Abstract
The gene xylBADP1 from Acinetobacter baylyi ADP1 (gene annotation number ACIAD1578), coding for a putative aryl alcohol dehydrogenase, was heterologously expressed in Escherichia coli BL21(DE3). The respective aryl alcohol dehydrogenase was purified by fast protein liquid chromatography to apparent electrophoretic homogeneity. The predicted molecular weight of 39,500 per subunit was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. According to the native Mw as determined by gel filtration, the enzyme forms dimers and therefore seems to be XylB related. The enzyme showed the highest activity at 40°C. For both the reduction and the oxidation reactions, the pH for optimum activity was 6.5. The enzyme was NADH dependent and able to reduce medium- to long-chain n-alkylaldehydes, methyl-branched aldehydes, and aromatic aldehydes, with benzaldehyde yielding the highest activity. The oxidation reaction with the corresponding alcohols showed only 2.2% of the reduction activity, with coniferyl alcohol yielding the highest activity. Maximum activities for the reduction and the oxidation reaction were 104.5 and 2.3 U mg−1 of protein, respectively. The enzyme activity was affected by low concentrations of Ag+ and Hg2+ and high concentrations of Cu2+, Zn2+, and Fe2+. The gene xylBADP1 seems to be expressed constitutively and an involvement in coniferyl alcohol degradation is suggested. However, the enzyme is most probably not involved in the degradation of benzyl alcohol, anisalcohol, salicyl alcohol, vanillyl alcohol, cinnamyl alcohol, or aliphatic and isoprenoid alcohols.
INTRODUCTION
Members of the genus Acinetobacter belonging to the Gammaproteobacteria are ubiquitously distributed in the environment. They are Gram negative and oxidase negative and form nonmotile coccoid rods. Acinetobacter species are strictly aerobic and nonfermentative bacteria exhibiting an extremely versatile range of substrates (3), including alkanes, aliphatic alcohols, dicarboxylic and fatty acids, amino acids, and a few sugars, as well as a variety of aromatic compounds (13). This makes these strains increasingly interesting for bioremediation purposes. In recent years Acinetobacter baylyi ADP1 (32), formerly known as Acinetobacter sp. strain ADP1, which is an unencapsulated mutant of strain BD4 (11), became the focus of research due to its high natural competence (12). Detailed studies have been done on the molecular organization and evolution of genes involved in aromatic compound degradation (5, 9, 19), on the metabolism of storage lipids (14, 24), and on the genome sequence of this interesting strain (1).
Because it is a hydrocarbonoclastic bacterium, aliphatic and aromatic molecules are important carbon sources for A. baylyi ADP1, and, therefore, it requires a wide spectrum of degradation pathways. Benzyl esters, for example, are cleaved and degraded by enzymes encoded by genes of the areABC operon (9). First, the benzyl ester is cleaved hydrolytically by AreA (9), followed by the NAD+-dependent oxidations of benzyl alcohol to benzaldehyde and finally to benzoate conducted by AreB and AreC, respectively (17). In several further reactions benzoate is converted into catechol by the benABCD-encoded enzymes (18). In a last step, aromatic compounds are degraded via the β-ketoadipate pathway, in which the aromatic rings of the formed catechol or protocatechuate, which result from the degradation of hydroxycinnamyl alcohols, are cleaved by enzymes encoded by catA or pcaGH, respectively, with the formation of an intradiol being the most important step. The degradation products are finally channeled into the tricarboxylic acid cycle as succinyl coenzyme A (succinyl-CoA) and acetyl-CoA (5).
So far, only areB within the areABC operon has been described to be involved in aryl alcohol degradation in Acinetobacter calcoaceticus NCIB 8250 (6, 17) and A. baylyi ADP1 (9), but according to genome sequence data, a putative second operon, including an aldehyde dehydrogenase (ACIAD1577) and an aryl alcohol dehydrogenase (ACIAD1578) homologous to areB, is present within the ADP1 genome. The aryl alcohol dehydrogenase (ACIAD1578) coded in the second operon shares homologies to benzyl alcohol dehydrogenases from Pseudomonas putida (49% homology) and Caulobacter crescentus (48% homology) (1).
Under nitrogen-limiting conditions, acetyl-CoA derived from carbohydrate and hydrocarbon degradation, in turn, is used by many Acinetobacter strains to synthesize neutral lipids like triacylglycerols (TAG) or wax esters (WE) as carbon storage compounds. Coenzyme A-activated fatty acids are therein reduced by the NADPH-dependent acyl-CoA reductase Acr1 (annotation number ACIAD3383) (23) to fatty aldehydes, which are proposed to be further reduced to the corresponding fatty alcohol. However, the enzyme responsible for the latter step in A. baylyi ADP1 has not been identified yet, although Tani et al. (31) suggested the NADPH-dependent alcohol dehydrogenase AlrA (ACIAD3612) to be responsible for this reaction in A. baylyi, and aldehyde-reducing enzymes have been recently identified in Marinobacter aquaeolei VT8 (33). Finally, the fatty alcohol is condensed with the fatty acyl moiety of the CoA derivative by the promiscuous wax ester synthase/diacylglycerol:acetyltransferase (WS/DGAT) (ACIAD0832) to form a wax ester. Additionally, the WS/DGAT is involved in the formation of the majority of TAGs accumulated by A. baylyi ADP1 (14).
In this study, the second aryl alcohol dehydrogenase from recombinant Escherichia coli was purified and characterized, and its potential function concerning aryl and acyl alcohol degradation and wax ester formation was investigated.
MATERIALS AND METHODS
Chemicals and nucleotides.
Aliphatic and aromatic alcohol and aldehydes were purchased from Sigma-Aldrich-Fluka (Deisenhofen, Germany) or TCI Europe (Brussels, Belgium). NAD(P)+ and NAD(P)H were obtained from Gerbu Biotechnik GmbH (Wieblingen, Germany). Primers were synthesized by MWG (Ebersberg, Germany).
Organisms and culture conditions.
For cloning purposes, cells of Escherichia coli Mach1-T1 or E. coli BL21(DE3) were cultivated in lysogeny broth (LB) medium (25) in the presence of ampicillin (100 μg ml−1) at 37°C overnight on a rotary shaker at 200 rpm. Protein overexpression in strains of E. coli BL21(DE3) was induced 3 h after inoculation by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cultivation was continued for another 3 h prior to cell harvest.
Cells of A. baylyi ADP1 (ATCC 33305) (32) or related mutants were grown in LB medium or mineral salts medium (MSM) (26) containing 1% (wt/vol) sodium gluconate.
Growth and degradation experiments.
A. baylyi ADP1 was investigated for growth on several substrates at 30°C in 100 ml Klett flasks containing 20 ml of mineral salts medium with hexadecanol (15 mM) or benzyl alcohol (5 mM) as single carbon sources. Growth was followed by measuring the changes of the optical density with a Klett photometer (Manostat Corp., New York) for at least 14 h. Since vanillyl alcohol, anisalcohol, salicyl alcohol, cinnamyl alcohol, geraniol, and coniferyl alcohol, turned out to inhibit cell growth even at low concentrations, the ability of A. baylyi ADP1 cells to degrade these substances was investigated. Mineral salts medium containing a 1 mM concentration of the respective alcohol was inoculated with an overnight preculture. The initial optical density at 600 nm (OD600) was adjusted to 0.125, 1-ml samples were withdrawn after 0, 4, 8, and 24 h, and the residual concentrations of the alcohols were determined by high-pressure liquid chromatography (HPLC) or gas chromatography (GC) analysis, respectively. Cinnamyl alcohol-containing medium was preincubated for 4 h on a horizontal shaker to allow cinnamyl alcohol to dissolve properly in the medium. All cultures were incubated on a horizontal shaker at 105 rpm.
Determination of residual alcohol concentrations.
The concentrations of residual vanillyl alcohol, anisalcohol, salicyl alcohol, cinnamyl alcohol, and coniferyl alcohol were determined directly from cell-free culture supernatants by high-performance liquid chromatography using an UltiMate 3000 HPLC system (Dionex GmbH, Idstein, Germany) equipped with a 250- by 2.1-mm Acclaim 120 C8 reversed-phase column (particle size, 5 μm; Dionex GmbH, Idstein, Germany) and a multiple-wavelength detector (MWD-3000; Dionex, Idstein, Germany). Samples of 5 μl each were injected, and substances were separated by applying a gradient of 0.1% (vol/vol) formic acid (eluant A) and acetonitrile (eluant B) in a range from 26 to 100% (vol/vol) eluant B, starting at a flow rate of 0.1 ml min−1 and increasing to 0.3 ml min−1 after 16 min when eluant B reached 100%. Compounds were identified by comparing their retention times and their absorption at a specific wavelength (vanillyl alcohol, anisalcohol, and salicyl alcohol, 280 nm; cinnamyl alcohol and coniferyl alcohol, 259 nm) with those of standard substances. Data were evaluated with Chromeleon 6.8 Chromatography Data Systems software (Dionex GmbH, Idstein, Germany).
Geraniol was detected by gas chromatography. One milliliter of cell-free culture supernatant was extracted with 0.5 ml of chloroform by vortexing vigorously for 1 min. The organic phase was analyzed using an Agilent 6850 series GC system (Agilent, Waldbronn, Germany) equipped with a BP21 capillary column (50 m by 0.22 mm; film thickness, 0.25 μm; SGE, Darmstadt, Germany) and a flame ionization detector (Agilent, Waldbronn, Germany). A 2-μl portion of sample was analyzed after split injection (1:15); hydrogen was used as the carrier gas, with a constant flow of 0.6 ml min−1. The temperatures of the injector and detector were 250°C and 275°C, respectively. The following temperature program was applied: 120°C for 5 min, increase of 3°C min−1 to 180°C, and increase of 10°C min−1 to 220°C, which was held for 31 min. Geraniol was identified by comparison of the retention time with that of an authenticated standard substance.
Plasmids and genetic manipulations.
Standard molecular biological techniques were applied as described by Sambrook et al. (25). For disruption of areB (ACIAD1429), alrA (ACIAD3612), and the gene with annotation number ACIAD1578 (referred to here as xylBADP1), the genes were amplified from isolated genomic DNA of A. baylyi ADP1 using the primers CATATGACAAAGTTTACCGAAATCAC (5′ end) and CTCGAGTTAACCAATTTTTAAAATGGG (3′ end) for areB, GGATCCAAGGAGGTTCTGTCATGACAACTAATGTGAT (5′ end) and GTCGACTTAAAAATCGGCTTTAAGTACAA (3′ end) for alrA, and CATATGAAAATTACAGCAGCAGTGGC (5′ end) and CTCGAGCTAATGATGGGCATGTTC (3′ end) for xylBADP1 using Taq DNA polymerase (Invitrogen, Carlsbad, CA). The amplification products were subcloned in the vector pGEM-T Easy (Promega, Madison, WI). Subsequently, the gentamicin resistance cassette isolated from pKSsymΩGm (20) by SmaI restriction was ligated into the unique Eco72I restriction site of areB, and the kanamycin resistance cassette from SmaI-restricted pKSsymΩKm (20) was ligated into the unique MlsI restriction site of alrA, while xylBADP1 was disrupted by the insertion of a streptomycin resistance gene into the unique SspI restriction site. The streptomycin resistance gene was amplified from vector pCDF-Duet1 (Novagen, Merck KgaA, Darmstadt, Germany) using the primers CCATGGCTCACGCCCGGAGCGTAGCGACC (5′ end) and CCATGGAACGACCCTGCCCTGAACCGACG (3′ end) and using Pfx DNA polymerase (Invitrogen, Carlsbad, CA). The disrupted genes were amplified again, and the PCR products were used to transform A. baylyi ADP1 as described by Palmen et al. (21). Homologous recombination with a double crossover resulted in the generation of disruption mutants. In total, the following single, double, and triple disruption mutants of alrA, areB, and xylBADP1 were generated: A. baylyi ADP1alrAΩKm, A. baylyi ADP1areBΩGm, A. baylyi ADP1xylBADP1ΩSm, A. baylyi ADP1alrAΩKm areBΩGm, A. baylyi ADP1alrAΩKmxylBADP1ΩSm, A. baylyi ADP1areBΩGmxylBADP1ΩSm, and A. baylyi ADP1alrAΩKm areBΩGmxylBADP1ΩSm. Correct replacement of areB, alrA, and xylBADP1 by the disrupted genes was confirmed by analytic PCR of isolated genomic DNA.
For overexpression of XylBADP1 the amplified gene was ligated to the expression vector pET23a (Novagen, Madison, WI), which was then cloned into E. coli BL21(DE3) (Novagen), resulting in strain E. coli BL21(DE3)(pET23a::xylBADP1).
Thin-layer chromatography (TLC) analysis of A. baylyi ADP1 disruption mutants.
A 15-mg portion of lyophilized cell mass was extracted with 1 ml chloroform-methanol (2:1, vol/vol) for 3 h at room temperature. Each extract (80 μl), and 25 μg triolein and oleyl oleate as standards, was applied to a Silica 60 TLC plate (Merck, Darmstadt, Germany). The TLC plate was developed using hexane-diethyl ether-acetic acid (90:7.5:1 [vol/vol/vol]) as the solvent system, and spots were later visualized with sublimated iodine.
Preparation of cell-free crude extracts and soluble protein fractions.
Cells of E. coli BL21(DE3) pET23a::xylBADP1 were cultivated and protein expression was induced as described above. In total, cells from three 1,000-ml cultures were collected and washed once with sodium phosphate buffer (20 mM, pH 7.4). All subsequent steps were performed at 4°C. After the cell pellet was resuspended in precooled sodium phosphate buffer, resulting in a final volume of 45 ml, cells were disrupted by three passages in a French press. The crude extract obtained by centrifugation at 5,000 × g for 15 min was separated into an insoluble and a soluble protein fraction by 120 min of ultracentrifugation at 100,000 × g. Protein concentrations were determined by the method of Bradford (2).
Purification of aryl alcohol dehydrogenase XylBADP1.
All purification steps described were performed at 4°C. During all chromatographic purification steps, the A280 was measured with a LKB UV-M II UV monitor (Pharmacia), and conductivity was measured with a Pharmacia conductivity meter.
(i) Ion exchange chromatography (IEX).
The soluble protein fraction resulting from ultracentrifugation was applied to a 45-ml Q Sepharose high-pressure (HP) column (10 by 2.6 cm) at a flow rate of 1 ml min−1. After the column was washed with 2 volumes of sodium phosphate buffer at 2 ml min−1, the enzyme was eluted at a NaCl concentration of 100 mM. Fractions of 5 ml were collected and analyzed for NADH-dependent aldehyde reducing activity with octanal as the substrate. Fractions with significant activity were pooled, yielding 90 ml of protein solution.
(ii) Hydrophobic interaction chromatography (HIC).
The (NH4)2SO4 concentration of the Q-Sepharose pool was adjusted to 1 M by careful addition of solid (NH4)2SO4. The solution was then applied to a 30-ml octyl Sepharose HP column (6 by 2.6 cm) at a flow rate of 1 ml min−1. Bound proteins were washed with 2 volumes of sodium phosphate buffer containing 0.25 M (NH4)2SO4. Tightly bound enzymes, including XylBADP1, were eluted with (NH4)2SO4-free sodium phosphate buffer. Again, 5-ml fractions were collected and analyzed for NADH-dependent aldehyde-reducing activity. Fractions with significant activity were pooled.
(iii) Ultrafiltration.
The protein solution obtained during HIC was subsequently concentrated 10-fold in a Vivaspin 20 ultrafiltration tube (10-kDa cutoff; Sartorius, Göttingen, Germany) to concentrate the proteins and remove any remaining salts and was then rediluted to 40 ml with 20 mM sodium phosphate buffer.
(iv) Affinity chromatography.
A 55-ml Cibacron blue F3GA Sepharose CL6B column (10.5 × 2.6 cm) was loaded with the ultrafiltration fraction at 1 ml min−1. After the flow was halted for 30 min, the column was washed with 2 bed volumes of 20 mM sodium phosphate buffer (pH 7.4) at a flow rate of 2 ml min−1. The flowthrough containing the aldehyde-reducing activity was collected, and proteins were eluted with a NaCl concentration of 1 M in fractions of 5 ml. Active fractions were pooled, concentrated using a Vivaspin 20 ultrafiltration tube (10-kDa cutoff; Sartorius, Göttingen, Germany), and charged with 10% (vol/vol) glycerol to yield a protein concentration of 1.25 mg ml−1. The enzyme was stable for several months when stored at −70°C.
Determination of the native molecular weight of XylBADP1 by gel filtration.
The Mw of the purified protein was estimated by gel filtration on a 25-ml Superdex 200 column (22 by 1.2 cm) equilibrated with 20 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl. A 100-μl aliquot of the protein solution was applied to the column and separated with equilibration buffer at a flow rate of 0.75 ml min−1. Fractions of 0.5 ml were collected and tested for aldehyde-reducing activity. The elution volume of the most active fraction was used to estimate the native molecular weight by means of a calibration curve based on the elution volume of following standard proteins: ferritin (molecular weight, 440,000), aldolase (158,000), conalbumin (75,000), and ovalbumin (44,000). Native molecular masses were determined from measurements taken four times.
Spectrophotometric assays.
Spectrophotometric assays were performed in quartz cuvettes with a path length of 1 cm at 30°C in an Evolution 100 spectrophotometer (Thermo Fisher Scientific, Dreieich, Germany), which detected the decrease of the absorption of NADH at a wavelength of 340 nm. All substrates were dissolved in 20 mM sodium phosphate buffer (pH 7.4) prior to use and were then mixed to yield a concentration of 0.1 mM NADH and a 1 mM concentration of the respective aldehyde or alcohol. For substrates exhibiting a high rate of absorption at 340 nm, the concentrations were reduced to 0.1 mM, and extinction coefficients were determined in sodium phosphate buffer and added to the extinction coefficient of 6,220 M−1 cm−1 reported earlier for NADH (35) to calculate specific activities. These substrates were coniferyl aldehyde (ε340 = 23,955 M−1 cm−1), salicyl aldehyde (8,554 M−1 cm−1), and vanillin (18,015 M−1 cm−1). Water-insoluble substrates were dispersed in the buffer by sonication for 30 s (50% intensity) using a UW200 microtip sonicator (Bandelin Electronics, Berlin, Germany). An appropriate amount of protein (from 2 μg ml−1 for purified enzyme to up to 250 μg ml−1 for crude extracts) was added to the assay mixture, and the reaction was started by the addition of the aldehyde after the reaction had been run for at least 1 min with NADH and was monitored for at least 3 min or until a steady state was reached. Control assays without aldehyde were performed, and the determined background NADH-dependent activity was subtracted from the aldehyde-reducing activity. One unit of enzyme activity was defined as 1 μmol of substrate converted per min at 30°C. For tests with NADPH, NADP+, and NAD+ as the cofactors, NADH was replaced with one of these compounds. In case of tests with NAD(P)+, the increase of absorption at 340 nm was measured. For inhibition studies, a maximum concentration of the potential inhibitor of 1 mM was added to the reaction mixture 5 min prior to the start of the reaction by adding NADH and the substrate.
Determination of pH optima and temperature optimum.
To test the effects of different pH values on enzyme activity, 20 mM solutions of the following buffers were prepared and if necessary adjusted to the indicated pH with acetic acid, HCl, or NaOH, respectively: sodium acetate (pH 4.5 to 5.5), bis-Tris-Cl (pH 5.5 to 7.0), sodium phosphate (pH 7.0 to 8.0), Tris-Cl (pH 8.0 to 9.0), and glycine NaOH (pH 9.0 to 10.0). All necessary solutions for photometrical enzyme tests were prepared in the respective buffer except for the protein solution.
For determination of the temperature optimum the reaction mixtures were preincubated at the respective temperatures for 5 min prior to the addition of enzyme and substrate.
Polyacrylamide gel electrophoresis.
Separation of proteins was performed under denaturing conditions in 11% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gels as described by Laemmli (15). Separated proteins were stained unspecifically with Coomassie brilliant blue as described by Weber and Osborn (34). Denatured molecular masses were determined by 6-fold measurements of the Rf values of the denatured protein and by calculating the molecular mass based on the Rf values of standard proteins contained in the Amersham LMW calibration kit (GE Healthcare, Little Chalfont, United Kingdom).
RNA isolation and RT-PCR.
For the isolation of RNA from bacterial cells, an RNeasy minikit (Qiagen, Hilden, Germany) was used. Digestion of contaminating DNA was done with RNA-free DNase I (Roche Diagnostics, Mannheim, Germany). Nucleic acid concentrations were determined with a Nanodrop device (Peqlab Biotechnologie GmbH, Erlangen, Germany). Finally the RT-PCRs were performed with a OneStep RT-PCR kit (Qiagen, Hilden, Germany) following the recommendations of the manufacturer. Internal primers for the amplification of xylBADP1 from DNA were GCCCAGATGGAAGCTCTACGC (5′ end) and GTAAGATATTGCGGATGTGTTCGG (3′ end), which amplified a 405-bp internal fragment of xylBADP1. A concentration of 7.5 ng μl−1 of RNA was used in each reaction. As a standard DNA ladder, the GeneRuler 1-kb DNA ladder (Fermentas, St. Leon-Rot, Germany) was used.
RESULTS
Purification of the NADH-dependent enzyme XylBADP1.
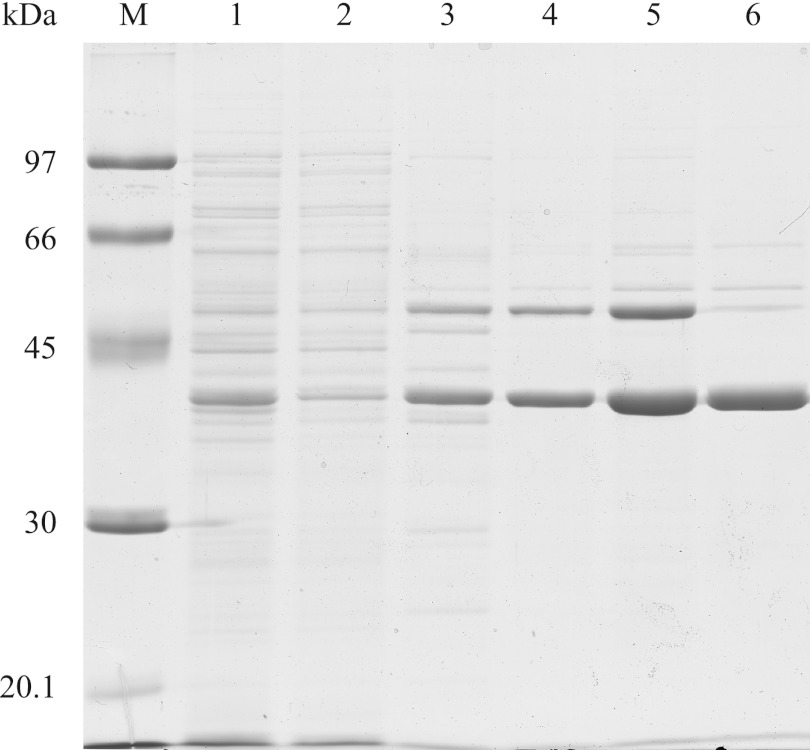
The enzyme was synthesized in cells of a recombinant E. coli BL21(DE3) strain harboring pET23a::xylBADP1 after induction with 0.5 mM IPTG. Formation of inclusion bodies was not observed during cultivation, and the enzyme was found exclusively in the soluble cell fraction. While the enzyme could be easily eluted from Q-Sepharose HP at a NaCl concentration of only 100 mM, it tightly bound to Octyl Sepharose from which it could not be completely eluted with (NH4)2SO4-free buffer which caused some protein losses. It turned out that XylBADP1 did not bind to a Cibacron blue F3GA Sepharose CL6B matrix. However, since significant amounts of remaining contaminating proteins were retained on this matrix, contaminating proteins could be removed to a large extent. In summary, the enzyme could be purified 13.2-fold, the specific activity of the purified enzyme was 104.5 U mg−1 of protein with benzaldehyde as the substrate, and the yield was 35.3% (Table 1). SDS-polyacrylamide gel electrophoresis (PAGE) revealed that the enzyme was purified to apparent homogeneity (Fig. 1).
Table 1.
Purification of recombinant aryl-alcohol dehydrogenase XylBADP1 found in A. baylyi ADP1 from E. coli BL21(DE3) pET23::xylBADP1
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U min−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 8,520 | 1,975 | 4.1 | 1.0 | 100.0 |
| Soluble fraction | 8,674 | 1,883 | 4.7 | 1.1 | 101.8 |
| Q-Sepharose | 4,630 | 386 | 12.0 | 2.9 | 54.3 |
| Octyl Sepharose | 2,822 | 121 | 23.3 | 5.6 | 33.1 |
| Ultrafiltration | 2,576 | 111 | 23.2 | 5.6 | 30.2 |
| Cibacron F3GA Sepharose CL6-B | 3,010 | 55.4 | 54.4 | 13.2 | 35.3 |
Fig 1.
SDS-PAGE of samples from all purification steps of the A. baylyi ADP1 aryl alcohol dehydrogenase XylBADP1. Each lane contained 10 μg of protein. Lane M, molecular weight markers; lane 1, cell crude extract; lane 2, soluble protein fraction; lane 3, Q-Sepharose pool; lane 4, octyl Sepharose pool; lane 5, ultrafiltration concentrate; lane 6, Cibacron blue F3GA Sepharose CL6B flowthrough.
Temperature and pH optima, molecular mass, and subunit structure.
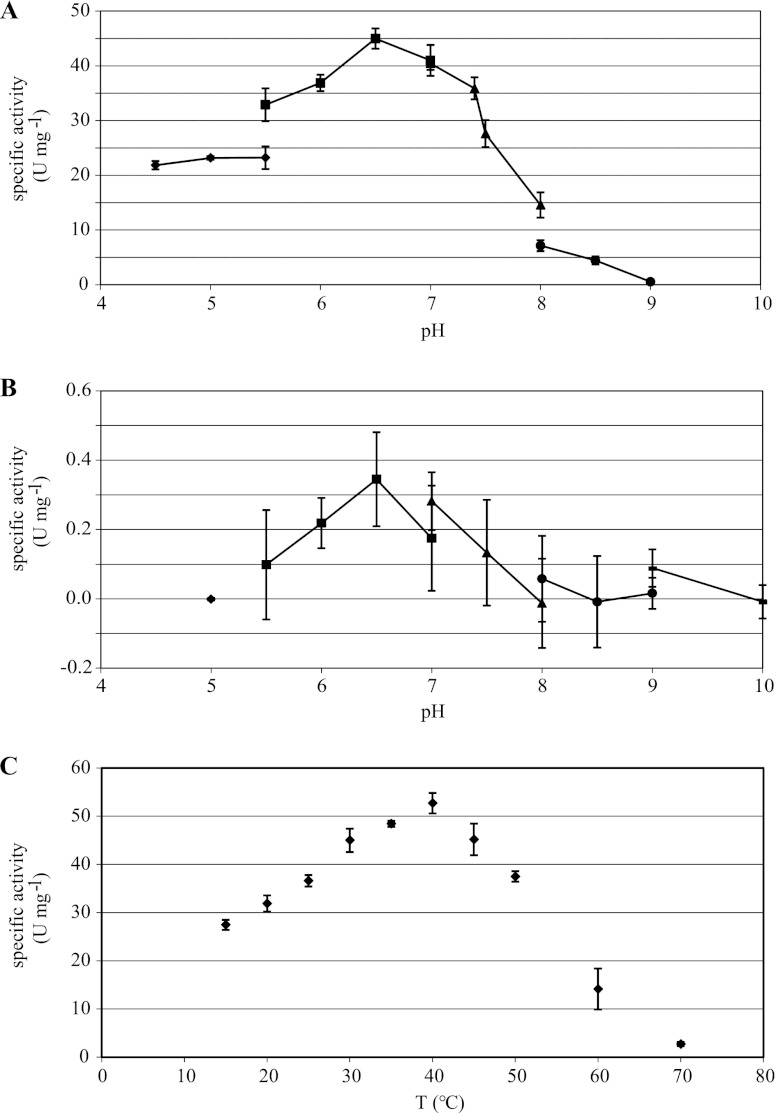
At pH 6.5, maximum activities for the reduction of benzaldehyde as well as for the oxidation of benzyl alcohol were found. Neither oxidation nor reduction activities were found at a pH of 9 or higher (Fig. 2).
Fig 2.
Effect of pH and temperature on enzyme activity. (A) Effect of pH on aldehyde reduction activity with benzaldehyde and NADH. (B) Effect of pH on alcohol oxidation activity with benzyl alcohol and NAD+. Buffers used include sodium acetate (◆), bis-Tris-Cl (■), sodium phosphate (▲), Tris-Cl (●), and glycine-NaOH ( ). (C) Effect of temperature on enzyme activity with benzaldehyde and NADH. All measurements were carried out in triplicate.
). (C) Effect of temperature on enzyme activity with benzaldehyde and NADH. All measurements were carried out in triplicate.
The enzyme was not thermostable, since it showed its highest activity at a temperature of 40°C. Whereas it reached 52% of its maximum activity at 15°C, it had lost 94% of its activity after only 2 min of incubation at 70°C (Fig. 2).
As calculated from the retention factors of molecular mass standard proteins, the molecular mass of XylBADP1 was estimated to be 40 ± 0.4 kDa by SDS-PAGE (Fig. 1) and 72.6 ± 2.2 kDa by gel filtration (data not shown), indicating a dimeric enzyme structure.
Substrate specificity.
The most specific substrate for the enzyme catalyzed reduction reaction was benzaldehyde, but the enzyme also catalyzed the reduction of aromatic aldehydes such as anisaldehyde and cinnamaldehyde, with specific activities of up to 72.2% compared to benzaldehyde. Additionally, a broad range of medium- and long-chain n-alkylaldehydes (C6 to C16) showing maximum activities of 36.5% with octanal and methyl branched aldehydes such as citronellal (21.8%) were used as the substrates. Some aromatic aldehydes (coniferyl aldehyde, vanillin, and salicyl aldehyde) had to be tested at nonsaturating concentrations of 0.1 mM due to their high extinction coefficient at 340 nm, making comparisons with the other investigated aldehydes difficult. The slightly modified enzyme test using these aromatic aldehydes showed that activities obtained with coniferyl aldehyde (96.4%) were comparable to those for benzaldehyde, while activities with salicyl aldehyde and vanillin were below 10%. Short-chain aldehydes (C2 to C4), glutaraldehyde, ketones, and aldoses did not serve as substrates (Table 2). NADH was the cofactor necessary for the reduction reaction and could not be replaced by NADPH (data not shown). The specific activities observed for the NAD+-dependent oxidation of alcohols were much lower than the activities with aldehydes and reached a maximum of only 2.2% of the activity found with benzaldehyde. As expected, the enzyme again showed the highest activities to aromatic substrates, but in contrast to the reduction reactions, coniferyl alcohol was the most specific substrate tested for the oxidation reaction, whereas benzyl alcohol accounted for only 12% of the activity with coniferyl alcohol. While diverse other aromatic alcohols like anisalcohol (82.6% of activity with coniferyl alcohol), cinnamyl alcohol (62.4%), and vanillyl alcohol (4.3%), several primary medium-chain aliphatic alcohols (e.g., hexanol [4.3%] and octanol [1.9%]), and methyl branched alcohols (e.g., geraniol [17.7%] and nerol [7.5%]) were converted to their respective aldehydes, no significant conversion of short-chain alcohols, 2-propanol, glycerol, citronellol, and salicyl alcohol was observed (Table 3).
Table 2.
Relative substrate specificity of XylBADP1 with different aldehydes compared to benzaldehydea
| Substrate | Concentration (mM) | Relative activity (%) |
|---|---|---|
| Acetaldehyde | 1 | >0.1 |
| Propanal | 1 | 0.6 |
| Butanal | 1 | 0.3 |
| Hexanal | 1 | 6.4 |
| Octanal | 1 | 36.5 |
| Decanal | 1 | 19.7 |
| Dodecanal | 1 | 8.9 |
| cis-11-Hexadecenal | 1 | 4.9 |
| Citral | 1 | 7.8 |
| Citronellal | 1 | 21.8 |
| Benzaldehyde | 1 | 100.0 |
| Anisaldehyde | 1 | 72.2 |
| Cinnamaldehyde | 1 | 58.7 |
| Glutaraldehyde | 1 | 0.6 |
| Glyoxal | 1 | 0.7 |
| Glucose | 1 | 0.2 |
| Mannose | 1 | >0.1 |
| 2-Decanone | 1 | 1.5 |
| Benzaldehyde | 0.1 | 100.0 |
| Vanillin | 0.1 | 4.0 |
| Salicyl aldehyde | 0.1 | 9.0 |
| Coniferyl aldehyde | 0.1 | 96.1 |
| Retinal | 0.1 | 0.1 |
Each value is an average of three independently determined values.
Table 3.
Relative substrate specificity of XylBADP1 with 1 mM concentrations of various alcohols compared to coniferylaldehydea
| Substrate | Relative activity (%) |
|---|---|
| Ethanol | 1.0 |
| Propanol | 0.7 |
| Butanol | 1.5 |
| Isopropanol | 1.2 |
| Hexanol | 4.1 |
| Octanol | 1.9 |
| Nerol | 7.5 |
| Geraniol | 17.7 |
| Citronellol | 0.6 |
| Benzyl alcohol | 12.1 |
| Anisalcohol | 82.6 |
| Cinnamyl alcohol | 62.4 |
| Coniferyl alcohol | 100.0 |
| Vanillyl alcohol | 4.3 |
| Salicyl alcohol | 0.7 |
| Glycerol | 0.5 |
Each value is an average of three independently determined values.
Kinetic parameters.
The apparent Km value for the reaction with benzaldehyde was 0.28 ± 0.04 mM, and Vmax was 19.6 ± 1.5 U mg−1. The apparent Km value for the reaction with NADH was found to be 0.017 ± 0.2 mM, while Vmax was 13.4 ± 0.44 U mg−1. Both reactions followed Michaelis-Menten kinetics (data not shown). The turnover numbers (kcat) were calculated to be 12,940 ± 1,010 s−1 for benzaldehyde and 8,920 ± 215 s−1 for NADH. The specificity constant (kcat/Km) was calculated to be 4.67 ± 0.5 × 107 s−1 M−1 for benzaldehyde and 5.28 ± 0.07 × 108 s−1 M−1 for NADH, respectively.
Inhibition of XylBADP1 activity.
Metal ions, iodoacetamide, N-ethylmaleimide, ethylenediamine tetraacetic acid (EDTA), and 1,10-phenanthroline were added to the reaction mixture at various concentrations to analyze their inhibitory effects. The enzyme was inhibited by Hg2+ (99.9% inhibition) and Ag+ (99.8%) at concentrations of 1 μM and 10 μM, respectively, and by Fe2+ (78.3%), Zn2+ (98.8%), and Cu2+ (100%) at concentrations of 1 mM. Enzyme activity was not affected negatively by Ca2+, Co2+, Mg2+, Mn2+, or Ni2+ even at concentrations as high as 1 mM, with Mn2+ maximally enhancing the activity by 28%. At concentrations of 1 mM, the enzyme was not susceptible to EDTA or iodoacetamide, but it was slightly inhibited by 1,10-phenanthroline (14.5% inhibition) and strongly inhibited by N-ethylmaleimide (42.6%) (Table 4).
Table 4.
Effects of metal ions, chelators, and thiol-blocking reagents on the activity of XylBADP1 with benzaldehyde and NADH as the substratea
| Inhibitor | Concn (μM) | Residual activity (%) |
|---|---|---|
| None (control) | 100.0 | |
| Ag+ | 1 | 100.1 |
| 10 | 0.2 | |
| 100 | 0 | |
| Ca2+ | 1,000 | 119.3 |
| Co2+ | 1,000 | 115.2 |
| Cu2+ | 1 | 105.8 |
| 10 | 33.4 | |
| 100 | 1.3 | |
| 1,000 | 0 | |
| Fe2+ | 1 | 109.9 |
| 10 | 113.4 | |
| 100 | 62.6 | |
| 1,000 | 21.7 | |
| Hg2+ | 1 | 0.1 |
| Mg2+ | 1,000 | 112.9 |
| Mn2+ | 1,000 | 128.0 |
| Ni2+ | 1,000 | 107.4 |
| Zn2+ | 1 | 105.8 |
| 10 | 64.5 | |
| 100 | 20.5 | |
| 1,000 | 1.2 | |
| EDTA | 1,000 | 120.9 |
| 1,10-Phenanthroline | 1,000 | 85.5 |
| Iodoacetamide | 1,000 | 123.5 |
| N-Ethylmaleimide | 1 | 97.5 |
| 10 | 79.0 | |
| 100 | 63.4 | |
| 1,000 | 57.4 |
Each value is an average of three independently determined values. Residual activity was measured relative to the activity in an inhibitor-free assay with benzaldehyde and NADH.
Expression of xylBADP1.
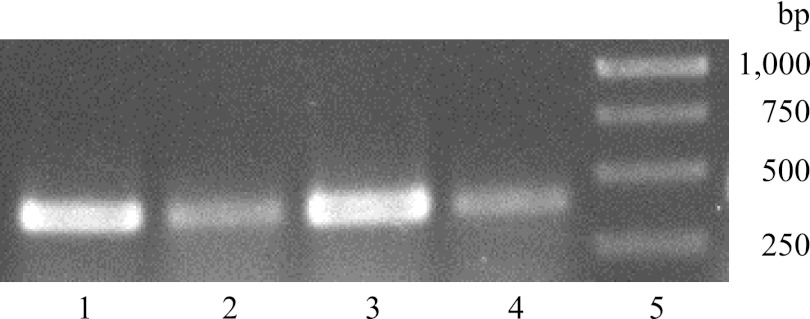
RNA was isolated from A. baylyi ADP1 cells grown in MSM with sodium gluconate under nitrogen-saturating and nitrogen-limiting conditions as well as with benzyl alcohol or coniferyl alcohol in nitrogen-saturated MSM. By RT-PCR analysis of the isolated RNA, a 405-bp DNA fragment was observed under all cultivation conditions, indicating constitutive expression of this gene (Fig. 3).
Fig 3.
Expression of xylBADP1 in A. baylyi ADP1 cells assessed by RT-PCR analysis using 37.5 ng RNA from cells incubated in MSM medium as the template in the presence of 5 mM benzyl alcohol for 3 h (lane 1), 1 mM coniferyl alcohol for 3 h (lane 2), 1% (wt/vol) sodium gluconate under nitrogen-saturated conditions (0.1% [wt/vol] NH4Cl) for 5 h (lane 3), and 1% (wt/vol) sodium gluconate under nitrogen-limited conditions (0.01% [wt/vol] NH4Cl) for 5 h (lane 4). Lane 5, 1-kb DNA ladder; bands correspond to 120 ng DNA (1,000 bp) or 50 ng DNA (750, 500, and 250 bp).
Growth and alcohol degradation behavior of disruption mutants.
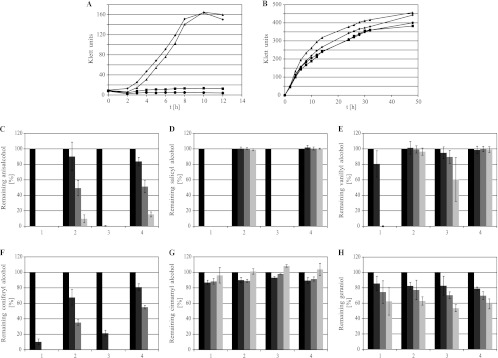
XylBADP1 exhibited its highest oxidating activities with aryl alcohols, e.g., benzyl alcohol and coniferyl alcohol, as well as with aliphatic and methyl branched alcohols, as substrates. To test the possible influence of a XylBADP1 disruption on the growth of wild-type A. baylyi ADP1 as well as xylBADP1 and areB single and double disruption mutants, the growth of these strains with benzyl alcohol and hexadecanol was investigated. areB disruption mutants were included in these experiments, since the enzymatic activity of AreB for aryl alcohols such as benzyl alcohol was described previously (9). Growth experiments revealed growth of wild-type A. baylyi ADP1 and the xylBADP1 disruption mutant to nearly identical cell densities within 12 h with 5 mM benzyl alcohol, while the areB and areB xylBADP1 disruption mutants showed no growth at all (Fig. 4A). With 15 mM hexadecanol as the sole carbon source, all four strains grew to nearly identical cell densities within 14 h (Fig. 4B).
Fig 4.
Effect of xylBADP1 disruptions on growth with several aromatic, aliphatic, and isoprenoid alcohols. (A) Growth of A. baylyi ADP1 strains in MSM with 5 mM benzaldehyde as the substrate. (B) Growth of A. baylyi ADP1 strains in MSM with 15 mM hexadecanol as the substrate. ◆, A. baylyi ADP1; ■, A. baylyi ADP1areBΩGm; ▲, A. baylyi ADP1xylBADP1ΩSm; ●, A. baylyi ADP1areBΩGmxylBADP1ΩSm. (C to H) Relative degradation of 1 mM anisalcohol (C), 1 mM salicyl alcohol (D), vanillyl alcohol (E), coniferyl alcohol (F), 1 mM cinnamyl alcohol (G), and 1 mM geraniol (H) by A. baylyi ADP1 strains in MSM within 0 (black), 4 (dark gray), 8 (gray), and 24 (light gray) h. Percentage degradation values are based on the alcohol contents of the culture medium after 0 h. (1) A. baylyi ADP1; (2) A. baylyi ADP1areBΩGm; (3) A. baylyi ADP1xylBADP1ΩSm; (4) A. baylyi ADP1areBΩGmxylBADP1ΩSm. All experiments were done in triplicate.
Vanillyl alcohol, anisalcohol, salicyl alcohol, cinnamyl alcohol, coniferyl alcohol, and geraniol were used as the substrates at a maximum concentration of 1 mM, since it turned out that they or their catabolites inhibited cell growth. Growth with such low concentrations of substrate would yield only very low cell densities that would not allow us to distinguish accurately between the growth rates of the investigated strains. Thus, a possible involvement of XylBADP1 in the oxidation of the tested alcohols was instead determined by HPLC following the degradation of the respective alcohol in the medium by a defined amount of cells.
Anisalcohol was degraded by all four strains. While it was completely degraded by wild-type A. baylyi ADP1 and the xylBADP1 disruption mutant within the first 4 h of incubation, degradation was much slower in both the areB and the double disruption mutant, and even after 24 h small amounts of anisalcohol could be detected in these cultures (Fig. 4C). Interestingly in all four cultures 4-methoxybenzoic acid was accumulated (data not shown).
Salicyl alcohol was degraded by wild-type A. baylyi ADP1 and xylBADP1 disruption mutant cells within 4 h after inoculation. No degradation of this substrate was observed for cultures of the areB single disruption mutant or of the areB xylBADP1 double disruption mutant (Fig. 4D).
Cells of A. baylyi ADP1 and of the xylBADP1 disruption mutant were able to oxidize vanillyl alcohol. However, while the wild type degraded vanillyl alcohol completely within 8 h, only about 40% of the initial vanillyl alcohol amount was oxidized by the xylBADP1 disruption mutant after 24 h of incubation. No degradation was observed in cultures of the single areB disruption mutant or the areB xylBADP1 double disruption mutant (Fig. 4E).
All four strains were able to reduce coniferyl alcohol. While the wild type of A. baylyi ADP1 and the xylBADP1 disruption mutant converted 90% and 79%, respectively, of the initially present coniferyl alcohol within the first 4 h and degraded it completely within 8 h, the areB and areB xylBADP1 disruption mutants reduced only 32% and 19%, respectively, of the alcohol within 4 h and 65% and 45% within 8 h (Fig. 4F).
While geraniol was degraded by all four strains at low and comparable rates (Fig. 4H), cinnamyl alcohol was not oxidized at all by these strains (Fig. 4G).
Wax ester synthesis.
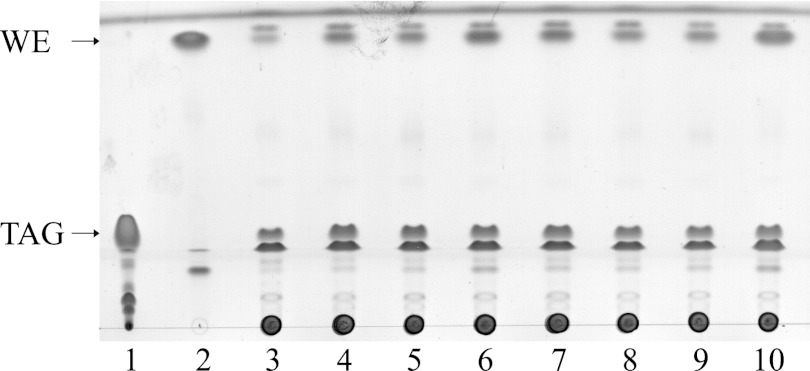
Tani et al. (31) suggested in their study the involvement of an NADPH-dependent aldehyde reductase (AlrA) in wax ester synthesis of Acinetobacter. Due to their biochemical characteristics, the enzymes AreB (9) and XylBADP1, which was characterized in this study, might also have such a function, and thus, TLC analyses of wild-type A. baylyi ADP1 cells, three single disruption mutants (alrA, areB, and xylBADP1), three double disruption mutants, and a triple disruption mutant of these genes were done to test the wax ester synthesis ability of these strains if grown with gluconate as the sole carbon source. All strains were grown in MSM with gluconate for the first 24 h and were then transferred into fresh nitrogen-limited MSM with gluconate for additional 24 h. The separation of the extracted lipids by TLC revealed that all tested strains of A. baylyi ADP1 were able to synthesize wax esters using gluconate as the carbon source (Fig. 5).
Fig 5.
TLC analysis of lipids extracted from lyophilized cells of wild-type A. baylyi ADP1 or its disruption mutant. Cells were grown with gluconate in MSM for 24 h and were then transferred to nitrogen-limited MSM with gluconate for additional 24 h. Reference substances were 25 μg triolein (lane 1) and 25 μg oleyl oleate (lane 2). Extracts from 1.2 mg lyophilized cell mass were applied to the lanes as follows: 3, A. baylyi ADP1 wild type; 4, A. baylyi ADP1alrAΩKm; 5, A. baylyi ADP1areBΩGm; 6, A. baylyi ADP1xylBADP1ΩSm; 7, A. baylyi ADP1alrAΩKmareBΩGm; 8, A. baylyi ADP1alrAΩKmxylBADP1ΩSm; 9, A. baylyi ADP1areBΩGmxylBADP1ΩSm and 10, A. baylyi ADP1alrAΩKm areBΩGmxylBADP1ΩSm.
DISCUSSION
The NAD+-dependent enzyme XylBADP1 from A. baylyi ADP1 exhibiting enzymatic activity for aliphatic and aromatic alcohols and aldehydes was purified from a recombinant E. coli strain and characterized. It had the highest homology (60%) to a putative aryl alcohol dehydrogenase from Sphingobium chlorophenolicum L-1 (GenBank accession no. CP002799.1); however, except for the protein sequence, no data are available for this enzyme. Additionally, XylBADP1 is 45% to 57% homologous to many other aryl or benzyl alcohol dehydrogenases from various bacteria known to be able to degrade aliphatic or aromatic compounds. Among many others, these include the aryl alcohol dehydrogenase XylB from Rhodococcus opacus TKN14 (54%) (22) and from the P. putida TOL plasmid (48%) (28). Furthermore, XylBADP1 is homologous to AreB from A. baylyi ADP1 (45%) (9), and its coding gene is located in the neighborhood of a putative aldehyde dehydrogenase, just as areB is (1). From all these facts, it was assumed that XylBADP1 might be involved in the degradation of aryl alcohols or related substances. Interestingly, none of the other Acinetobacter strains sequenced so far (e.g., several A. baumannii strains, A. lwoffii SH145, A. junii SH205, A. radioresistens SH164, A. haemolyticus ATCC 19194, A. johnsonii SH046, and A. calcoaceticus RUH2202) possesses a gene corresponding to xylBADP1 from A. baylyi ADP1, indicating an involvement in a special metabolic function exhibited only by strain ADP1.
Like its homologues from other genera, the amino acid sequence of XylBADP1 exhibits all the characteristics of a group I long-chain zinc-dependent alcohol dehydrogenases (ADH) as it is typified by horse liver ADH (4). In XylBADP1 the catalytic Zn2+ may be bound by the conserved residues Cys40, His61, and Cys168, while the four Cys residues at positions 90, 93, 96, and 104 may bind the structural Zn2+. The enzyme also contains the characteristic zinc-dependent alcohol dehydrogenase motif GHEXXGXXXXXGXXV (23) at residues 60 to 74. In XylBADP1, as in AreB from A. baylyi ADP1, XylB from R. opacus TKN14, and XylB from the P. putida TOL plasmid, His51 (according to the position in the horse liver ADH), which is conserved in almost all of the long-chain zinc-dependent ADH and which is believed to act as a general base during the catalytic reaction, is replaced by a hydrophobic residue, which is in this case Val45. However, it has been suggested by Inoue et al. (7) that an adjacent His residue might take over this role in XylB from P. putida. Based on this assumption, the adjacent residue His41 might function as the necessary base in XylBADP1.
Although most NAD(P)+-dependent alcohol dehydrogenases preferentially catalyze the oxidation of alcohols into aldehydes (23), few enzymes have been described which catalyze the reverse reaction at a higher rate (31, 36). However, XylBADP1 from A. baylyi ADP1 shows a high preference for aldehydes, while its specific activities for the oxidation reactions are very low. Therefore, XylBADP1 might be better designated an aldehyde reductase than an alcohol dehydrogenase. Nevertheless, its affinity for its preferred substrate benzaldehyde is relatively low, since the Km value for benzaldehyde of 0.28 mM is 10 times higher than, e.g., that for XylB from the P. putida TOL plasmid (27). Additionally, the enzyme exhibits a very low reaction velocity compared to XylB from Pseudomonas, as Vmax of XylB is 45 times higher than that of XylBADP1. As was reported for many aryl alcohol dehydrogenases (e.g., AreB from A. baylyi ADP1 and XylB from P. putida), for XylBADP1 the highest activities were detected with benzaldehyde, cinnamyl aldehyde, or their substituted derivatives, indicating an enzymatic function during the degradation of aromatic alcohols. On the other hand, the specificity for aliphatic medium-chain aldehydes was more similar to that of AlrA (25% homology) (31), supporting the possible involvement of XylBADP1 in storage lipid metabolism or in the detoxification of degradation products harboring aldehyde groups.
While AreB from Acinetobacter was found to be a homotetramer like the ADH from Saccharomyces cerevisiae (16) and most bacterial ADH (23), its purified homologue in strain ADP1, XylBADP1, is a homodimer, like the mammalian ADH from horse liver (10) and the XylB enzymes from R. opacus TKN14 and P. putida, strongly supporting the prediction of a XylB-like protein presented earlier by Barbe et al. (1). The many similarities of XylBADP1 with XylB enzymes, which themselves are more closely related to mammalian ADH than to bacterial ones (23), indicate that its evolution in A. baylyi ADP1 was independent from that of AreB.
The optimum pH of most prokaryotic and eukaryotic alcohol dehydrogenases for the oxidative reaction has been reported to be about 8.5 to 10. Contrary to that finding, the optimum pH for XylBADP1 determined in this study was 6.5, and to our knowledge, such a low optimum pH for the oxidation of alcohols has not been reported previously for bacterial ADH. The reduction of aldehydes by XylBADP1 was also optimum at pH 6.5, which is close to the optimum pH of 5.7 reported for XylB from the P. putida TOL plasmid (27) and to the optimum pH of 7 described for AlrA from Acinetobacter (31).
XylBADP1 was particularly sensitive to inactivation by Hg2+, Ag+, Cu2+, and Zn2+ and to a lower extent also to Fe2+, which inhibited the enzyme activity completely or at least significantly. The complete inhibition of a zinc-containing enzyme by Zn2+ was somehow unexpected, but was previously also reported for AlrA from Acinetobacter (31) and ADH from Thermococcus guaymasensis (37). Nevertheless, the inhibition of the enzyme activity by the mentioned metal ions suggests the importance of sulfhydryl groups for the activity of the enzyme which is also supported by the sensitivity of XylBADP1 to N-ethylmaleimide, a thiol-blocking reagent that is known to inhibit all bacterial aromatic alcohol dehydrogenases as well as mammalian and yeast alcohol dehydrogenases (17). In contrast to many other alcohol dehydrogenases, XylBADP1 showed no susceptibility to iodoacetamide. Although XylBADP1 belongs to the zinc-containing alcohol dehydrogenases, it was not or only slightly susceptible to inhibition by metal ion chelators like EDTA or 1,10-phenanthroline, respectively, a property it shares with, for example, AlrA and AreB from Acinetobacter (17, 31), XylB from the P. putida TOL-plasmid (27), several other bacterial aromatic alcohol dehydrogenases (8, 29), and the archaeal ADH from T. guaymasensis (37). Contrary to this, the zinc-containing yeast and horse liver alcohol dehydrogenases are sensitive to metal ion chelators (30), indicating a general difference in resistance of the zinc ions to chelation between prokaryotic and eukaryotic alcohol dehydrogenases.
It was demonstrated that xylBADP1 is expressed during growth on benzyl and coniferyl alcohol as well as under nitrogen-saturated and nitrogen-limited conditions in the presence of sodium gluconate as a carbon source. This finding indicates that (i) ACIAD1578 definitively fulfills some enzymatic function within the bacterial cell and (ii) it might be constitutively expressed.
An involvement in the degradation of benzyl alcohol and salicyl alcohol, which are both degraded via the catechol pathway, could be excluded, since a lack of growth on benzyl alcohol and the observed inability to degrade salicyl alcohol, respectively, could be attributed to an areB disruption, while xylBADP1 disruptions had no influence on growth or alcohol degradation (Fig. 4A, D, and G). Of the three alcohols tested, which are degraded via the protocatechuate pathway, only an involvement of XylBADP1 in the oxidation of coniferyl alcohol is implied by the obtained data, since the degradation of coniferyl alcohol is reduced significantly in the xylBADP1 single disruption mutant compared to wild-type A. baylyi ADP1 or in the areB xylBADP1 double disruption mutant compared to A. baylyi ADP1 areBΩGm. However, most of the coniferyl alcohol is degraded by AreB and at least one additional coniferyl alcohol-oxidating enzyme, since even the areB xylBADP1 mutant is still able to degrade coniferyl alcohol to some extent (Fig. 4F). Nevertheless, no involvement of XylBADP1 in the oxidation of vanillyl alcohol and anisalcohol was observed. In fact, vanillyl alcohol degradation in A. baylyi ADP1 was found to be solely dependent on AreB, since areB disruption mutants show no vanillyl alcohol degradation. Although the degradation of vanillyl alcohol is delayed in the xylBADP1 disruption mutant, an involvement of XylBADP1 in this reaction is unlikely, since the areB mutant, which possesses an active XylBADP1, showed no signs of vanillyl alcohol degradation. The delayed vanillyl alcohol degradation by A. baylyi ADP1xylBADP1ΩSm cannot be explained as yet (Fig. 4E). Anisalcohol is oxidized by at least two enzymes, including AreB (Fig. 4C). XylBADP1 is not involved in that reaction. Interestingly, 4-methoxybenzoic acid was accumulated during anisalcohol degradation by all four strains, but we did not investigate whether this accumulation was due to a lack of 4-methoxybenzoic acid demethylase activity in A. baylyi ADP1 or a toxic effect of the catabolite on bacterial cells.
Apart from the involvement of XylBADP1 in coniferyl alcohol degradation and the general possibility of an involvement in basal cellular metabolism, a possible role of XylBADP1 during wax ester metabolism was supported by the relatively high specificity of the enzyme for the reduction of medium- and long-chain aliphatic aldehydes to their corresponding alcohols, which might then be used for synthesis of wax esters by WS/DGAT (14), and its low activity with alcohol substrates. However, the probability that XylBADP1 is part of the wax ester synthesis pathway in Acinetobacter appears to be low, since all mutants with single, double, and triple disruptions of known alcohol dehydrogenases or aldehyde reductases (AlrA, AreB, and XylBADP1) investigated in this study were still able to synthesize wax esters from gluconate as the sole source of carbon. This would not have been observed if one or all of the investigated enzymes were involved in the reduction of fatty aldehydes (Fig. 5). This finding strongly suggests (i) that none of the tested enzymes is involved in fatty aldehyde reduction on its own, (ii) that a combined activity of the tested enzymes can be excluded, and (iii) that, hence, at least one additional still unknown enzyme capable of reducing fatty aldehydes must be involved in A. baylyi ADP1 wax ester synthesis.
Furthermore, it was shown that neither areB nor xylBADP1 disruption mutants were impaired in their ability to grow with hexadecanol as the sole carbon source or to oxidize geraniol (Fig. 4B and H) and that therefore a catabolic role of XylBADP1 in long-chain alcohol oxidation or isoprenoid alcohol oxidation, respectively, is also unlikely.
Recombinant bacterial strains expressing XylBADP1 might be suitable tools for reduction reactions in biotechnological applications involving aliphatic or aromatic aldehydes. For example, the recombinant enzyme might be used in specialty chemistry for the synthesis of alcohols such as cinnamyl alcohol or geraniol or the synthesis of precursors for the production of jojoba-like wax esters which are all used in the pharmaceutical, food, and cosmetics industries. Furthermore, it may be possible to synthesize the mentioned wax esters from cheap carbon sources like simple sugars if XylBADP1 is coexpressed with a suitable acyl-CoA reductase and a wax ester-synthesizing enzyme.
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Barbe V, et al. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 3. Brisou J, Prevot AR. 1954. Etudes de systematique bacterienne. X. Revision des especes reunies dans le genre Achromobacter. Ann. Inst. Pasteur 86:722–728 [PubMed] [Google Scholar]
- 4. Eklund H, Samama JP, Wallen L, Branden CI. 1981. Structure of a triclinic ternary complex of horse liver alcohol dehydrogenase. J. Mol. Biol. 146:561–587 [DOI] [PubMed] [Google Scholar]
- 5. Fischer R, Bleichrodt FS, Gerischer UC. 2008. Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154:3095–3103 [DOI] [PubMed] [Google Scholar]
- 6. Gillooly DJ, Robertson AGS, Fewson CA. 1998. Molecular characterization of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II of Acinetobacter calcoaceticus. Biochem. J. 330:1375–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoue J, Tomioka N, Itai A, Harayama S. 1998. Proton transfer in benzyl alcohol dehydrogenase during catalysis: alternate proton-relay routes. Biochemistry 37:3305–3311 [DOI] [PubMed] [Google Scholar]
- 8. Jaeger E, Eggeling L, Sahm H. 1981. Partial purification and characterization of a coniferyl alcohol dehydrogenase from Rhodococcus erythropolis. Curr. Microbiol. 6:333–336 [Google Scholar]
- 9. Jones RM, Collier LS, Neidle EL, Williams PA. 1999. areABC genes determine the catabolism of aryl esters in Acinetobacter sp. strain ADP1. J. Bacteriol. 181:4568–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jørnvall H. 1970. Horse liver alcohol dehydrogenase. The primary structure of the protein chain of the ethanol-active isoenzyme. Eur. J. Biochem. 16:25–40 [DOI] [PubMed] [Google Scholar]
- 11. Juni E. 1972. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112:917–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Juni E, Janik A. 1969. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum). J. Bacteriol. 98:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juni E. 1978. Genetics and physiology of Acinetobacter. Annu. Rev. Microbiol. 32:349–371 [DOI] [PubMed] [Google Scholar]
- 14. Kalscheuer R, Steinbüchel A. 2003. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278:8075–8082 [DOI] [PubMed] [Google Scholar]
- 15. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 16. Leskovac V, Trivić S, Peričin D. 2002. The three zinc-containing alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 4:481–494 [DOI] [PubMed] [Google Scholar]
- 17. MacKintosh RW, Fewson CA. 1988. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Biochem. J. 255:653–661 [PMC free article] [PubMed] [Google Scholar]
- 18. Neidle EL, Shapiro MK, Ornston LN. 1987. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J. Bacteriol. 169:5496–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ornston LN, Neidle EL. 1991. Evolution of genes for the β-ketoadipate pathway in Acinetobacter calcoaceticus, p 201–237 In Towner K, Bergogne-Berezin E, Fewson CA. (ed), The biology of Acinetobacter. Plenum Press, New York, NY [Google Scholar]
- 20. Overhage J, Priefert H, Rabenhorst J, Steinbüchel A. 1999. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl. Microbiol. Biotechnol. 52:820–828 [DOI] [PubMed] [Google Scholar]
- 21. Palmen R, Vosman B, Buijsman P, Breek CK, Heilingwerf KJ. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295–305 [DOI] [PubMed] [Google Scholar]
- 22. Peng X, et al. 2006. Characterization of four Rhodococcus alcohol dehydrogenase genes responsible for the oxidation of aromatic alcohols. Appl. Microbiol. Biotechnol. 71:824–832 [DOI] [PubMed] [Google Scholar]
- 23. Reid MF, Fewson CA. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 20:13–56 [DOI] [PubMed] [Google Scholar]
- 24. Reiser S, Somerville C. 1997. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179:2969–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Schlegel HG, Kaltwasser H, Gottschalk G. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209–222 [PubMed] [Google Scholar]
- 27. Shaw JP, Harayama S. 1990. Purification and characterization of TOL plasmid-encoded benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase of Pseudomonas putida. Eur. J. Biochem. 191:705–714 [DOI] [PubMed] [Google Scholar]
- 28. Shaw JP, Rekik M, Schwager F, Harayama S. 1993. Kinetic studies on benzyl alcohol dehydrogenase encoded by TOL plasmid pWW0: a member of the zinc-containing long chain alcohol dehydrogenase family. J. Biol. Chem. 268:10842–10850 [PubMed] [Google Scholar]
- 29. Suhara K, Takemori S, Katagiri M. 1969. The purification and properties of benzyl alcohol dehydrogenase from Pseudomonas sp. Arch. Biochem. Biophys. 130:422–429 [DOI] [PubMed] [Google Scholar]
- 30. Sund H, Theorell H. 1963. Alcohol dehydrogenase, p 25–83 In Boyer PD, Lardy H, Myrbäck K. (ed), The enzymes, 2nd ed, vol 7 Academic Press, New York, NY [Google Scholar]
- 31. Tani A, Sakai Y, Ishige T, Kato N. 2000. Thermostable NADP1-dependent medium-chain alcohol dehydrogenase from Acinetobacter sp. strain M-1: purification and characterization and gene expression in Escherichia coli. Appl. Environ. Microbiol. 66:5231–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vaneechoutte M, et al. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wahlen BD, Oswald WS, Seefeldt LC, Barney BM. 2009. Purification, characterization, and potential bacterial wax production role of an NADPH-dependent fatty aldehyde reductase from Marinobacter aquaeolei VT8. Appl. Environ. Microbiol. 75:2758–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weber K, Osborn M. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406–4412 [PubMed] [Google Scholar]
- 35. Windholz M. (ed). 1983. The Merck index, 10th ed, p 910 Merck & Co. Inc., Rahway, NJ [Google Scholar]
- 36. Yamada H, Shimizu S, Kataoka M, Sakai H, Miyoshi T. 1990. A novel NADPH-dependent aldehyde reductase, catalyzing asymmetric reduction of β-keto acid esters, from Sporobolomyces samonicolor: purification and characterization. FEMS Microbiol. Lett. 70:45–48 [Google Scholar]
- 37. Ying X, Ma K. 2011. Characterization of a zinc-containing alcohol dehydrogenase with stereoselectivity from hyperthermophilic archaeon Thermococcus guaymasensis. J. Bacteriol. 193:3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]