Abstract
Four custom Axiom genotyping arrays were designed for a genome-wide association (GWA) study of 100,000 participants from the Kaiser Permanente Research Program on Genes, Environment and Health. The array optimized for individuals of European race/ethnicity was previously described. Here we detail the development of three additional microarrays optimized for individuals of East Asian, African American, and Latino race/ethnicity. For these arrays, we decreased redundancy of high-performing SNPs to increase SNP capacity. The East Asian array was designed using greedy pairwise SNP selection. However, removing SNPs from the target set based on imputation coverage is more efficient than pairwise tagging. Therefore, we developed a novel hybrid SNP selection method for the African American and Latino arrays utilizing rounds of greedy pairwise SNP selection, followed by removal from the target set of SNPs covered by imputation. The arrays provide excellent genome-wide coverage and are valuable additions for large-scale GWA studies.
Keywords: Microarray, Genome-wide association study, Coverage, Imputation, Single nucleotide polymorphism, Throughput
1. Introduction
Genome-wide association (GWA) studies have produced a large number of replicated novel genetic variants [1–4] for many diseases for which no variants had been previously found. The success of these studies has been a result of high-throughput genotyping platforms assaying hundreds of thousands to a million SNPs, with large sample sizes leading to an increased number of replicated associations [5,6]. Many of these have focused on common genetic variation (MAF (minor allele frequency) of 0.10 or greater), based on the HapMap catalog [7]. Sequencing projects, particularly the 1000 Genomes Project (KGP) (http://www.1000genomes.org), are developing larger catalogs which can be leveraged to design arrays that assay lower frequency variants, further enabling discovery of disease-associated genetic variations.
Here we describe the development of three new microarrays for the Axiom Genotyping Solution tailored to individuals of East Asian, African American, and Latino race/ethnicity. These are the remaining three of four custom microarrays developed for the genome-wide genotyping analysis of 100,000 participants in the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH). A description of the genotyping project and RPGEH cohort is included in [8]. Axiom arrays are limited to approximately 700,000 SNPs when SNPs are tiled with two replicates, which is the standard. Budget constraints for this project allowed for the genotyping of either a single array on 100,000 individuals or two arrays (up to 1.4 million SNPs) on 50,000 individuals. We opted to genotype 100,000 individuals with a single array. As a consequence, however, we chose to design four different arrays to maximize genome-wide coverage, especially for lower frequency variants, in each of the major US race/ethnicity groups (African Americans, East Asians, Latinos and Whites) represented in the RPGEH cohort.
The design of the first array in the series, optimized for US whites (designated EUR), has been described [8]. The East Asian (EAS) array was designed for individuals of East Asian ancestry, although we also included SNPs to provide coverage of European-specific variants to accommodate some RPGEH subjects with mixed East Asian/European ancestry. The target set for the African American (AFR) array included both West African and European variants, recognizing the mixed ancestry of African Americans. Because Latinos have ancestry from three continents, we targeted SNPs common and specific to Europeans, West Africans and Native Americans for the Latino (LAT) array. These arrays were developed to maximize the number of high resolution SNPs for genome-wide coverage; to saturate regions previously identified as disease associated from prior GWA studies for both replication and fine mapping; to improve coverage of both common and uncommon variants by making use of data from the low pass and high pass phases of the KGP; and to incorporate redundant coverage of SNPs with known strong disease associations [8]. For the EAS, AFR and LAT arrays, we used several approaches to enhance the overall genome-wide coverage, including modification to the SNP selection algorithm and reduction of the number of replicates for some SNPs on the array to create more space for additional SNPs.
There have been several methods proposed for SNP selection, starting with a greedy pairwise correlation (“tagging”) algorithm [9]. There have also been efforts to extend pairwise tagging to tagging using multi-marker correlations to increase efficiency [10]. However, to our knowledge, less has been done with imputation for tagging, aside from using it to tag singleton SNPs [8].
Imputation has played a major role in the analysis of genome-wide association data [11]; here we explore its use in the design of genotyping microarrays. Imputation of missing SNPs using HapMap reference samples can lead to an overall increase in power of up to 10% [12], and is becoming possible with larger sequenced reference panels, e.g., from the KGP. Simulations show that imputation is potentially the most beneficial for rare variants, which are harder to tag with a single marker [13]. Several papers that imputed all variants in HapMap found significant associations with imputed SNPs that would not have been found by analyzing only the SNPs on the GWA array [14]. Motivated by this analysis strategy of imputing all variants from a reference panel, in this paper, we describe a novel hybrid design method for selection of SNPs for genotype microarrays. The method uses alternating rounds of SNP selection based on pairwise tagging followed by rounds of target set coverage calculations based on imputation r2 values, which enables removal from the target set of SNPs that can be covered by imputation but were not covered by pairwise tagging. Using this approach, we were able to increase genome-wide coverage with the same fixed number of SNPs on the designed array.
The three new custom arrays described here utilize the Axiom Genotyping Solution (http://media.affymetrix.com/support/technical/datasheets/axiom_genotyping_solution_datasheet.pdf). Briefly, it is a two-color ligation-based assay utilizing 30-mer oligonucleotide probes synthesized in situ on a microarray substrate with automated parallel processing of 96 samples per plate, with a total of ~1.38 million features available for experimental content. In the design of the EUR array, every SNP was represented by at least 2 features (2-rep); some high-value SNPs that had poor resolution were tiled on the array with more than two representations, and hence required more than 2 features (e.g., 4 features or 8 features). As a consequence, the EUR array contains a total of 674,518 SNPs. At the time of design of the EAS, AFR and LAT arrays, it became apparent through analysis of the two representations on the EUR array that the highest resolution SNPs could be tiled on the array with a single feature with only a very small reduction in call rate. We therefore increased the genome-wide coverage of these arrays by tiling some of the highest resolution SNPs with only a single feature (1-rep), enabling greater SNP content on the arrays.
At the time of design of the AFR and LAT arrays, Affymetrix introduced a new reagent kit, Axiom Reagent Kit 2.0. An increased number of SNPs were validated by Affymetrix on the new kit, providing a larger sample of candidate SNPs for the design of these two arrays. The benefits were two-fold: more of the primary, secondary and tertiary SNPs could be directly tiled onto the arrays, and a wider choice of high resolution SNPs were available for selection for genome-wide coverage.
2. Results
2.1. Genome-wide coverage algorithm comparison
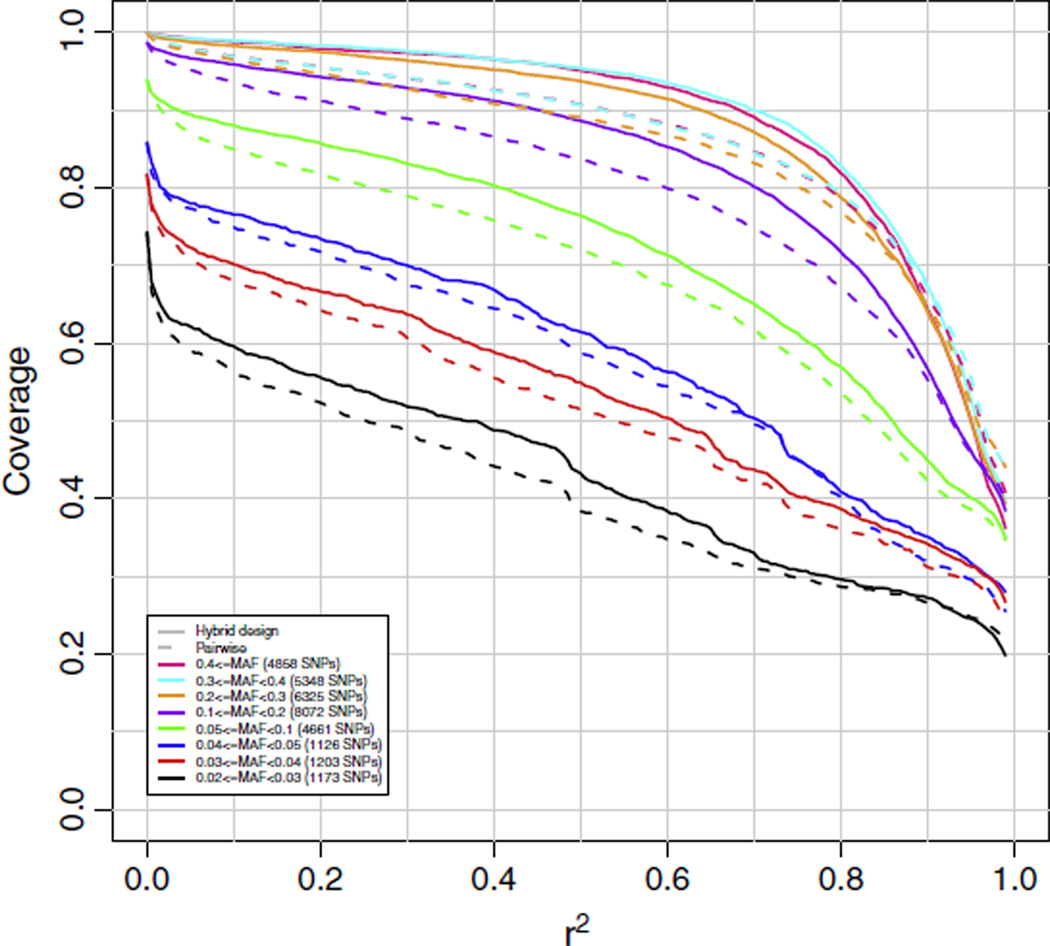
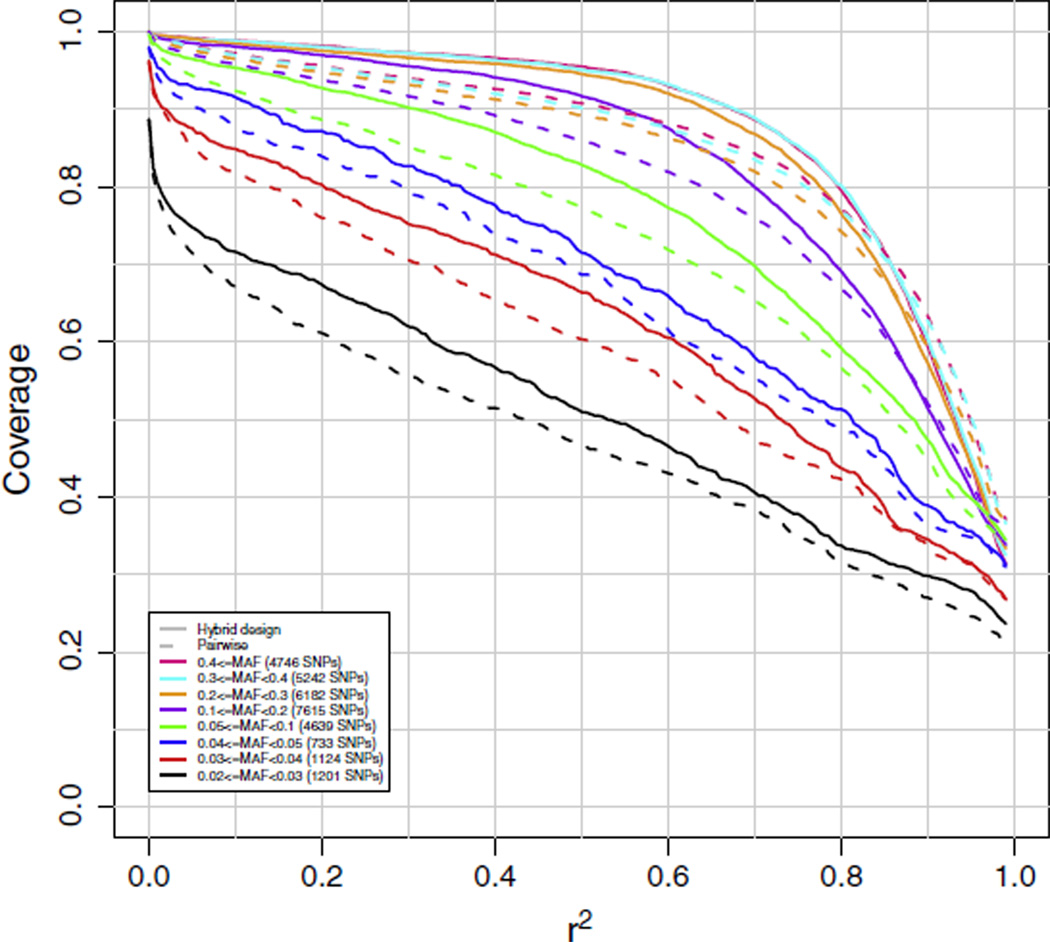
Results in Fig. 1 for the HapMap sample African Ancestry in Southwest USA (ASW) and Fig. 2 for the Luhya in Webuye, Kenya (LWK) compare coverage for a hypothetical array designed in the Yoruba in Ibadan population (YRI) by hybrid SNP selection to one designed by pairwise tagging (on chromosome 21), and show that the hybrid SNP selection algorithm outperforms the pairwise tagging selection algorithm by an average of about 5% on all coverage curves. During the course of hybrid SNP selection in creating this hypothetical array, we noted that the number of SNPs marked as covered by the imputation piece of the algorithm was 28,788 (87%), compared with 19,293 (58%) marked as covered by simple pairwise tagging. A separate analysis of chromosome 20 produced similar results. As a consequence, we proceeded to design both the AFR and LAT arrays using the hybrid SNP selection strategy described in Section 4.1. The EAS array, which required fewer SNPs for genome wide coverage, was designed by traditional pairwise tagging SNP selection.
Fig. 1.
Chromosome 21 coverage of the African Ancestry in Southwest USA (ASW) population based on two hypothetical arrays, one designed by pairwise tagging and the other by hybrid SNP selection for the Yoruba in Ibadan (YRI) population. Coverage was based on imputation using the YRI population as reference. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range.
Fig. 2.
Chromosome 21 coverage of the Luhya in Webuye, Kenya (LWK) population based on two hypothetical arrays, one designed by pairwise tagging and the other by hybrid SNP selection for the Yoruba in Ibadan (YRI) population. Coverage was based on imputation using the YRI population as reference. The numbers in parentheses are the numbers of markers in the target set in each particular minor allele frequency range.
2.2. Array statistics
The four arrays developed for the genotyping project on the Kaiser Permanente RPGEH were optimized for individuals of varying ancestries. The design of the EUR array is given elsewhere [8]; the design of the remaining 3 arrays is given below in Section 4.3.2. The collection of SNPs on the four arrays differed, although there was considerable overlap. It was part of the design algorithm to maximize the overlap of SNP content between the arrays. Table 1 provides a description of SNP content for the four arrays, including a breakdown by type (autosomal, X-linked, Y-linked or mitochondrial), the number of 1-rep SNPs on each of the arrays (the SNPs tiled with one representation were only those selected for genome-wide coverage), and the number of overlapping SNPs between the different arrays. Among the four arrays, 804,385 SNPs were unique to a single array; 403,981 were shared by two arrays; 156,270 were shared by three arrays; and 254,438 were shared by all four arrays. In total, 1,619,074 unique SNPs were included on at least one array.
Table 1.
The number of SNPs that are common to each of the arrays, broken down by type, and the number with a single feature (1-rep). SNPs in the pseudoautosomal region are included in the count for the X chromosome.
| Overlap with other arrays | Type breakdown | Features | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Array | EUR | EAS | AFR | LAT | Mitochondrial | Y | X | Autosomal | 1-rep |
| EUR | 674,518 | 386,841 | 384,966 | 434,028 | 116 | 289 | 13,123 | 660,990 | 0 |
| EAS | 712,950 | 303,850 | 314,794 | 83 | 158 | 13,385 | 699,324 | 65,473 | |
| AFR | 893,631 | 574,940 | 98 | 234 | 26,264 | 867,035 | 429,451 | ||
| LAT | 817,810 | 123 | 234 | 25,397 | 792,056 | 282,901 | |||
The design of each array, as described for the EUR array [8], involved selection of SNPs from a preselect set that was prioritized for inclusion and a target set for which SNPs were selected for coverage. The preselect set consisted of three tiers of the most important SNPs described below in Section 4.3.1 (e.g., related to disease) that were directly tiled on the array before other rounds of SNP selection began [8]. The EAS array had 258 SNPs in the primary tier; 9764 in the secondary; and 43,908 in the tertiary. The AFR array had 270 primary; 16,669 secondary; and 43,398 tertiary. Lastly, the LAT array had 279 primary; 20,020 secondary; and 43,398 tertiary. The increasing number of secondary SNPs on the AFR and LAT arrays is primarily a result of the availability of more validated SNPs at the time the array was designed, due to the greater numbers of SNPs available for use with the new Affymetrix Axiom Reagent Kit 2.0 and to the increasing SNP requirement due to decreased LD in African ancestry populations for imputing missing SNPs.
2.3. Genome-wide coverage of the arrays
Coverage was computed for each array against an appropriate target population by calculating imputation r2 values. To obtain an unbiased estimate of coverage, we used chromosome 2 sequence data of the 1000 Genomes Project interim June 2011 release data (KG2011) (http://1000genomes.org) consisting of 1094 individuals of 14 race/ethnicities: 61 ASW, 87 Utah residents with ancestry from Northern and Western Europe from Centre d'Etude du Polymorphisme Humain (CEU), 97 Han Chinese in Beijing (CHB), 100 Han Chinese South (CHS), 60 Colombian in Medellin, Colombia (CLM), 93 Finnish individuals from Finland (FIN), 89 British individuals from England and Scotland (GBR), 14 Iberians in Spain (IBS), 89 Japanese (JPT), 97 LWK, 66 HapMap Mexican individuals from Los Angeles, California (MXL), 55 Puerto Ricans in Puerto Rico (PUR), 98 Toscani in Italia (TSI), and 88 YRI. To compute coverage, all subjects other than the target group were included in the reference set. For example, for the 97 target Chinese in Beijing (CHB) individuals, we used all other populations except CHB in the reference sample. Imputation accuracy is affected by the size of the reference sample, among other things [15]. Our reference and target panels vary slightly amongst the different populations; however, the sizes are sufficiently large and similar that results are comparable.
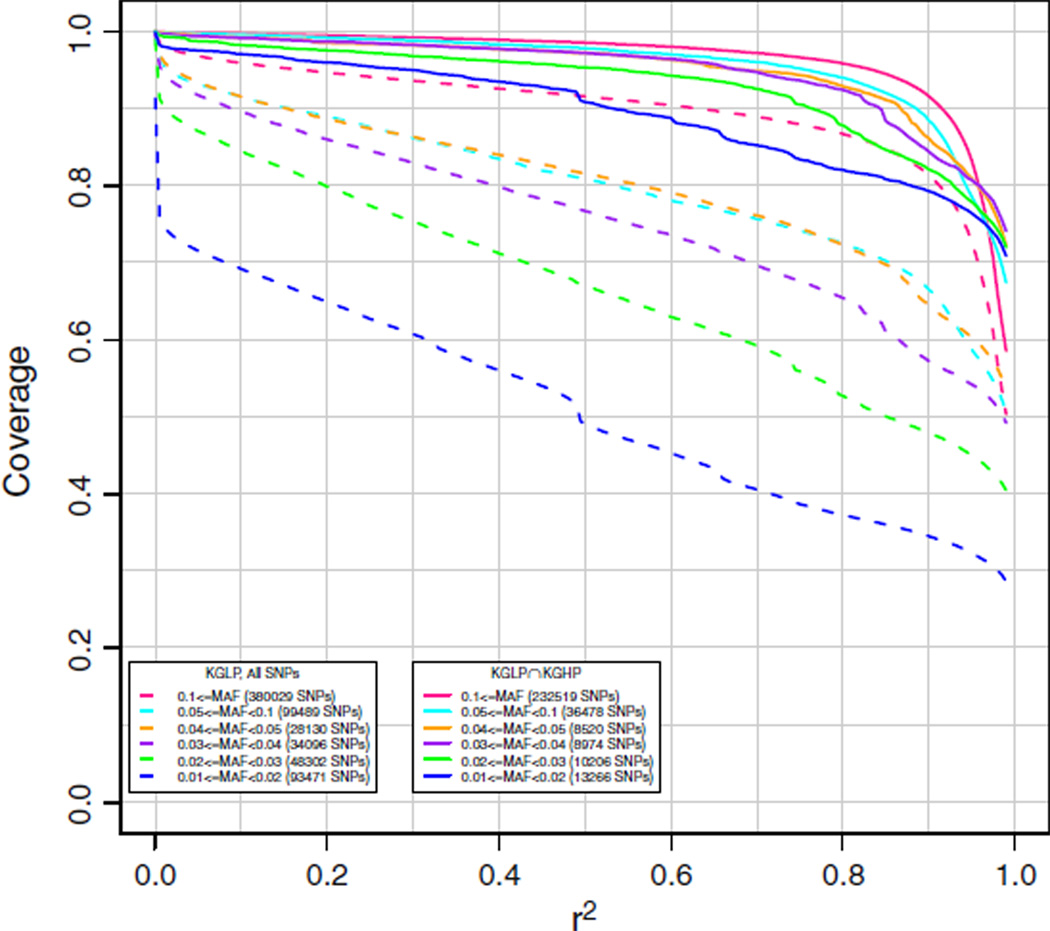
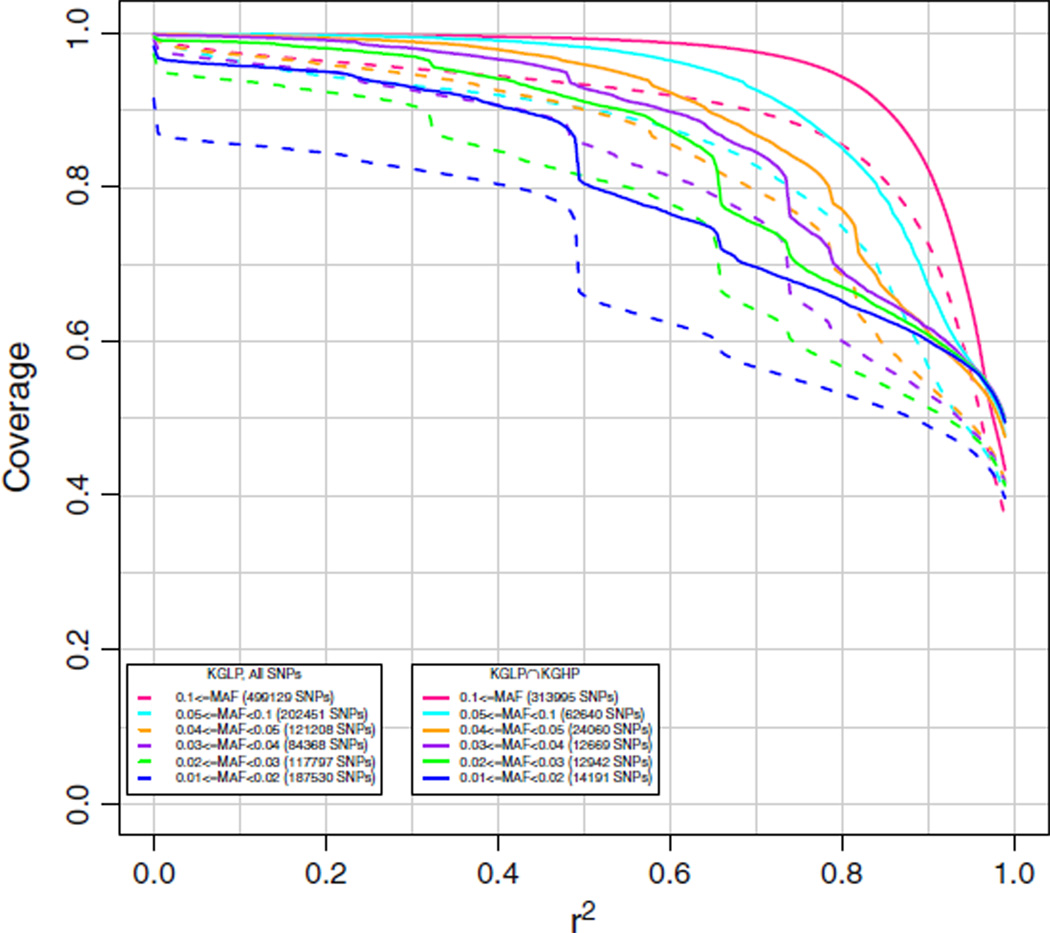
The EAS array was designed to cover SNPs from the ASI population (up to 90 CHB and 89 JPT unrelated HapMap 3 individuals [16] using HapMap and Axiom validated dbSNP and KGP SNPs) with MAF ≥ 0.02, and SNPs from the CEU population (up to 116 unrelated HapMap 3 CEU individuals [16], using HapMap and Axiom validated dbSNP and KGP SNPs) with MAF ≥ 0.10. To obtain an unbiased estimate of genome-wide coverage, we used the KG2011 data for the imputation calculation, as described above. Results are given in Fig. 3 for CHB. This dataset was sequenced at a low (average ~5×) coverage [17,18], and some genotype calls for these subjects were improved through imputation from HapMap 3 data [19]. Hence, because of potential noise in the low pass sequencing phase of the KGP, in Fig. 3 we also display coverage of the subset of SNPs also found in the 1000 Genomes High Pass (KGHP) data (coverage of 20–60×). Because these high quality SNPs were derived from sequencing only two trios, they are biased towards more common allele frequencies. However, this set still contains low frequency variants, and we have stratified the results based on MAF ranges found in the ASI. As can be seen in the figure, coverage is excellent down to a MAF of 0.01. We note that coverage of the subset of KGHP SNPs is considerably better than for the KG2011 SNPs as a whole. As we have reported before [8], this is likely due, at least in part, to the low coverage sequence data containing inaccurate genotype calls and false positive SNPs due to the low pass sequencing. Results were nearly identical for imputation coverage in CHS using all other individuals except CHS, as well as in JPT using all individuals but JPT (results not shown).
Fig. 3.
Chromosome 2 coverage by the new AX_KP_UCSF_EAS array of the 1000 Genomes interim June 2011 (KG2011) Han Chinese in Beijing (CHB) genotypes. Coverage was based on imputation of the target CHB set using all other individuals except CHB. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range. Subsets refer to SNPs identified in the 1000 Genomes High Pass (KGHP) sequencing (indicated by solid lines with “KGLP ∩ KGHP”) versus all SNPs (indicated by dashed lines with “KGLP, All SNPs”).
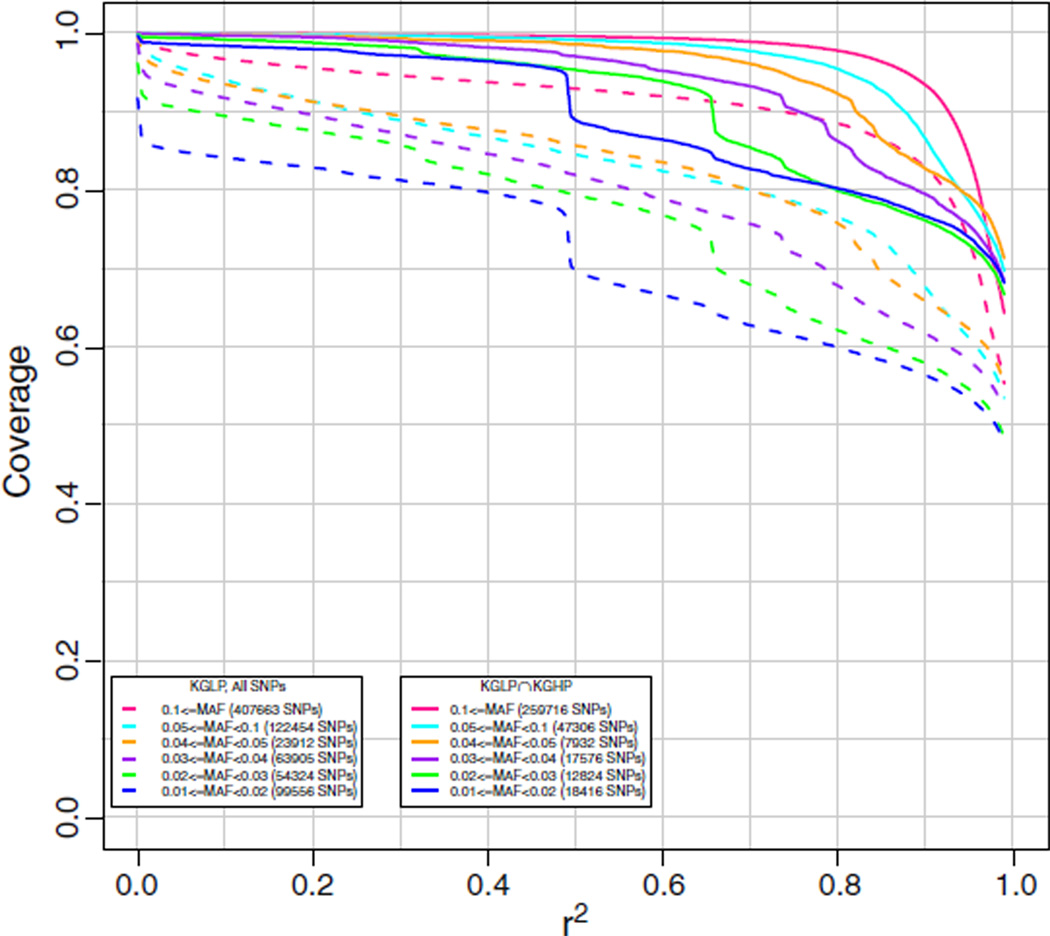
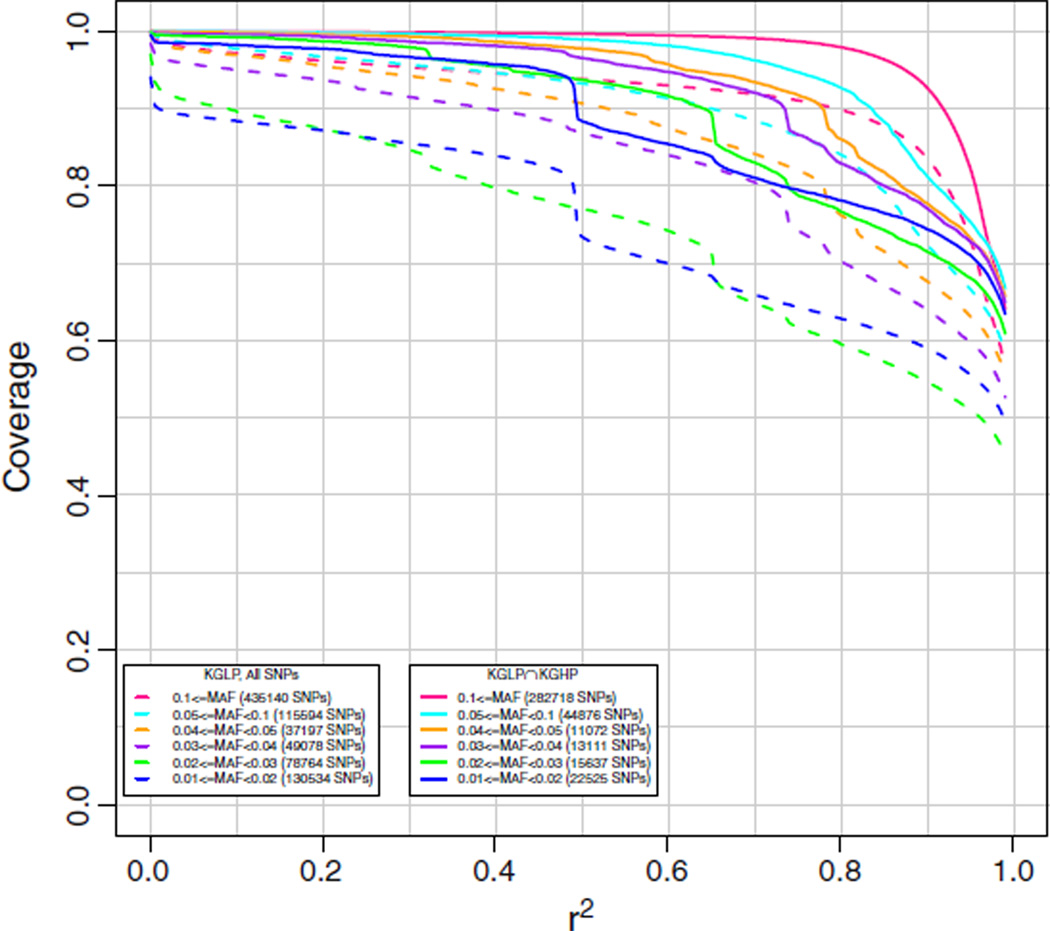
The AFR array was designed to cover SNPs from the YRI population (up to 116 unrelated HapMap 3 individuals [16] using HapMap and Axiom validated dbSNP and KGP SNPs) with MAF ≥ 0.02, and SNPs from the CEU population (same genotypes as in EAS array) with MAF ≥ 0.10. Coverage results for the 61 KG2011 ASW individuals are given in Fig. 4. Coverage is excellent for MAF of 0.04 or greater at an r2 of 0.8, and still good for MAF of 0.01 or greater. This reflects the increased genetic variation and decreased linkage disequilibrium observed in the YRI population.
Fig. 4.
Chromosome 2 coverage by the new AX_KP_UCSF_AFR array of the 1000 Genomes interim June 2011 release (KG2011) African Ancestry in Southwest USA (ASW) genotypes. Coverage was based on imputation of the target ASW set using all other individuals except ASW. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range. Subsets refer to SNPs identified in the 1000 Genomes High Pass (KGHP) sequencing (indicated by solid lines with “KGLP ∩ KGHP”) versus all SNPs (indicated by dashed lines with “KGLP, All SNPs”).
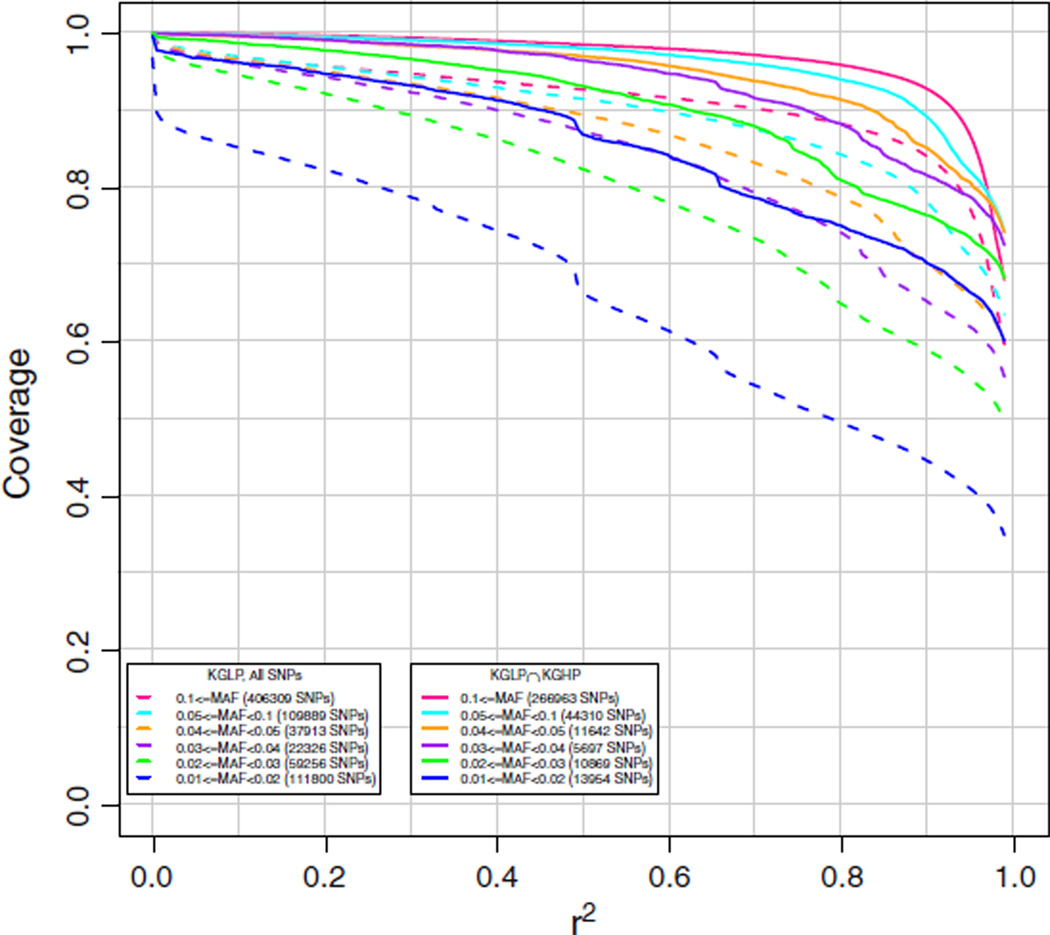
The LAT array was designed to cover YRI SNPs with a MAF ≥ 0.10, CEU SNPs with a MAF ≥ 0.03, in addition to a set of projected Native American-specific SNPs (see Section 4.3.2.3). Results are shown for MXL and PUR in Fig. 5 and Fig. 6, respectively. Coverage is excellent for both populations for a MAF greater than 0.01. The LAT array was designed for Latino populations with higher amounts of African ancestry, such as the PUR individuals; individuals from Mexico have only a modest degree of African ancestry, on average [20]. Coverage, however, is excellent for both populations for a MAF greater than 0.01.
Fig. 5.
Chromosome 2 coverage by the new AX_KP_UCSF_LAT array of the 1000 Genomes interim June 2011 release (KG2011) Mexicans in Los Angeles, CA (MXL) genotypes. Coverage was based on imputation of the target MXL set using all other individuals except MXL. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range. Subsets refer to SNPs identified in the 1000 Genomes High Pass (KGHP) sequencing (indicated by solid lines with “KGLP ∩ KGHP”) versus all SNPs (indicated by dashed lines with “KGLP, All SNPs”).
Fig. 6.
Chromosome 2 coverage by the new AX_KP_UCSF_LAT array of the 1000 Genomes interim June 2011 release (KG2011) Puerto Rican in Puerto Rico (PUR) genotypes. Coverage was based on imputation of the target PUR set using all other individuals except PUR. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range. Subsets refer to SNPs identified in the 1000 Genomes High Pass (KGHP) sequencing (indicated by solid lines with “KGLP ∩ KGHP”) versus all SNPs (indicated by dashed lines with “KGLP, All SNPs”).
Finally, we previously reported the coverage of CEU subjects by the EUR array [8], using the 1000 Genomes Pilot Phase I data, and cross validation imputation coverage using 60 individuals. Because of the small reference sample size, coverage for the array was underestimated. Fig. 7 shows the coverage of the EUR array on the CEU population using the much larger reference sample described above. The coverage is excellent down to a MAF of 0.01. Results were nearly identical for imputation coverage of FIN, GBR, and TSI (results not shown).
Fig. 7.
Chromosome 2 coverage by the AX_KP_UCSF_EUR array of the 1000 Genomes interim June 2011 release (KG2011) Utah residents with ancestry from Northern and Western Europe (CEU) genotypes. Coverage was based on imputation of the target CEU set using all other individuals except CEU. The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range. Subsets refer to SNPs identified in the 1000 Genomes High Pass (KGHP) sequencing (indicated by solid lines with “KGLP ∩ KGHP”) versus all SNPs (indicated by dashed lines with “KGLP, All SNPs”).
3. Discussion
While genotyping arrays with millions of SNPs that offer universal coverage are clearly optimal for GWA studies of multi-racial and multi-ethnic cohorts, the production time and expense associated with such arrays was prohibitive for a very large scale project such as ours. Also, we felt that a single array platform with up to 700,000 SNPs universally applied to individuals of all racial/ethnic backgrounds was not optimal, because it would provide less coverage of lower frequency variation overall. Hence, our compromise was to design race/ethnicity specific arrays, which could provide coverage of both common and rare variation in multiple racial/ethnic groups. Several advances during the design of the four arrays in this project led to enhanced coverage. First, the reagent kits developed by Affymetrix improved by the time of design of the AFR and LAT arrays, affording us a wider choice of Axiom validated SNPs to tile onto those arrays. Second, we developed a novel hybrid SNP selection scheme which enhanced the ultimate coverage of the AFR and LAT arrays over what they would have been had SNP selection been based simply on pairwise tagging. Third, we determined that high performing SNPs could be tiled with a single representation on the Axiom arrays without significant loss of genotype quality. These latter three developments were most critical for the AFR and LAT arrays, where African ancestry required both a larger number of SNPs and improved SNP selection.
One priority for SNP selection on all three arrays described here was overlap with the first designed array, the EUR array [8]. As a consequence, over 250,000 SNPs are overlapping on all four arrays. These SNPs represent common variation found in all race/ethnicity groups. By contrast, across all four arrays, there are over 1.6 million SNPs represented. This large number reflects both common and lower frequency variation that is race/ethnicity specific. Many of the SNPs in this collection of 1.6 million are polymorphic or high frequency in only one or a few race/ethnicity groups, and monomorphic in others. One disadvantage of a universal array is that for a given race/ethnicity group, many of the SNPs on that array will be monomorphic (in particular if the cost in time or money is a factor). On the other hand, for SNPs that are polymorphic in two or more race/ethnicity groups, non-overlap of SNPs on the various arrays means that imputation must be used to create a set of SNPs common to the arrays. While imputation may be accurate for many of the SNPs, it may not be accurate for all.
Each array demonstrates good to excellent genome-wide coverage for the datasets that they were designed to cover. As expected, coverage of the EAS, LAT, and CEU arrays are very high, with the AFR array modestly less. While coverage is substantially greater for SNPs that appeared in the KG high pass as well as low pass data in general, the difference is more dramatic for comparisons based on the EAS array and East Asian populations. The reason for this is likely due to the number of minor alleles observed in the reference sample, which can have a strong influence on imputation coverage for that SNP. The high pass data were derived from one trio of European ancestry and one trio of African ancestry. Hence, SNPs found in the high pass data are likely to occur in the imputation reference sample at higher frequency than SNPs found in the low pass data that were not found in the high pass data (e.g., SNPs that are specific to East Asians). This bias has less impact on other arrays and populations. Although our present coverage only uses chromosome 2, we expect the coverage to be similar to the genome-wide coverage, as we have seen in other datasets.
We believe that the cost and throughput of next-generation genotyping arrays in conjunction with imputation from dense next generation sequencing data, will aid in the discovery of novel common and low frequency disease-associated variants, especially when used in large scale, well phenotyped populations. It is likely that genome-wide genotyping arrays will continue to be higher throughput and less expensive than whole genome sequencing. In particular, we look forward to the identification of novel variants associated with a variety of diseases and traits using the data from these arrays in the Kaiser Permanente RPGEH.
4. Materials and methods
4.1. The hybrid SNP selection algorithm
A novel hybrid SNP selection algorithm was based on cycles alternating greedy SNP selection based on pairwise tagging with imputation coverage calculations. The candidate set of SNPs corresponds to SNPs validated by Affymetrix for use with the Axiom Genotyping Solution that are available for tiling on the array. The target set of SNPs refers to the set of SNPs for which coverage is attempted (typically larger than the candidate set and limited to SNPs passing a MAF cutoff). The selected set of SNPs is the collection of SNPs chosen for tiling on the array after each cycle of SNP selection. The selected set of SNPs increases with each cycle, and the target set of SNPs is reduced after each cycle.
At each cycle of the hybrid SNP selection, a certain number of SNPs are selected from the candidate set based on greedy pairwise coverage and added to the selected set. The selection step is then followed by a coverage step, wherein the selected set of SNPs is used to calculate imputation-based coverage for each SNP remaining in the target set. SNPs in the target set with an imputation r2 greater than a given threshold are removed from the target set. The two steps constitute a single cycle. For computational reasons, imputation was done without cross validation. Imputation coverage was calculated as the correlation between dosages of true genotypes and expected dosages from imputed genotype probabilities [8,15].
The number of SNPs selected was determined based on the total number of SNPs that could be covered and the total number of rounds of SNP selection. In general, more iterations resulted in more efficient SNP selection (smaller number of selected SNPs to reach the same coverage), but with greatly increased computational time (primarily due to the imputation coverage step).
4.1.1. Comparison of hybrid SNP selection to pairwise tagging
We compared the novel hybrid SNP selection strategy to the standard greedy SNP selection algorithm [8] which is based on pairwise linkage disequilibrium in terms of expected genome-wide coverage for the design of the AFR array. We created a hypothetical array under both strategies to tag 31,119 SNPs from HapMap 3 and some Affymetrix internal screens with MAF ≥ 0.02 in YRI on chromosome 21, using an imputation r2 cutoff of 0.9 and pairwise r2 cutoff of 0.8. For the hybrid array, we allowed the hybrid SNP selection algorithm to run until all markers were covered that could be, which resulted in selecting 7544 markers. To compare imputation with greedy SNP selection, we used the first 7544 markers chosen by the algorithm for the second hypothetical array.
We then compared genome-wide coverage of the two hypothetical arrays by imputing all genotypes for two HapMap samples: the ASW and the LWK. For each hypothetical array, imputation was based on the SNPs present on that array. The reference genotype sample for imputation was the HapMap YRI. Imputation coverage for a SNP was calculated as the square of the correlation (r2) of the expected dosages derived from imputation to the dosages derived from the true genotypes using Beagle version 3.3.0 [21]. Only SNPs with at least 50 genotypes available in both the reference and target sample were included in these analyses. Genome-wide coverage was calculated as the proportion of SNPs in the target sample with a given imputation r2 value or greater.
4.2. Cluster separation
Cluster separation for a SNP with alleles A and B was assessed by a Fisher's Linear Discriminant-related Score (FLD Score) [8,22] which is defined as the minimum of two linear discriminants as follows:
FLD Score = mini=AA,BB [(MAB − Mi)/SAA,AB,BB]
where MAB is the center of the heterozygous cluster in the log ratio dimension, MAA and MBB are the centers of the two respective homozygous clusters in the log ratio dimension, and SAA,AB,BB is the standard deviation of the clusters pooled across all three distributions. SNPs with higher FLD Score values are very highly correlated with tighter clusters and higher call rates.
4.3. SNP selection for inclusion on the arrays
Many of the initial strategies for designing the EAS, AFR, and LAT arrays were the same as those used for the EUR array, explained in detail elsewhere [8]. We describe these again briefly here, with an extended discussion of the modifications to them.
SNPs that were considered for tiling on the array (candidate set) were selected based on having good cluster separation (high FLD Score), a minimum of 3 observed examples of the minor allele (unless in the primary set as described below), and good accuracy (concordance with HapMap when possible, reproducibility, and consistency with Mendelian inheritance). SNP selection proceeded progressively through tiers of importance; SNPs comprising the tiers were updated during each successive array design. All SNPs, aside from those in the primary set, were filtered to have allele frequencies above a certain threshold (discussed below on an array-wise basis).
4.3.1. The preselect set
Primary SNPs were based on strongly confirmed disease associations from literature and online databases [23,24]. Most were directly tiled on the array with redundant coverage based on SNPs chosen for imputation coverage. When adding coverage/redundant coverage to the primary SNPs, we first selected tag SNPs based on the population of greatest relevance. If a single tag SNP with an r2 greater than 0.8 with the target SNP was not available, we selected coverage SNPs by imputation. This entailed greedily adding SNPs so long as it improved the imputation r2 by more than 0.03. Then, additional SNPs were selected, if necessary, for tagging the same target SNP in other relevant populations. Redundant coverage of the same target SNP was obtained by repeating the imputation tagging process with a new set of candidate SNPs.
The secondary set consisted of SNPs that were suggestive of association with disease or traits of interest but were not as strongly replicated as the primary SNPs. This group derived from a variety of sources [23–26]. When these SNPs could not be directly tiled, coverage was obtained by selection of tagging SNPs based on imputation. This group was not provided with redundant coverage.
The tertiary set consisted of SNPs that were mined from various database sources for potential functional significance (e.g., miRNA, splice site, MHC, coding, etc., SNPs). When possible (i.e. an Affymetrix Axiom validated probeset was available), these SNPs were directly tiled on the array, and they were also included in the first target set for greedy pairwise SNP selection. This first target set also included “gene-enrichment” SNPs in coding regions, adjacent introns and upstream and downstream UTR regions of approximately 5000 genes of interest [8].
4.3.2. Genome-wide coverage
The genome-wide coverage algorithm differed amongst the four arrays. The EAS array followed a simple greedy SNP selection paradigm similar to that used for the EUR array, whereas the AFR and LAT arrays utilized hybrid SNP selection. All 3 of the new arrays described here tiled some SNPs with a single representation to increase the total number of SNPs on the array, although their numbers varied among arrays. Only the highest resolution SNPs not in the preselect set were tiled with a single representation.
4.3.2.1. Design of the EAS array
The EAS array was designed primarily to cover common and rare polymorphisms in East Asians. However, because some individuals in the RPGEH have mixed East Asian and European ancestry, we also wanted to optimize this array for such mixed-ancestry individuals. Therefore, coverage included SNPs not polymorphic in East Asians but polymorphic in Europeans down to a frequency of 0.10. There were 3 rounds of coverage, as follows:
- Round 1: To cover tertiary and gene enrichment SNPs in addition to coverage achieved with the preselect set
- Target sets
- For ASI: Tertiary and gene enrichment SNPs with MAF ≥ 0.01
- For CEU: Tertiary and gene enrichment SNPs with MAF ≥ 0.10 in CEU and absent in the ASI target set
- Candidate set: Axiom validated SNPs with no more than 2 features
- SNP selection algorithm: Greedy pairwise. The coverage contribution of a candidate SNP in ASI was the primary factor of consideration. However, other factors, including coverage contribution in CEU, overlap with the EUR array, and expected genotyping performance were also considered.
- r2 cutoff for pairwise coverage: 0.8
- Termination criterion: Maximal pairwise coverage in both ASI and CEU is reached
- # SNPs selected: 34,742
- Round 2: Additional coverage for tertiary and gene enrichment SNPs
- Target sets: Same as Round 1
- Candidate set: Same as Round 1, except only SNPs in the top tier of performance
- SNP selection algorithm: Same as Round 1
- r2 cutoff for pairwise coverage: 0.6
- Termination criterion: Same as Round 1
- # SNPs selected: 1,825
- Round 3: Genome-wide coverage in addition to coverage achieved with selected SNPs
- Target sets
- For ASI: genome-wide SNPs with MAF ≥ 0.02
- For CEU: genome-wide SNPs with MAF ≥ 0.10 in CEU and absent in the ASI target set
- Candidate set: Same as Round 1
- SNP selection algorithm: Same as Round 1
- r2 cutoff for pairwise coverage: 0.8
- Termination criterion: Same as Round 1
- # SNPs selected: 617,168
In round 3, the total number of selected SNPs and features was greater than could fit on a single array. Therefore, in order to fit all selected SNPs into a single array, 10% of the SNPs were included using 1 feature instead of 2 features. These SNPs all have a FLD Score ≥ 8.5 and were chosen from the end of the ranked selected SNP list.
4.3.2.2. Design of the AFR array
Design of the AFR array took into account the mixed continental ancestry of African Americans, so that both African and European SNPs were considered. However, the lower MAF threshold for the two ancestries was different. We assumed that for an African American population with approximately 20% European ancestry [27], it was sufficient to include European-specific SNPs with a MAF of 0.10 or greater, as this would translate into a MAF of 0.02 or greater in an African American sample. There were two rounds of coverage, as follows:
- Round 1: To cover gene enrichment SNPs in addition to coverage achieved with the preselect set
- Target sets
- For YRI: Gene enrichment SNPs with MAF ≥ 0.01
- For CEU: Gene enrichment SNPs with MAF ≥ 0.10 in CEU and absent in the YRI target set
- Candidate set: Axiom validated SNPs with no more than 2 features
- SNP selection algorithm: Greedy pairwise. Candidate SNPs with FLD Score ≥ 7 were selected before SNPs with FLD Score < 7. Other factors considered during SNP selection include coverage in YRI, CEU, overlap with the EUR array, and expected genotyping performance.
- r2 cutoff for pairwise coverage: 0.8
- Termination criterion: Maximal pairwise coverage in both YRI and CEU is reached
- # SNPs selected: 136,615
- Round 2: Genome-wide coverage in addition to coverage achieved with selected SNPs
- Target sets
- For YRI: Genome-wide SNPs with MAF ≥ 0.02
- For CEU: Genome-wide SNPs with MAF ≥ 0.10 in CEU and absent in the YRI target set
- Candidate set: Same as Round 1
- SNP selection algorithm: Hybrid, 9–11 cycles (done on a per chromosome basis, the number of cycles depended on the chromosome). A quota for SNPs with FLD Score < 7 was enforced so that the total number of SNPs with FLD Score < 7 would not exceed 321 k. Once the allowed quota was reached, only SNPs with FLD Score ≥ 7 were selected. Factors considered during greedy SNP selection are the same as Round 1.
- r2 cutoff for pairwise coverage: 0.8
- r2 cutoff for imputation coverage: 0.8
- Termination criterion: Maximum space on the array with tiling SNPs with FLD Score ≥ 7.5 with 1 feature and FLD Score < 7.5 with 2 features (~170 K SNPs that were covering only singleton YRI SNPs were removed from the maximum pairwise coverage of YRI and CEU list)
- # SNPs selected: 695,048
4.3.2.3. Design of the LAT array
Design of the LAT array was the most complex, because it needed to take into account three different continental ancestries—African, European and Native American. Adding to the complexity, Latino populations differ considerably in their relative proportions of these 3 ancestries [20]. Therefore, we started with coverage in the YRI population, assuming the target Latino population had up to about 40% African ancestry, on average, for example as has been observed in Dominicans [28]. We started with SNPs that had been selected for the AFR array, and then removed and added additional SNPs according to coverage characteristics for the other two ancestries. Coverage of European SNPs was based on recent sequence data. Coverage of Native American SNPs, i.e., those polymorphic in Native Americans but absent or of low frequency in other race/ethnicity groups, was complicated by the fact that at the time of design, no such sequence data for Native Americans or Latinos was available. Therefore, for coverage of Native American variation, we needed to rely instead on sources of genotype data for SNPs previously identified. One of these sources was a sample of 92 Latinos from Kaiser Permanente Northern California that was genotyped for approximately 5 million SNPs by Affymetrix specifically to assist SNP selection for the LAT array. There were 5 rounds of SNP selection, as follows:
- Round 1: Choosing SNPs from the AFR array
- SNP selection method: Less important SNPs from the AFR array for covering SNPs in Latino populations were removed from the AFR array. These SNPs included those selected during hybrid SNP selection, with MAF < 0.10 in YRI, and that did not cover any CEU target SNPs at the time of SNP selection; in addition, those selected in hybrid SNP selection after the end of cycle 5 that did not tag any CEU target SNPs at the time of SNP selection were removed.
- # SNPs selected: 543,858
- Round 2: Genome-wide coverage of CEU in addition to coverage achieved with selected SNPs
- Target set
- For CEU: Genome-wide SNPs with MAF ≥ 0.03
- Candidate set: Axiom validated SNPs with no more than 2 features
- SNP selection algorithm: Hybrid, consider factors including coverage in CEU, overlap with the EUR array, and expected genotyping performance
- r2 cutoff for pairwise coverage: 0.8
- r2 cutoff for imputation coverage: 0.8
- Termination criterion: When 244,548 SNPs are selected in 6 cycles
- # SNPs selected: 244,548.
- Round 3: Coverage of “Native American” SNPs via HapMap MXL
- Target set
- For MXL: 19,368 SNPs with MAF ≥ 0.05 in MXL and MAF < 0.02 in both CEU and YRI (to ascertain SNPs with an increased likelihood of being specifically increased in frequency in Native Americans)
- Candidate set: All Axiom validated SNPs
- SNP selection algorithm: Greedy pairwise, consider factors including pairwise coverage in MXL, number of features a SNP requires, and expected genotyping performance
- r2 cutoff for pairwise coverage: 0.8
- Termination criterion: Maximal pairwise coverage in MXL is reached
- # SNPs selected: 9,643
- Round 4: Coverage of “Native American” SNPs via KPNC Latino
- Target set
- For the KPNC Latino (described above): 34,953 SNPs with MAF ≥ 0.05 in Latino and MAF < 0.02 in both CEU and YRI
- Candidate set: All Axiom validated SNPs
- SNP selection algorithm: Greedy pairwise, consider factors including pairwise coverage in KPNC Latino, number of features a SNP requires, and expected genotyping performance
- r2 cutoff for pairwise coverage: 0.8
- Termination criterion: Maximal pairwise coverage in KPNC Latino is reached
- # SNPs selected: 20,365
- Round 5: Native American Ancestry Informative Markers
- SNP selection method: Choose Axiom-validated SNPs from 2120 Native American ancestry informative markers (AIMs) from [29]
- # SNPs selected: 1840
Rounds 3–5 produced some overlapping SNPs. Removing the overlap, the 3 rounds resulted in a total of 28,047 unique “Native American” SNPs added to the LAT array. All SNPs carried over from the AFR array for the LAT array were tiled with the same number of features as in the AFR array. Native American SNPs (described above) were tiled at half the original number of features when their FLD Score was at least 7.5. SNPs selected during hybrid SNP selection were also tiled with a single feature instead of 2 features when their FLD Score was at least 7.5.
4.4. Estimating genome-wide coverage of the final arrays
Genome-wide coverage of the EAS, AFR and LAT arrays was evaluated by calculating imputation r2 values for all SNPs in the appropriate target set, as described previously [15] and above. For each array, the target set included SNPs obtained in the KGHP sequencing effort, but using the KG2011 genotype data derived for those SNPs from sequencing the samples. When computing the coverage of specific racial/ethnic groups for a given array, we used one population from that racial/ethnic group in the target, and all other individuals from populations of that racial/ethnic group plus other racial/ethnic groups in the reference. As described above, we used the program Beagle version 3.3.0 [21] when designing the array, but final coverage estimates for the array were calculated using the program Impute2 version 2.1.2 [30] which we found had slightly higher accuracy, as has been shown before [11].
Acknowledgments
This work was supported by grant RC2 AG036607 from the National Institutes of Health, grants from the Robert Wood Johnson Foundation, the Ellison Medical Foundation, the Wayne and Gladys Valley Foundation, Kaiser Permanente, and NIH postdoctoral training grant R25 CA112355. We are grateful to the Kaiser Permanente Northern California members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment and Health.
Abbreviations
- GWA
genome-wide association
- MAF
minor allele frequency
- KGP
1000 Genomes Project
- RPGEH
Research Program on Genes, Environment and Health
- EUR
European and West Asian
- EAS
East Asian
- AFR
African American
- LAT
Latino
- 2-rep
2 features
- 1-rep
1 feature
- ASW
African Ancestry in Southwest USA
- CEU
Utah residents with ancestry from Northern and Western Europe from Centre d'Etude du Polymorphisme Humain
- CHB
Han Chinese in Beijing
- CHS
Han Chinese South
- CLM
Colombian in Medellin, Colombia
- Fin
Finnish from Finland
- GBR
British individuals from England and Scotland
- IBS
Iberians in Spain
- JPT
Japanese in Tokyo
- LWK
Luhya in Webuye Kenya
- MXL
Mexican in Los Angeles, CA
- PUR
Puerto Rican in Puerto Rico
- TSI
Toscani in Italia
- YRI
Yoruba in Ibadan, Nigeria
- KGHP
1000 Genomes High Pass
- KPNC
Kaiser Permanente Northern California
- AIMs
Ancestry Informative Markers
- KG2011
1000 Genomes interim June 2011 release
Contributor Information
Thomas J. Hoffmann, Email: hoffmannt@humgen.ucsf.edu.
Andrea Finn, Email: andrea_finn@affymetrix.com.
Catherine Schaefer, Email: cathy.schaefer@kp.org.
Pui-Yan Kwok, Email: pui.kwok@ucsf.edu.
Neil Risch, Email: rischn@humgen.ucsf.edu.
References
- 1.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte JS. Genome-wide association studies and beyond. Annu. Rev. Public Health. 2010;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–891. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 11.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 12.Spencer CCA, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000477. e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 14.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burchard EG, Borrell LN, Choudhry S, Naqvi M, Tsai H-J, Rodriguez-Santana JR, et al. Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am. J. Public Health. 2005;95:2161–2168. doi: 10.2105/AJPH.2005.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher RA. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936;7:179–188. [Google Scholar]
- 23.Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. [Accessed January 12, 2010]; http://www.genome.gov/gwastudies.
- 24.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 25.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroetz DL, Yee SW, Giacomini KM. The pharmacogenomics of membrane transporters project: research at the interface of genomics and transporter pharmacology. Clin. Pharmacol. Ther. 2010;87:109–116. doi: 10.1038/clpt.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, Jorgenson E, Gadde M, Kardia SLR, Rao DC, Zhu X, et al. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum. Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 28.Peralta CA, Li Y, Wassel C, Choudhry S, Palmas W, Seldin MF, et al. Differences in albuminuria between Hispanics and whites: an evaluation by genetic ancestry and country of origin: the multi-ethnic study of atherosclerosis. Circ. Cardiovasc. Genet. 2010;3:240–247. doi: 10.1161/CIRCGENETICS.109.914499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, et al. A genome-wide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000529. e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]