety in the virus sample. As these frequencies are likely to be inflated at the extremities of each gene due to the low coverage of wild-type reads in these regions (in particular at the 3′ terminus) (Fig. S2), we also show the rates over just the ORFs of each gene. These values indicate that RNA recombination is an abundant source of mutagenesis and is comparable in magnitude to that of mismatch mutation (Table 2).
Table 2.
Mutation frequency across the FHV genome
| Recombination at 5′ strand | Recombination at 3′ strand | Mismatch mutation | |
|---|---|---|---|
| FHV RNA1 | 29.9 × 10− 4 | 4.8 × 10− 4 | 14.4 × 10− 4 |
| (ORF only) | (28.3 × 10− 4) | (4.9 × 10− 4) | (12.9 × 10− 4) |
| FHV RNA2 | 11.0 × 10− 4 | 73.3 × 10− 4 | 10.5 × 10− 4 |
| (ORF only) | (0.8 × 10− 4) | (7.8 × 10− 4) | (16.7 × 10− 4) |
| VLP RNA2 | 0.087 × 10− 4 | 0.2 × 10− 4 | 4.4 × 10− 4 |
| (ORF only) | (0.095 × 10− 4) | (0.15 × 10− 4) | (4.6 × 10− 4) |
As we are using RNAseq with single short reads, we are unable to detect recombination events that occur between two homologous templates but at identical sites as this would result in the conservation of the local nucleic acid sequence. However, the high rate of RNA recombination that we do detect suggests that such ‘silent recombination’ will also be abundant. Indeed, such a process would be highly important by allowing for the mating of homologous templates, potentially removing deleterious mutations, or by allowing the reshuffling of advantageous mutations that have occurred on separate RNA molecules.
Conclusions
NGS has proven itself to be a valuable tool in assessing the mutational landscape of a viral genome. NGS has been used to map the positions of single-nucleotide polymorphisms in a viral population and to measure the frequency of mismatch mutation. Both of these are a source of considerable diversity within the ‘genetic cloud’ of a viral genome and are used to characterize the quasi-species present in a virus sample. Here, we have laid out an approach that extends these capabilities to include RNA recombination. By mapping the position and frequency of every possible junction within the genome of FHV, we present a highly detailed and complex landscape of the numerous recombination events that occur during viral RNA replication.
The strategy laid out in this article could equally be applied to a wide range of virus samples, including DNA viruses, and could add to our understanding of their diversity and evolution. The frequency of RNA recombination is known to vary widely between viral species, even among different positive-sense RNA virus, and could be assessed using NGS. The frequency of RNA recombination could also be compared between different preparations of the same virus, for example, during the course of an infection, or when amplified in different host cells for viruses that have a broad host range (e.g., dengue virus) or when viruses are exposed to antiviral therapies (e.g., ribavirin treatment for hepatitis C virus). Additionally, it would also be possible to search for recombination between different viral species during co-infections. This would be important for understanding the role of RNA recombination in the evolution of new viruses.
Our approach could also be used to discover and characterize novel defective RNAs and DI-RNAs potentially present in a variety of infectious viruses. The generation of DI-RNAs has been proposed to be a critical stage in the transition of acute to chronic viral infections43 and DI-RNAs have been found in patients persistently infected with measles virus,44 dengue virus,45 and hepatitis C virus.46 Characterizing DI-RNAs present even in very low titers may improve our understanding of viral infections and help identify variations of such elements between individuals or host organisms or during the progression of a viral infection. Additionally, characterizing what portions of the virus genome are present in the DI-RNAs will help us understand which components of the genome are necessary for replication by the viral polymerases. Similarly, by analyzing the nucleic acids packaged inside viruses, we may potentially find which components of the genome are required for successful encapsidation. Finally, therapeutic applications could be envisioned as discovering the identity of DI-RNAs may allow for their exploitation for treatment or prevention of acute viral infections. Such applications have been demonstrated, for example, in the form of deliverable vaccines47 or through the transgenic expression of DI-RNA-like molecules.48
Materials and Methods
Virus and VLP preparation
For authentic FHV production, cultured S2 cells were grown in Schneider's media (Sigma) containing 15% fetal bovine serum (Gibco) using standard laboratory procedures. Cells were concentrated to 4 × 107 cells/ml, infected with FHV at a multiplicity of infection of 1 and rocked for 1 h at room temperature. The cells were then diluted with Schneider's insect media to a final concentration of 8 × 106 cells/ml and incubated at 27 °C in a rotary shaker. Cells were harvested 2 days postinfection. For the production of VLPs of FHV, Sf21 cells were cultured in TC-100 media (Invitrogen) supplemented with 10% fetal bovine serum (Gibco) using standard laboratory procedures. FHV RNA2 was expressed from the pBacPAK9 baculovirus vector as previously described.22 Cells were harvested 3 days posttransformation.
Authentic virions and VLPs were purified using a series of sucrose gradients as is well established30 in 50 mM Hepes, pH 7.0. Clarified cell lysates were spun at 40,000 RPM for 2.5 h onto a 30% sucrose cushion. The pellet was resuspended and then applied to a 10–40% sucrose gradient and spun at 40,000 RPM for 1.5 h. Fractions from the sucrose gradient were removed and analyzed by SDS-PAGE. Fractions containing only viral capsid proteins were pooled. This sample was then supplemented with 10 × DNase I reaction buffer (NEB), 20 U of DNase I (NEB), and 0.5 μg of RNase A (Roche) and incubated at room temperature for 2 h to remove any non-encapsidated co-purified DNA or RNA. The samples were then extensively washed with 50 mM Hepes, pH 7.0, on a 100-kDa molecular mass cutoff centrifugal concentrator. This sample was then applied to the top of a second 10–40% sucrose gradient and spun at 40,000 RPM for 1.5 h. Again, fractions from the sucrose gradient were removed and analyzed by SDS-PAGE. Fractions containing only viral capsid proteins were pooled and extensively washed on a 100-kDa molecular mass cutoff centrifugal concentrator. After this extensive purification, no cellular proteins could be detected by Coomassie stain on an SDS-PAGE gel and no RNA or DNA could be detected on a native agarose gel when loading 3 μg of virus.
RNA preparation
Purified FHV or VLPs were disrupted at room temperature by incubation in 0.1% SDS and 0.1 M NaCl for 15 min. RNA was extracted from the disrupted particles using an equal volume of acid phenol followed by three washes with 100% chloroform. RNA was then ethanol precipitated in the presence of 100 mM sodium acetate, pH 5.3. RNA pellets were washed in 70% ethanol, dried, and resuspended in pure water.
Directional RNAseq
RNA (0.4 μg) was prepared for NGS using a modified version of the Illumina protocol† where 12 cycles of PCR were performed and standard TruSeq adapters and TruSeq barcoded primers were used. A final size selection was performed by native agarose gel electrophoresis to yield a library of inserts approximately 200 bases in length suitable for 100-base single-read sequencing. The library was extracted from the agarose gel using standard oligo purification columns. The prepared library was then loaded onto an Illumina HiSeq v3 single-read flowcell, standard cluster generation was performed on a Cbot, and sequencing was performed for 100 bases of the insert and 7 bases of the index read using standard HiSeq sequencing reagents on an Illumina HiSeq 2000 instrument. Reads were processed using CASAVA 1.8.2 and demultiplexed based on index sequences.
Read quality filtering
Reads containing any fragment of the 3′ TruSeq adapter were detected and trimmed using cutadapt‡ 49 with default settings. Reads containing any base with a PHRED score < 20 were discarded using the FASTX toolkit§ . The quality of the reads in the data set was assessed using the FastQC package‖ . This revealed a poor average base-calling quality in the final 5 nt of each read. Consequently, each read was trimmed down from the 3′ end to a total of 95 nt in length. Shorter reads were discarded.
Read mapping
The D. melanogaster reference genome r5.22 was downloaded from the FlyBase repository¶ , and the mRNA refSeq library was obtained from the University of California Santa Cruz genome browser websitea . ESTs from Spodoptera frugiperda cell lines were downloaded from ‘Spodobase’b .50 Sequences for the FHV RNA2 (NC_004144), FHV RNA1 (NC_004146), the Attacus ricini 45 s rDNA (AF463459), the baculovirus genome (NC_001623), and defective RNAs (GU393238 and GU393241GU393238GU393241) were obtained from the National Center for Biotechnology Information. Reads were aligned to the host genome reference using the Bowtie alignment package version 0.12.731 c in –v mode, tolerating up to three mismatched nucleotides per 95-base read. Alignment files were processed using SAMtools51 d and alignments were visualized and inspected using Tablet.52
Reads were mapped to the FHV genome using Bowtie parameters -v 2 --best. Junctions were detected by alignment of the remaining reads using Bowtie parameters -v 2 --best to a library of sequences corresponding to all possible recombination events in the FHV genome as described in the main text. Reads that mapped with mismatches and that mapped to the edges of the reference sequences were removed from the alignment (from the .sam file). This is because the mismatch tolerance can allow a read to map with fewer than the required 20 nt at either the donor or acceptor strand by claiming mismatching at the junction site of an adjacent but incorrect reference.
Accession numbers
The FHV-RNAseq and VLP-RNAseq data sets are available online at the National Center for Biotechnology Information Small Reads Archive with accession number SRP013296.
Acknowledgements
We thank Steven Head for advice with NGS. We thank Madan Babu for advice and discussions. We thank David Veesler and Tatiana Domitrovic for proofreading and discussions. We thank Andrew Ball and Anette Schneemann for critically reading the manuscript. This work was funded by National Institutes of Health grant R37-GM034220 to J.E.J. A.R. is supported by a European Molecular Biology Organization Long-Term Fellowship, ALTF 573-2010.
Edited by J. Weissman
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.10.005
Appendix A. Supplementary Data
Supplementary materials
References
- 1.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chetverin A.B., Chetverina H.V., Demidenko A.A., Ugarov V.I. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallei A., Pankraz A., Thiel H.J., Becher P. RNA recombination in vivo in the absence of viral replication. J. Virol. 2004;78:6271–6281. doi: 10.1128/JVI.78.12.6271-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Ball L.A. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 1993;67:3854–3860. doi: 10.1128/jvi.67.7.3854-3860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarvis T.C., Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oprisan G., Combiescu M., Guillot S., Caro V., Combiescu A., Delpeyroux F., Crainic R. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 2002;83:2193–2200. doi: 10.1099/0022-1317-83-9-2193. [DOI] [PubMed] [Google Scholar]
- 9.Worobey M., Rambaut A., Holmes E.C. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl Acad. Sci. USA. 1999;96:7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes E.C., Worobey M., Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 1999;16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- 11.Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White K.A., Morris T.J. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 1994;68:14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak K.B., Nagy P.D. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses. 2009;1:895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jovel J., Schneemann A. Molecular characterization of Drosophila cells persistently infected with Flock House virus. Virology. 2011;419:43–53. doi: 10.1016/j.virol.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball L.A., Li Y. cis-Acting requirements for the replication of flock house virus RNA 2. J. Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D., Lott W.B., Lowry K., Jones A., Thu H.M., Aaskov J. Defective interfering viral particles in acute dengue infections. PLoS One. 2011;6:e19447. doi: 10.1371/journal.pone.0019447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesko K.N., Fitzpatrick K.A., Ryan E.M., Shi P.Y., Zhang B., Lennon N.J. Internally deleted WNV genomes isolated from exotic birds in New Mexico: function in cells, mosquitoes, and mice. Virology. 2012;427:10–17. doi: 10.1016/j.virol.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 19.Odegard A., Banerjee M., Johnson J.E. Flock house virus: a model system for understanding non-enveloped virus entry and membrane penetration. Curr. Top. Microbiol. Immunol. 2010;343:1–22. doi: 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta R., Free H.M., Zietlow S.L., Paskewitz S.M., Aksoy S., Shi L. Replication of flock house virus in three genera of medically important insects. J. Med. Entomol. 2007;44:102–110. doi: 10.1603/0022-2585(2007)44[102:rofhvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Routh A., Domitrovic T., Johnson J.E. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc. Natl Acad. Sci. USA. 2012;109:1907–1912. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneemann A., Dasgupta R., Johnson J.E., Rueckert R.R. Use of recombinant baculoviruses in synthesis of morphologically distinct viruslike particles of flock house virus, a nodavirus. J. Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C., Mitsuya Y., Gharizadeh B., Ronaghi M., Shafer R.W. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordey S., Junier T., Gerlach D., Gobbini F., Farinelli L., Zdobnov E.M. Rhinovirus genome evolution during experimental human infection. PLoS One. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright C.F., Morelli M.J., Thebaud G., Knowles N.J., Herzyk P., Paton D.J. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J. Virol. 2011;85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Salzberg S.L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan K., Wang L., Wang J., Ittmann M.M., Li W., Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc. Natl Acad. Sci. USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bass A.J., Lawrence M.S., Brace L.E., Ramos A.H., Drier Y., Cibulskis K. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher C.A., Kumar-Sinha C., Cao X., Kalyana-Sundaram S., Han B., Jing X. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneemann A., Marshall D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 1998;72:8738–8746. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Q., Luo Y., Lu R., Lau N., Lai E.C., Li W.X., Ding S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl Acad. Sci. USA. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onions D., Cote C., Love B., Toms B., Koduri S., Armstrong A. Ensuring the safety of vaccine cell substrates by massively parallel sequencing of the transcriptome. Vaccine. 2011;29:7117–7121. doi: 10.1016/j.vaccine.2011.05.071. [DOI] [PubMed] [Google Scholar]
- 34.Li T.C., Scotti P.D., Miyamura T., Takeda N. Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 2007;81:10890–10896. doi: 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venter P.A., Krishna N.K., Schneemann A. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 2005;79:6239–6248. doi: 10.1128/JVI.79.10.6239-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barabasi A.L., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 37.Luscombe N.M., Qian J., Zhang Z., Johnson T., Gerstein M. The dominance of the population by a selected few: power-law behaviour applies to a wide variety of genomic properties. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0040. RESEARCH0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorzer I., Guelly C., Trajanoski S., Puchhammer-Stockl E. The impact of PCR-generated recombination on diversity estimation of mixed viral populations by deep sequencing. J. Virol. Methods. 2010;169:248–252. doi: 10.1016/j.jviromet.2010.07.040. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 40.Shinde D., Lai Y., Sun F., Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 2003;31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albers C.A., Lunter G., MacArthur D.G., McVean G., Ouwehand W.H., Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang A.S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 44.Cattaneo R., Schmid A., Eschle D., Baczko K., ter Meulen V., Billeter M.A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aaskov J., Buzacott K., Thu H.M., Lowry K., Holmes E.C. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- 46.Noppornpanth S., Smits S.L., Lien T.X., Poovorawan Y., Osterhaus A.D., Haagmans B.L. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 2007;81:12496–12503. doi: 10.1128/JVI.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mann A., Marriott A.C., Balasingam S., Lambkin R., Oxford J.S., Dimmock N.J. Interfering vaccine (defective interfering influenza A virus) protects ferrets from influenza, and allows them to develop solid immunity to reinfection. Vaccine. 2006;24:4290–4296. doi: 10.1016/j.vaccine.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Lyall J., Irvine R.M., Sherman A., McKinley T.J., Nunez A., Purdie A. Suppression of avian influenza transmission in genetically modified chickens. Science. 2011;331:223–226. doi: 10.1126/science.1198020. [DOI] [PubMed] [Google Scholar]
- 49.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 50.Negre V., Hotelier T., Volkoff A.N., Gimenez S., Cousserans F., Mita K. SPODOBASE: an EST database for the lepidopteran crop pest Spodoptera. BMC Bioinformatics. 2006;7:322. doi: 10.1186/1471-2105-7-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials