Abstract
Since 1997, cases of Vibrio parahaemolyticus-related gastroenteritis from the consumption of raw oysters harvested in Washington State have been higher than historical levels. These cases have shown little or no correlation with concentrations of potentially pathogenic V. parahaemolyticus (positive for the thermostable direct hemolysin gene, tdh) in oysters, although significant concentrations of tdh+ V. parahaemolyticus strains were isolated from shellfish-growing areas in the Pacific Northwest (PNW). We compared clinical and environmental strains isolated from the PNW to those from other geographic regions within the United States and Asia for the presence of virulence-associated genes, including the thermostable direct hemolysin (tdh), the thermostable-related hemolysin (trh), urease (ureR), the pandemic group specific markers orf8 and toxRS, and genes encoding both type 3 secretion systems (T3SS1 and T3SS2). The majority of clinical strains from the PNW were positive for tdh, trh, and ureR genes, while a significant proportion of environmental isolates were tdh+ but trh negative. Hierarchical clustering grouped the majority of these clinical isolates into a cluster distinct from that including the pandemic strain RIMD2210633, clinical isolates from other geographical regions, and tdh+, trh-negative environmental isolates from the PNW. We detected T3SS2-related genes (T3SS2β) in environmental strains that were tdh and trh negative. The presence of significant concentrations of tdh+, trh-negative environmental strains in the PNW that have not been responsible for illness and T3SS2β in tdh- and trh-negative strains emphasizes the diversity in this species and the need to identify additional virulence markers for this bacterium to improve risk assessment tools for the detection of this pathogen.
INTRODUCTION
Vibrio parahaemolyticus is a naturally occurring Gram-negative pathogen that is responsible for the majority of bacterial infections from consumption of seafood in the United States (39). Infection is usually acquired by consumption of raw or undercooked shellfish, particularly oysters, and results in self-limiting, mild to severe gastroenteritis. Wound infections are less frequent but can occur when an open wound is exposed to seawater (9, 25).
The bacterium was first identified in Japan in 1950 and has long been recognized as a cause of sporadic seafood-related illness in Asian countries. Sporadic V. parahaemolyticus infections and smaller outbreaks from consumption of raw or undercooked seafood have also been reported in the United States since the 1970s (12). The species is very diverse, and strains that carry the gene for the thermostable direct hemolysin (tdh) and/or the thermostable-related hemolysin (trh) are considered to be pathogenic (42, 55). Strains of V. parahaemolyticus belong to different serogroups based on differences in their lipopolysaccharide (O) and capsular (K) antigens (27). A new and unique clone of serotype O3:K6 emerged in India in 1996 (37, 48). This clone, referred to as the pandemic clone, has spread globally from southeast Asia to Europe, Africa, South America, and the United States, and it has been responsible for the majority of V. parahaemolyticus-related infections worldwide (49).
The first reported outbreak of V. parahaemolyticus O3:K6-related infections in the United States occurred in 1998 and was associated with oysters harvested from Galveston Bay, TX (13). The outbreak affected 416 people in 12 states and was attributed to higher than normal water temperature and salinity. Later that same year, another multistate outbreak attributed to O3:K6 was reported from New York, New Jersey, and Connecticut and was linked to oysters from Long Island, NY (7). During the same period when O3:K6 V. parahaemolyticus was first recognized in the United States, a large multistate outbreak affecting more than 200 people was linked to oysters harvested from the coasts of Oregon, Washington, California, and British Columbia, Canada, areas where only sporadic cases of V. parahaemolyticus-related illnesses had been reported previously (8). Until that point, water temperatures and environmental conditions were never considered to be conducive for large outbreaks. However, during that outbreak, illnesses were attributed to strains with a different serotype, O4:K12 (12).
The abundance of V. parahaemolyticus in environmental samples varies considerably depending on the geographic location. Studies conducted in the United States suggest a seasonal and regional variation in populations of total and potentially pathogenic V. parahaemolyticus in oysters with reported concentrations of total V. parahaemolyticus between 101 and >104/g in oysters (15, 30, 31). Potentially pathogenic strains (tdh+ or trh+) have been isolated less frequently from the environment and appear to constitute from 1 to 10% of the population (10, 11, 16, 18, 24, 55). When detected, concentrations of the tdh+ strain have been reported to be <13/g (10, 16, 30). Low densities of tdh+ trh+ strains have also been reported from Japan and Chile (2, 20). However, significantly higher densities have been reported in oysters from one study location in India where tdh was detected in 10.2% of strains and trh in 60% of strains isolated from oysters (14).
Over the past decade, there have been increasing reports of V. parahaemolyticus-related gastroenteritis from the consumption of raw oysters harvested in Washington State (Washington State Department of Health [WDOH], personal communication). Initially these increases were speculated to be a result of higher than normal water temperatures. However, extensive monitoring and analyses of oysters by the WDOH have shown little or no correlation between water temperature at time of harvest, concentrations of potentially pathogenic (tdh+) V. parahaemolyticus, and illnesses. The continued persistence of illnesses has prompted public health officials and the oyster industry in Washington State to implement stricter postharvest handling regimens to reduce the potential increase in concentrations of V. parahaemolyticus in oysters after harvest. Nevertheless, in spite of these postharvest temperature control measures, higher than normal levels of illness have continued to occur (WDOH, personal communication).
Environmental investigations conducted in Washington State following outbreaks in 1997 and 1998 noted a wide variation in densities of V. parahaemolyticus in oyster tissue over a period of a month (August to September), with total populations of V. parahaemolyticus between <3 and 46,000/g (16). However, potentially pathogenic (tdh+ and/or trh+) strains either were not detected in oyster and water samples or their concentrations, when detected, were <10/g (16). In the same study, tdh+ strains also were not detected in oysters from two other study sites, Galveston Bay, TX, and Long Island, NY. In a separate study conducted by our laboratory in 2007, we detected a significantly high concentration (>50% in some samples) of tdh+ V. parahaemolyticus strains in water and net tow samples (23 and unpublished results). Concentrations of tdh+ strains were low or nondetectable in oysters harvested from these same growing areas, although V. parahaemolyticus-related illnesses continued to occur during this time period from consumption of raw oysters. This apparent lack of a correlation between concentrations of tdh+ isolates in water and oysters and the occurrence of illness suggests that additional factors contribute to the virulence of these strains.
In an effort to assess the similarities and differences between clinical and environmental V. parahaemolyticus isolates from the PNW and the O3:K6 pandemic strains, we compared the distribution of tdh, trh, and other putative virulence-related genes, including genes associated with type 3 secretion systems (T3SS1 and T3SS2), as well as pandemic markers in clinical and environmental isolates from the PNW and other geographic regions. Additional information on differences in the genetic makeup of V. parahaemolyticus strains from the Pacific Northwest is necessary for improved risk assessment of this pathogen.
MATERIALS AND METHODS
Bacterial strains.
The sources of the V. parahaemolyticus strains chosen for this study are shown in Table S1 in the supplemental material. The clinical strains were obtained from the Washington State Department of Health, Public Health Laboratories, Shoreline, WA, and were isolated from patients with gastroenteritis that was traced to the consumption of raw oysters between 1997 and 2007. Clinical strains that included strains isolated in the United States and other regions worldwide were also obtained from the Gulf Coast Seafood Laboratory, Food and Drug Administration (FDA), Dauphin Island, AL. The environmental strains were isolated in our laboratory from water, oysters, and net tow samples of plankton by direct plating on thiosulfate-citrate bile salt sucrose agar (BD, Franklin Lakes, NJ) and identified by multiplex PCR using published protocols (3). The majority of environmental samples were from oyster-growing areas in Hood Canal and the Washington coast during 2007. During the period of sampling, several illnesses were traced to oysters harvested from these areas. Additional strains isolated from shellfish and water from diverse geographical locations were obtained from the FDA Pacific Regional Laboratory, Bothell, WA. Strains were grown in Trypticase soy broth (BBL, BD, Franklin Lakes, NJ) with an additional 1.5% NaCl. Genomic DNA was extracted using the Qiagen QIAamp DNA minikit (Qiagen, Valencia, CA) and stored at −20°C.
Analysis of clinical and environmental V. parahaemolyticus strains.
V. parahaemolyticus strains were initially analyzed for the presence of the hemolysins tdh and trh (3), two pandemic group-specific markers, toxRS (new) and orf8 (22, 37, 41), urease (ureR) (29), the homolog of the gene encoding the N-acetylglucosamine (GlcNAc)-binding protein, gbpA, in V. cholerae (33), and selected genes encoding T3SS1 and T3SS2 (listed in Table S2 in the supplemental material) by PCR by following previously published protocols (see Table S2). PCR primers for the initial analysis were based on the sequenced strain RIMD2210633. Some strains in our study showed ambiguous results using the previously published protocols for trh (3). Therefore, the presence or absence of trh in all strains was subsequently verified using a second set of primers, trh53-F and trh531-R (see Table S2). For the detection of trh, each 25-μl reaction mixture included 5 μl of 5× buffer, 400 nM each of the primers, 0.2 mM each of the deoxynucleoside triphosphates (dNTPs), 1.5 mM MgCl2, 0.625 U of Go Taq Flexi DNA polymerase (Promega, Madison, WI), and 25 ng DNA template. Initial denaturation at 94°C for 2.5 min was followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C of 9.5 min. The expected PCR product is predicted to be 479 bp. The oligonucleotide primers VpgbpAF and VpgbpAR were used for detection of the putative gbpA (see Table S2). The PCR for amplification of gbpA was performed using 5 μl of 5× buffer, 1.5 mM MgCl2, 0.2 mM (each) dNTPs, 400 nM each of the primers, 0.625 U Go Taq polymerase (Promega Corporation, Madison, WI), and 25 ng DNA template. The amplification reactions were performed with an initial denaturation at 95°C for 1.5 min, 30 cycles of 95°C for 1 min, 57°C for 1 min, and 72°C for 1 min, and a final extension of 72°C for 7 min. V. parahaemolyticus RIMD2210633 (tdh+, trh negative) and V. parahaemolyticus HCR7sed (tdh negative, trh+) were used as positive controls in all reactions. V. parahaemolyticus strains that were negative for the presence of T3SS2 genes in the initial screen were retested using primers to differentiate the T3SS2α and T3SS2β genes by following the protocols by Okada et al. (46, 47). In addition, 32 strains representing the 4 different clusters (Fig. 1; also see Table S1 in the supplemental material) were selected for differentiation of the T3SS2α and T3SS2β genes using primers and PCR conditions for detection of vopCLP and vscC2T2R2D2 by following the protocols by Okada et al. (46, 47). In cases where the published protocols failed to yield a PCR product, the PCRs were optimized for each set of primers using the V. parahaemolyticus strains RIMD2210633 and HCR7sed as positive controls.
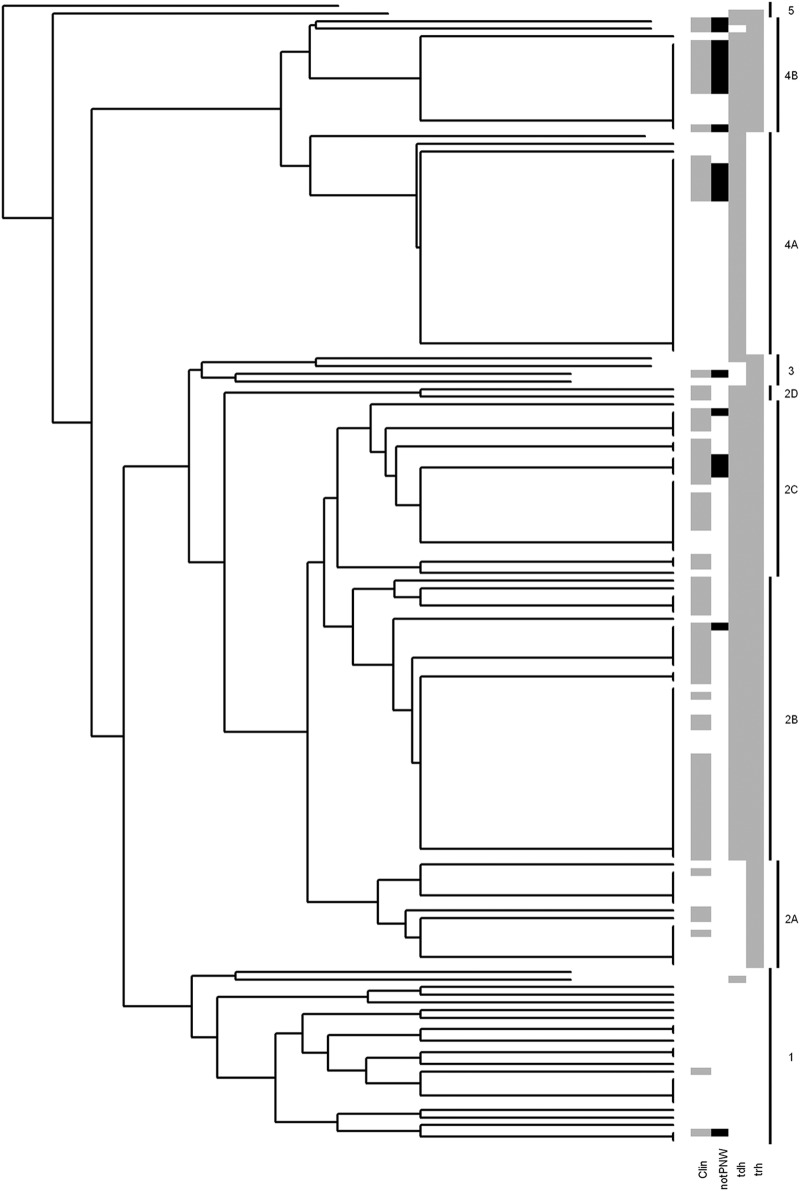
Fig 1.
Dendrogram of V. parahaemolyticus strains generated by hierarchical clustering analysis. The analysis separated the strains into 2 large clusters (2 and 4) and 3 smaller clusters (1, 3, and 5). The gray shading denotes the clinical isolates (Clin) in the clusters, while black denotes clinical isolates from other geographic regions (notPNW). The proportions of tdh+ and trh+ strains in each cluster are also denoted in gray.
Clustering analysis and random forest predictive model.
Hierarchical clustering was used to group samples based on the results of the PCR analysis of all 20 genes. The tree was constructed using the Agnes agglomerative clustering algorithm (28) as implemented in the statistical language R (53). The algorithm starts with each observation as a cluster, and clusters are merged by sequentially combining the two nearest clusters together until a single cluster is left. For this application, the Euclidean distance between each sample is used to measure distance (although the so-called Manhattan distance, being the sum of differences in this case, yields the same tree because this is binary data). The average distance between two clusters is the metric used to determine the closest two clusters in each step of the algorithm. Since many of the samples are identical, all of the closest groupings are groups with identical signatures for the genes tested.
We also built a predictive model to classify environmental and clinical samples using the random forest algorithm (5) as implemented in the randomForest package (34). Briefly, this involves constructing many classification trees and summarizing results into a single predictive model. The error rate that would occur if this model were applied to new data is estimated using the out-of-bootstrap error generated as a by-product of the algorithm. Because the samples were opportunistic and were collected using different methodologies, any predictions based on these models should be viewed only as suggestive or hypothesis generating.
RESULTS
Twenty genes from a total of 149 strains that included both clinical and environmental V. parahaemolyticus isolates were examined by PCR amplification by following protocols described in Materials and Methods. The random forest algorithm was applied to the results to predict the origin of the strains (clinical or environmental) based on the variation in the 20 genes examined. Using this model, the overall rate of prediction of strain origin was 76%, with the prediction rate for clinical strains at 80 and 73% for environmental strains. If only strains from PNW sources were included, the prediction rates were 90% for the clinical isolates and 81% for the environmental isolates, resulting in an overall error rate of 16%. The individual genes that provided the highest predictive power were trh and ure. For example, if trh alone is used as a predictor of virulence for all samples, 72% of the strains are correctly classified as either clinical or environmental (i.e., 28% error rate). Note that in all of the above analyses, more than half of the observed errors were due to groups of strains with identical signatures of the 20 genes examined where some of the strains were clinical isolates and others environmental.
We also applied hierarchical clustering to the same 20 genes. This grouped the majority of V. parahaemolyticus strains into two large clusters (76 strains in cluster 2 and 44 strains in cluster 4), each of which includes smaller clusters, 2A, 2B, 2C, 2D, 4A, and 4B, which are referred to as subgroups (Fig. 1). The remaining strains grouped into three smaller clusters (1, 3, and 5) with 23, 4, and 2 strains, respectively. The majority of strains isolated from clinical cases traced to oysters harvested from shellfish-growing areas in the PNW clustered in subgroups 2B, 2C, and 2D. Strains in cluster 4 were a mixture of largely environmental isolates from the PNW and clinical isolates from other geographic regions. Subgroup 4A also included the pandemic strain RIMD2210633. The majority of strains in cluster 1 were environmental isolates (21/23), while cluster 3 included 3 environmental isolates from the PNW and 1 clinical isolate from the Gulf of Mexico.
Distribution of putative virulence and pandemic markers.
In this study, we examined the distribution of the putative virulence-associated genes, tdh, trh, and ureR, as well as orf8 and toxRS (new), that have been used to differentiate the pandemic and nonpandemic strains. Strains were also screened for eight genes found in T3SS1 and six genes that are part of T3SS2. Also included was gbpA, a homolog of the gene encoding the chitin-binding protein in V. cholerae that plays a role in environmental survival and human infection (33) (Table 1).
Table 1.
Variation in targeted genes/ORFs among the clusters of V. parahaemolyticus strains
| Target gene/ORF | No. of genes present in clustera: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2A | 2B | 2C | 2D | 3 | 4A | 4B | 5 | |
| tdh | 1/23 | 0/14 | 37/37 | 23/23 | 2/2 | 3/4 | 29/29 | 15/15 | 1/2 |
| trh | 0/23 | 14/14 | 37/37 | 23/23 | 2/2 | 4/4 | 0/29 | 15/15 | 1/2 |
| toxRS (new) | 3/23 | 1/14 | 0/37 | 0/23 | 0/2 | 0/4 | 28/29 | 13/15 | 0/2 |
| orf8 | 4/23 | 0/14 | 0/37 | 3/23 | 0/2 | 1/4 | 29/29 | 15/15 | 2/2 |
| ureR | 0/23 | 14/14 | 37/37 | 23/23 | 2/2 | 4/4 | 0/29 | 15/15 | 1/2 |
| gbpA | 20/23 | 14/14 | 35/37 | 22/23 | 0/2 | 4/4 | 29/29 | 15/15 | 1/2 |
| VPA1339 | 8/23 | 6/14 | 0/37 | 23/23 | 0/2 | 0/4 | 29/29 | 14/15 | 1/2 |
| VPA1346 | 5/23 | 0/14 | 5/37 | 3/23 | 0/2 | 1/4 | 29/29 | 15/15 | 2/2 |
| VPA1338 | 23/23 | 14/14 | 31/37 | 23/23 | 0/2 | 3/4 | 29/29 | 15/15 | 1/2 |
| VPA1349 | 21/23 | 13/14 | 37/37 | 20/23 | 2/2 | 0/4 | 28/29 | 15/15 | 1/2 |
| VPA1354 | 22/23 | 14/14 | 37/37 | 21/23 | 2/2 | 3/4 | 28/29 | 15/15 | 1/2 |
| VPA1362 | 21/23 | 13/14 | 35/37 | 23/23 | 2/2 | 4/4 | 29/29 | 15/15 | 2/2 |
| C/E ratiob | 2/21 | 4/10 | 30/7 | 16/7 | 2/0 | 1/3 | 6/23 | 10/5 | 0/2 |
Clusters as outlined in Fig. 1 and Table S1 in the supplemental material. T3SS1 genes were detected in all strains. Data are number positive/total number tested.
Ratio of clinical to environmental strains.
The distribution of the hemolysin genes, tdh and trh, and of ureR, encoding urease, was distinct between the clusters. Strains encoding all three genes were clustered in subgroups 2B, 2C, 2D, and 4B (Fig. 1 and Table 1). All 62 strains from subgroups 2B, 2C, and 2D were tdh+, trh+, and ureR+. Forty-six of the strains (74%) from these subgroups were isolated from clinical cases associated with the consumption of raw oysters from the PNW, 13 (21%) were environmental isolates also from the PNW, while 3 (5%) were clinical isolates from other geographic regions (Fig. 1 and Table 1). All three genes were also detected in all 15 strains from subgroup 4B, 10 of which were clinical isolates from geographic locations other than the PNW, while 5 were environmental isolates from the PNW (Fig. 1; also see Table S1 in the supplemental material). Strains positive for tdh, trh, and ureR have been persistently isolated from the west coast of the United States since 1979 (1, 30, 32, 43, 48), and the presence of urease has been suggested as a predictor of potential pathogenic isolates in the PNW (29) and several other geographic locations (35, 45, 51, 56).
The two markers for the pandemic strains, orf8 and toxRS (new), were detected in the majority of strains (both clinical and environmental) from clusters 4 and 5 (Table 1). Genes for either toxRS (new) or orf8 (but not both) were detected in a few environmental strains from cluster 1 (4/23) and clinical strains from cluster 2C (2/23) and cluster 3 (1/6) (see Table S1). The clinical strains were not isolated from the PNW. The gene encoding gbpA was detected in the majority (140/149) of strains in this study. The nine exceptions were scattered across all clusters and are likely indicative of sequence variation.
Distribution of T3SS genes.
We screened the strains for variation in the genes encoding both type 3 secretion system gene clusters, T3SS1 and T3SS2. Strains were initially screened using primers based on homology to V. parahaemolyticus RIMD2210633 (tdh+, trh negative) as described in Materials and Methods. Two of the genes targeted in T3SS1 (VP1667 and VP1690) encode known or putative proteins that are part of the secretion apparatus, while the other six proteins (VP1656, VP1680, VP1686, VP1696, VP1670, and VP1659) are either known or putative translocators or effectors (4, 36, 50, 54, 58). All the clinical and environmental isolates were positive for all 8 T3SS1 genes, reconfirming that the T3SS1 apparatus is well conserved in V. parahaemolyticus strains (36, 40, 44, 52, and data not shown).
While the presence of T3SS1 has been reported in both clinical and environmental strains of V. parahaemolyticus, T3SS2 genes have been more commonly associated with strains carrying tdh+ but not trh (36), which were initially identified in V. parahaemolyticus RIMD2210633 as part of a large pathogenicity island (Vp-PA1) on chromosome 2 (36). Subsequently, another T3SS2 (T3SS2β) of a different lineage was identified in a tdh-negative, trh+ V. parahaemolyticus strain (46). Although the genetic organization of T3SS2β is similar to that of tdh+, trh-negative strains (referred to as T3SS2α), the homologies of individual genes within the secretion system vary significantly (46).
In order to assess strain variation in the clinical and environmental isolates, we initially screened all strains for the presence of T3SS2 genes based on homology to RIMD2210633 (which encodes T3SS2α proteins). Strains were screened by PCR for the presence of four genes encoding components of T3SS2, including a highly conserved inner membrane protein and putative ATPase (VPA1338), a putative outer membrane secretin (VPA1339), a putative C-ring component involved in translocation (VPA1349), and an inner membrane protein required for effector secretion (VPA1354) (40), as well as two putative secreted proteins, VopA/P (VPA1346) (57) and a homolog of EspD (VPA1362) (40, 58). Significant variation was noted in the detection of two of the six proteins, VPA1346 (VopA/P) and VPA1339, in the initial analyses (Table 1). VPA1339 was not detected in the majority of isolates from clusters 1, 2A, 2B, and 2C, while VPA1346 was not detected in the majority of the strains from cluster 1, 2, or 3 (Table 1). Strains from cluster one were largely environmental isolates (lacking both tdh and trh), while the majority of strains from cluster 2 are clinical isolates from the PNW. Cluster 3 includes environmental isolates from the PNW and 1 clinical isolate from the Gulf Coast (see Table S1 in the supplemental material). Strains with negative results from these analyses were subsequently tested using primers for the same genes based on homology to both T3SS2α and T3SS2β as described by Okada et al. (47). VPA1339 was detected in all but 2 environmental strains using primers based on homology to T3SS2β. VPA1346 was detected in all but 14 environmental strains, suggesting that sequences in the T3SS2 genes are even more variable than the presently observed differences in T3SS2α and T3SS2β (47). Similarly, T3SS2β sequences were detected for both VPA1338 (vscN2) and VPA1362 (vopB2) in all but 2 isolates. In our analyses, we detected VPA1362 (vopB2) in all but 5 of the 149 strains (clinical and environmental) tested in the initial analysis based on homology to RIMD2210633. However, these 5 strains tested positive using primers homologous to T3SSβ. The presence of vopB2 has previously been reported in tdh+, trh-negative clinical isolates but not in environmental isolates (44). Finally, primers based on T3SS2α or T3SS2β, or additional primers designed on available sequences for these genes, failed to amplify the genes in the remaining 12 strains for VPA1349 (vscQ2) and 5 strains for VPA1354 (vscU2), suggesting significant sequence variability or absence of these genes from these few strains (data not shown).
Our analysis indicated the presence of several T3SS2 genes in environmental V. parahaemolyticus strains for which the presence of tdh and trh is variable. To access the variability in strains from the different clusters, we screened a subset of strains that included strains from clusters 1, 2, and 4 that were tdh and trh negative, tdh+ trh+, tdh+ but trh negative, and tdh negative but trh+ using primers that have been shown to target and differentiate the gene clusters for T3SSα and T3SSβ (47). V. parahaemolyticus RIMD2210633 (tdh+, trh negative) was used as a positive control for the presence of T3SS2α, while V. parahaemolyticus HCR7sed (tdh negative, trh+), an environmental isolate from Washington State, was used as a positive control for T3SSβ. A total of 32 strains were screened for the presence of vscC2T2R2 and vopCPD2 by following protocols described by Okada et al. (47). The results are summarized in Table 2, and detailed results are in Table S3 in the supplemental material. Surprisingly, amplicons for all seven of the T3SS2β genes tested were detected in two tdh- and trh-negative environmental isolates from the PNW. One to five of the T3SS2β genes were also detected in eight other tdh- and trh-negative strains, while four T3SS2α genes were detected in one PNW environmental isolate (Table 2; also see Table S3). All trh-negative isolates were also urease negative. The detection of T3SS2β in tdh-, trh-negative V. parahaemolyticus isolates from the PNW suggests that these strains are part of a distinct population, since to our knowledge the presence of genes related to T3SS2 in tdh- and trh-negative strains has not been previously reported (46).
Table 2.
Distribution of T3SS2α and T3SS2β genes in select V. parahaemolyticus strains from this study
| C/E ratioa | Gene status | Detection (no. positive/total no. tested) of T3SS2 gene type: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α |
β |
||||||||||||||
| vopC | vscC2 | vscT2 | vscR2 | vopP | vopD2 | vopL | vopC | vscC2 | vscT2 | vscR2 | vopP | vopD2 | vopL | ||
| 0/11 | tdh negative, trh negative | 0/11 | 2/11 | 1/11 | 1/11 | 1/11 | 1/11 | 0/11 | 6/11 | 7/11 | 6/11 | 7/11 | 4/11 | 10/11 | 6/11 |
| 7/5 | tdh+, trh+ | 0/12 | 0/12 | 0/12 | 3/12 | 0/12 | 3/12 | 0/12 | 12/12 | 12/12 | 12/12 | 12/12 | 12/12 | 12/12 | 12/12 |
| 1/1 | tdh negative, trh+ | 0/2 | 1/2 | 0/2 | 0/2 | 0/2 | 1/2 | 0/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 |
| 0/5 | tdh+, trh negative | 5/5 | 2/5 | 4/5 | 5/5 | 3/5 | 4/5 | 1/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
Ratio of clinical to environmental strains.
Five of the strains tested were tdh+, trh-negative environmental isolates from the PNW that clustered with RIMD2210633 (cluster 4) in our analysis. All seven T3SS2α genes were detected only in RIMD2210633, while only one to six of the genes were detected in the five environmental isolates (Table 2; also see Table S3 in the supplemental material), again emphasizing the significant sequence diversity that might be present in these strains. The seven T3SS2β genes were detected in all 12 tdh+ trh+ isolates from the PNW (seven clinical and five environmental) and also from the two tdh-negative, trh+ isolates (one clinical and one environmental from the PNW), similar to previous reports (26).
DISCUSSION
Assessing the risk of V. parahaemolyticus-related illness from consumption of oysters has been particularly challenging in the PNW over the last decade. Current risk assessment methods are based primarily on water temperature at the time of harvest, the concentrations of total (tlh+) and tdh+ V. parahaemolyticus in oysters, as well as postharvest handling conditions, such as the time between harvest and refrigeration (19). Over the past decade, risk of illness from consumption of oysters harvested from Washington State has been difficult to estimate based on these criteria, as illnesses have occurred in the absence of significant concentrations of tdh+ V. parahaemolyticus strains (WDOH, personal communication).
In this study, we analyzed the variation in 20 virulence-associated genes by hierarchical cluster and random forest analysis. Hierarchical cluster analysis based on all 20 genes grouped strains into distinct clusters based on source of isolation, geographic location, and the presence/absence of putative virulence genes. Environmental isolates that lacked tdh and trh were clearly separated from the majority of strains (cluster 1), while the tdh+ trh+ clinical isolates from the PNW (cluster 2) grouped separately from the tdh+, trh-negative clinical isolates from other geographic regions (cluster 4). Cluster 4 also included the pandemic strain, RIMD2210633, and several other clinical isolates of serotype O3:K6 that have been responsible for the majority of V. parahaemolyticus-related illnesses worldwide, as well as tdh+, trh-negative environmental isolates from the PNW. This suggests that a significant proportion of environmental isolates from the PNW have a virulence-related gene profile similar to that of the pandemic strains, including the presence of tdh, the pandemic markers orf8 and toxRS (new), and components of the T3SS2. To date, however, strains with this profile have not been isolated from clinical cases in the PNW. Our results indicate that for V. parahaemolyticus strains isolated from the PNW, the presence of tdh, orf8, toxRS (new), and the T3SS2-related genes are not adequate markers for virulence. The high incidence of these genes in environmental isolates also suggests that these genes have a distinct advantage in survival in the PNW environment, as has been suggested by Matz et al. (38).
We detected sequence variation in two genes that are part of the T3SS2, VopA/P (VPA1346) and VPA1339, in clinical strains from the PNW (cluster 2). The secreted effector, VopA/P (VPA1346), inhibits the mitogen-activated protein kinase (MAPK) signaling pathway that may inhibit cytokine production, resulting in attenuation of the host defense response (40, 57). It is possible that sequence variation in this effector contributes to differences in virulence in the V. parahaemolyticus strains from the PNW. Sequences of the putative outer membrane protein, VPA1339 (vscC2), from cluster 2 were homologous to those identified in tdh-negative, trh+ V. parahaemolyticus strains that carry T3SS2β (46). Contrary to an earlier study by Noriea et al. (44), we detected the gene vopB2 (VPA1362) in all 149 strains, including the environmental isolates from the PNW, although with variation in sequence, underscoring the difference in strains from this region compared to those from other geographic regions.
Genes homologous to T3SS2β have been reported in tdh-negative, trh+ strains and tdh+ trh+ strains (26, 44, 46), while those homologous to T3SS2α have been identified in tdh+, trh-negative strains. In our study, we detected genes homologous to T3SS2-related genes (mainly T3SSβ and T3SSα in 2 strains) including genes for VopA/P, a secreted effector in environmental isolates from the PNW that were negative for both tdh and trh. The presence of these T3SS2-related genes in tdh-, trh-negative strains has not been previously reported, emphasizing the differences in strains isolated from the PNW and the potential for acquisition of new virulence-associated genes by environmental isolates.
Random forest modeling was a useful tool to allow the prediction of clinical and environmental strains based on the genes in this study. In our analysis, environmental strains could be classified correctly 81% of the time, while clinical strains were classified correctly 90% of the time. Differences in targeted genes could conceivably be used to monitor shifts in the genetic composition of strains in the environment.
Analysis and comparison of isolates from the PNW to those from the Atlantic or Gulf Coast based on serotyping, ribotyping, and multilocus sequence typing (MLST) has previously suggested that the V. parahaemolyticus population from the PNW is distinct from that of the Gulf and Atlantic coasts of the United States and the pandemic clonal complex that is distributed worldwide (17, 21). González-Escalona et al. identified a distinct clonal complex (CC36) of strains from the PNW that was geographically restricted but included multiple serotypes (21). These authors also note that genetic diversity in V. parahaemolyticus appears to be driven primarily by recombination rather than mutation.
Our analysis shows that a large proportion of the clinical isolates from the PNW carry the genes for both of the hemolysins, tdh and trh, are urease positive, and also carry T3SS2-related (T3SS2β) genes. However, a large proportion of the environmental isolates from the PNW were tdh+ and trh negative, and they grouped with the tdh+, trh-negative clinical isolates from other geographic regions, including the pandemic strain RIMD2210633. These strains encode some genes homologous to those of T3SS2 (T3SSα) and the pandemic markers toxRS (new) and orf8 but appear to be distinct from the strains responsible for illness in the PNW. The presence of pandemic markers in environmental V. parahaemolyticus has previously been reported and represents a population that could be of concern to public health (6). The detection of tdh in environmental isolates suggests that tdh alone is not an adequate marker for potentially virulent V. parahaemolyticus strains isolated from oysters in the PNW. However, the presence of trh and the gene encoding urease was a more definitive predictor of virulence, as has been reported in earlier studies (29, 56). While others have suggested the presence of T3SS2 defines potentially pathogenic strains, our data show that T3SS2 is widespread in both environmental and clinical isolates in the PNW. The clustering analysis has been useful as a preliminary method to differentiate strains, and it will allow us to focus on examining the differences in clinical and environmental strains from the PNW and those from other geographic regions by various genomic and genetic methods for the identification of additional virulence markers that could be useful for forecasting and risk assessment tools for this pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Strom, William Nilsson, and Gladys Yanagida for critical review of the manuscript. Thanks also to William Nilsson for technical assistance.
This work was supported by the NOAA West Coast Center for Oceans and Human Health (WCCOHH) as part of the NOAA Oceans and Human Health Initiative. The WCCOHH is part of the National Marine Fisheries Service's Northwest Fisheries Science Center, Seattle, WA.
Footnotes
Published ahead of print 5 October 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abbott SL, et al. 1989. Emergence of a restricted bioserovar of Vibrio parahaemolyticus as the predominant cause of Vibrio-associated gastroenteritis on the West Coast of the United States and Mexico. J. Clin. Microbiol. 27:2891–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alam MJ, Miyoshi S-I, Shinoda S. 2003. Studies on pathogenic Vibrio parahaemolyticus during a warm weather season in the Seto Inland Sea, Japan. Environ. Microbiol. 5:706–710 [DOI] [PubMed] [Google Scholar]
- 3. Bej AK, et al. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215–225 [DOI] [PubMed] [Google Scholar]
- 4. Bhattacharjee RN, et al. 2006. VP1686, a Vibrio type III secretion protein, induces toll-like receptor-independent apoptosis in macrophage through NF-κB inhibition. J. Biol. Chem. 281:36897–36904 [DOI] [PubMed] [Google Scholar]
- 5. Brieman L. 2001. Random forests. Machine Learning 45:5–32 [Google Scholar]
- 6. Caburlotto G, Gennari M, Ghidini V, Tafi M, Lleo MM. 2010. Serological and molecular characterization of Vibrio parahaemolyticus marine strains carrying pandemic genetic markers. ISME J. 4:1071–1074 [DOI] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 1999. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound–Connecticut, New Jersey, and New York, 1998. MMWR Morb. Mortal. Wkly. Rep. 48:48–51 [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention 1998. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters–Pacific Northwest, 1997. MMWR Morb. Mortal. Wkly. Rep. 47:457–462 [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention 2005. Vibrio illnesses after Hurricane Katrina–multiple states, August-September 2005. MMWR Morb. Mortal. Wkly. Rep. 54:928–931 [PubMed] [Google Scholar]
- 10. Cook DW, Bowers JC, DePaola A. 2002. Density of total and pathogenic (tdh+) Vibrio parahaemolyticus in Atlantic and Gulf coast molluscan shellfish at harvest. J. Food Prot. 65:1873–1880 [DOI] [PubMed] [Google Scholar]
- 11. Cook DW, et al. 2002. Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: a national survey from June 1998 to July 1999. J. Food Prot. 65:79–87 [DOI] [PubMed] [Google Scholar]
- 12. Daniels NA, et al. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J. Infect. Dis. 181:1661–1666 [DOI] [PubMed] [Google Scholar]
- 13. Daniels N, et al. 2000. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284:1541. [DOI] [PubMed] [Google Scholar]
- 14. Deepanjali A, Kumar HS, Karunasagar I, Karunasagar I. 2005. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71:3575–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DePaola A, Hopkins LH, Peeler JT, Wentz B, McPhearson RM. 1990. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl. Environ. Microbiol. 56:2299–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DePaola A, Kaysner CA, Bowers J, Cook DW. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DePaola A, et al. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. FDA 2005. Vibrio parahaemolyticus risk assessment. U.S. Food and Drug Administration, Washington, DC [Google Scholar]
- 20. Fuenzalida L, et al. 2006. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8:675–683 [DOI] [PubMed] [Google Scholar]
- 21. González-Escalona N, et al. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iida T, et al. 2001. Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerg. Infect. Dis. 7:477–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson CN, et al. 2012. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington, United States. Appl. Environ. Microbiol. 78:7249–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson C, et al. 2009. Genetic relatedness among tdh+ and trh+ Vibrio parahaemolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb. Ecol. 57:437–443 [DOI] [PubMed] [Google Scholar]
- 25. Johnson D, Weinberg L, Ciarkowski J, West P, Colwell R. 1984. Wound infection caused by Kanagawa-negative Vibrio parahaemolyticus. J. Clin. Microbiol. 20:811–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones JL, et al. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50:2343–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joseph S, Colwell R, Kaper J. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10:77–124 [DOI] [PubMed] [Google Scholar]
- 28. Kaufman L, Rousseeuw PJ. 1990. An introduction to cluster analysis. John Wiley & Sons, New York, NY [Google Scholar]
- 29. Kaysner CA, et al. 1994. Urea hydrolysis can predict the potential pathogenicity of Vibrio parahaemolyticus strains isolated in the Pacific Northwest. Appl. Environ. Microbiol. 60:3020–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaysner C, Abeyta C, Jr, Stott RF, Lilja JL, Wekell M. 1990. Incidence of urea-hydrolyzing Vibrio parahaemolyticus in Willapa Bay, Washington. Appl. Environ. Microbiol. 56:904–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelly MT, Stroh EM. 1988. Temporal relationship of Vibrio parahaemolyticus in patients and the environment. J. Clin. Microbiol. 26:1754–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelly MT, Stroh EM. 1989. Urease-positive, Kanagawa-negative Vibrio parahaemolyticus from patients and the environment in the Pacific Northwest. J. Clin. Microbiol. 27:2820–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866 [DOI] [PubMed] [Google Scholar]
- 34. Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22 [Google Scholar]
- 35. Magalhães M, Takeda Y, Magalhães V, Tateno S. 1992. Brazilian urease-positive strains of Vibrio parahaemolyticus carry genetic potential to produce the TDH-related hemolysin. Mem. Inst. Oswaldo Cruz 87:167–168 [DOI] [PubMed] [Google Scholar]
- 36. Makino K, et al. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749 [DOI] [PubMed] [Google Scholar]
- 37. Matsumoto C, et al. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matz C, Nouri B, McCarter L, Martinez-Urtaza J. 2011. Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 6:e20275 doi:10.1371/journal.pone.0020275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mead PS, et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meador CE, et al. 2007. Virulence gene and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 45:1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasu H, et al. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishibuchi M, Fasano A, Russell RG, Kaper JB. 1992. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60:3539–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nolan CM, et al. 1984. Vibrio parahaemolyticus gastroenteritis. An outbreak associated with raw oysters in the Pacific Northwest. Diagn. Microbiol. Infect. Dis. 2:119–128 [DOI] [PubMed] [Google Scholar]
- 44. Noriea NF, III, Johnson CN, Griffitt KJ, Grimes DJ. 2010. Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J. Appl. Microbiol. 109:953–962 [DOI] [PubMed] [Google Scholar]
- 45. Oberhofer TR, Podgore JK. 1982. Urea-hydrolyzing Vibrio parahaemolyticus associated with acute gastroenteritis. J. Clin. Microbiol. 16:581–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okada N, et al. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77:904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okada N, et al. 2010. Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 10:302–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Okuda J, Ishibashi M, Abbott SL, Janda JM, Nishibuchi M. 1997. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J. Clin. Microbiol. 35:1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okuda J, et al. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ono T, Park K-S, Ueta M, Iida T, Honda T. 2006. Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74:1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Osawa R, Okitsu T, Morozumi H, Yamai S. 1996. Occurrence of urease-positive Vibrio parahaemolyticus in Kanagawa, Japan, with specific reference to presence of thermostable direct hemolysin (TDH) and the TDH-related-hemolysin genes. Appl. Environ. Microbiol. 62:725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park K-S, et al. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. R Development Core Team 2011. R: a language and environment for statistical computing. R-project.org, Vienna, Austria [Google Scholar]
- 54. Shimohata T, et al. 2011. Vibrio parahaemolyticus infection induces modulation of IL-8 secretion through dual pathway via VP1680 in Caco-2 cells. J. Infect. Dis. 203:537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shirai H, et al. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suthienkul O, et al. 1995. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J. Infect. Dis. 172:1405–1408 [DOI] [PubMed] [Google Scholar]
- 57. Trosky JE, et al. 2004. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J. Biol. Chem. 279:51953–51957 [DOI] [PubMed] [Google Scholar]
- 58. Zhou X, Konkel ME, Call DR. 2010. Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 192:3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.