Abstract
meso-Diaminopimelate dehydrogenase (meso-DAPDH) is an NADP+-dependent enzyme which catalyzes the reversible oxidative deamination on the d-configuration of meso-2,6-diaminopimelate to produce l-2-amino-6-oxopimelate. In this study, the gene encoding a meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum was cloned and expressed in Escherichia coli. In addition to the native substrate meso-2,6-diaminopimelate, the purified enzyme also showed activity toward d-alanine, d-valine, and d-lysine. This enzyme catalyzed the reductive amination of 2-keto acids such as pyruvic acid to generate d-amino acids in up to 99% conversion and 99% enantiomeric excess. Since meso-diaminopimelate dehydrogenases are known to be specific to meso-2,6-diaminopimelate, this is a unique wild-type meso-diaminopimelate dehydrogenase with a more relaxed substrate specificity and potential for d-amino acid synthesis. The enzyme is the most stable meso-diaminopimelate dehydrogenase reported to now. Two amino acid residues (F146 and M152) in the substrate binding sites of S. thermophilum meso-DAPDH different from the sequences of other known meso-DAPDHs were replaced with the conserved amino acids in other meso-DAPDHs, and assay of wild-type and mutant enzyme activities revealed that F146 and M152 are not critical in determining the enzyme's substrate specificity. The high thermostability and relaxed substrate profile of S. thermophilum meso-DAPDH warrant it as an excellent starting enzyme for creating effective d-amino acid dehydrogenases by protein engineering.
INTRODUCTION
Not only do d-amino acids serve as specialized components of many types of machineries in living organisms, such as neural signaling (20), and bacterial cell walls (19), but they also are important components or building blocks in the production of pharmaceuticals and other fine chemicals (4, 5, 12). As such, many methods for the synthesis of d-amino acids and their derivatives have been developed. Just as l-amino acid dehydrogenases are useful for preparation of l-amino acids from the corresponding 2-keto acids (6, 7, 13, 21), the use of d-amino acid dehydrogenase (d-AADH) should offer a straightforward approach, in which the enzyme catalyzes the reductive amination of 2-keto acid to give d-amino acid. However, NAD(P)H-dependent d-amino acid dehydrogenase is much less abundant in nature than its l-amino acid counterpart and largely unexplored. The most-known d-AADH is meso-diaminopimelate dehydrogenase (meso-DAPDH; EC 1.4.1.16), which is a key enzyme in the lysine biosynthetic pathway and has been found in bacteria, such as Bacillus sphaericus (17) and Corynebacterium glutamicum (14). In addition, meso-DAPDH has also been isolated from plants, for example, soybeans (Glycine max) (24). meso-DAPDH is NADP+ dependent and catalyzes the reversible oxidative deamination on the d-configuration center of meso-2,6-diaminopimelate (meso-DAP) to yield l-2-amino-6-oxopimelate (17). However, previously reported meso-DAPDHs are generally specific toward meso-DAP and showed only very slight activities toward lanthionine or some 2-keto acids (2, 14, 16, 23). It is unusual that meso-DAPDH can distinguish the two configurations on the same symmetric molecule and act stereospecifically on the d-stereocenter. Because of this high stereospecificity to the d-center, meso-DAPDH could serve as the starting point for creating d-amino acid dehydrogenases by expanding its substrate specificity (1, 23). Recently, amino acid residues of the meso-DAPDH from C. glutamicum that interact with the substituents on the l-carbon of meso-DAP have been targeted for site-directed saturation mutagenesis in order to create d-amino acid dehydrogenase with activity toward substrates other than meso-DAP (23). Combined with two rounds of random mutagenesis over the entire gene, a mutant d-AADH enzyme was obtained and applied to the synthesis of d-cyclohexylalanine (23). A similar mutant d-AADH has been created by site-directed mutagenesis of Ureibacillus thermosphaericus DAPDH at the positions reported by Akita et al. (1).
Although a few d-AADHs with relatively broad substrate profiles have been obtained by enzyme engineering (1, 23), new d-AADHs with high activity and expanded substrate specificity are still highly required, considering their great potential in the synthesis of d-amino acids. Therefore, we have become interested in searching for new meso-DAPDHs as the entry to the desired d-AADHs. As such, a diaminopimelate dehydrogenase from an uncultivable thermophilic bacterium, Symbiobacterium thermophilum IAM14863 (22), was cloned and overexpressed in Escherichia coli. Interestingly, the purified enzyme showed activities toward d-alanine, d-valine, d-lysine, as well as meso-DAP and catalyzed the reductive amination of 2-keto acids, such as pyruvic acid, to generate d-amino acids. Two amino acid residues in the substrate binding sites of S. thermophilum meso-DAPDH were found to be different from the sequences of other meso-DAPDHs by amino acid sequence alignment. In order to investigate whether they are critical on the substrate spectrum of the enzyme, these two residues were replaced with the conserved amino acids in other meso-DAPDHs, and the activities of the resulting mutant enzymes were studied.
MATERIALS AND METHODS
Materials.
PrimeSTAR HS DNA polymerase and the DNA ligation kit were from TaKaRa (Japan). Restriction enzymes were purchased from Fermentas (Germany). The expression vector pET-32a (+) was purchased from Novagen (Merck, Germany). The plasmid extraction kit was from CWBIO (China). The gel extraction kit was from Tiangen Biotech (China). The gel filtration calibration kit was purchased from GE. All NAD, NAD(P)H, and NAD(P)+ were ordered from Codexis. 2-Keto acids and derivatives were purchased from Alfa Aesar or Tokyo Chemical Industry Co. All d-amino acids were from Chengdu Chengnuo New-Tech Co., Ltd. (China). All l-amino acids were ordered from Tianjin Guangfu Fine Chemical Research Institute (China).
Cloning of wild-type meso-DAPDH gene and site-directed mutagenesis.
The meso-diaminopimelate dehydrogenase gene (GenBank accession number AP006840.1) was synthesized chemically by Shanghai Xuguan Biotechnological Development Co., Ltd. (China). Primers were designed in order to fuse a 6× His tag at the N terminus of the wild type (see Table S1 in the supplemental material), and PCR was performed with PrimeSTAR HS DNA polymerase under the following conditions: denaturation at 94°C for 5 min, followed by 30 cycles of 98°C for 10 s, 67°C for 5 s, and 72°C for 65 s. The amplified DNA fragments were loaded on a 0.8% agarose gel containing a gel indicator (GelRed; Biotium, Inc., Canada) for visualizing the DNA and purified. The obtained DNA fragment flanked by NdeI and XhoI restriction sites was inserted into pUC19, which was digested by SmaI. E. coli DH5α was transformed with the ligated plasmid and plated onto an LB agar plate containing 100 mg · liter−1 ampicillin, 40 mg · liter−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Positive clones were incubated for plasmid extraction. The recombinant plasmid was digested by NdeI and XhoI restriction enzymes and ligated into pET-32a (+), which was digested with the same restriction enzymes. For site-directed mutagenesis, PCRs were performed by following the same protocols using the primers listed in Table S1 in the supplemental material and the wild-type expression vector as the template. Two single mutations and one double mutation were created.
Expression and purification.
The recombinant plasmid, wild type, and mutants were introduced into E. coli BL21(DE3). Expression of the gene was induced using 0.1 mM IPTG for 6 h at 37°C when the optical density at 600 nm (OD600) was 0.6 to 0.8. For purification, cells were harvested by centrifugation at 12,000 × g for 15 min (Thermo) and disrupted with a high-pressure homogenizer (APV, Germany). The recombinant native protein was centrifuged at 25,000 × g for 30 min, filtered through a 0.22-μm-pore-size filter, and then purified by Ni affinity chromatography (GE) with an Äkta purifier 10 (GE). The column was equilibrated with 20 mM Tris-HCl (pH 7.4) containing 250 mM NaCl, 5% glycerol and eluted with a buffer containing 20 mM Tris-HCl, 250 mM NaCl, 5% glycerol, 500 mM imidazole (pH 7.4). The protein concentration was determined with a bicinchoninic acid assay kit (CWBIO, China). The resulting enzyme solution was used for activity assay.
Activity assay and kinetic constant determination.
The assay system used for determination of oxidative deamination activity contained 5 mM meso-DAP, 0.5 mM NADP+, 200 mM Na2CO3-NaHCO3 buffer (pH 9.5), and enzyme in a final volume of 200 μl. The assay system used for reductive amination consisted of 20 mM 2-keto acids, 200 mM NH4Cl, 0.5 mM NADPH, 200 mM Na2CO3-NaHCO3 buffer (pH 9.0), and enzyme in a final volume of 200 μl. After incubation, reactions were performed in 96-well plates with a Spectramax M2e apparatus (Molecular Devices, USA) and initiated by addition of NADP(H). The OD340 was monitored at 30°C using a molar absorption coefficient of 6.22 mM−1 · cm−1. One unit was defined as the amount of enzyme producing or consuming 1 μmol NADPH per minute. In a blank, substrate or enzyme was replaced by buffer. All the experiments were conducted with three replicates.
Kinetic constant measurements for reductive amination and oxidative deamination were performed at 30°C. The amination reactions were performed in 100 mM Na2CO3-NaHCO3 buffer (pH 8.5) with pyruvic acid and NH4Cl as the substrates, while the final concentration of NADPH was 0.5 mM. When kinetic parameters for pyruvic acid were determined, the concentration of NH4Cl was 200 mM and the concentration of pyruvic acid was varied from 5 to 150 mM; for NH4Cl, the concentration of pyruvic acid was 50 mM and the concentration of NH4Cl was in the range of 25 to 1,000 mM. When deamination was investigated, native meso-DAP was chosen as the substrate and the reactions were performed in 100 mM Na2CO3-NaHCO3 buffer (pH 10.0) with 0.5 mM NADP. The concentration of meso-DAP was between 0.5 mM and 100 mM. All the experiments were conducted with three replicates. Km and Vmax values were calculated by nonlinear fitting.
Molecular mass determination.
The molecular mass of the subunit was determined by SDS-PAGE according to standard procedures using 12.5% polyacrylamide gels (9). The native molecular mass of the enzyme was estimated by gel filtration on Superdex 200 10/300 GL (GE). Elution was performed with 50 mM phosphate buffer (pH 7.2) containing 150 mM NaCl. Standard proteins, thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa), were used for calibration.
Effects of pH and temperature on enzyme activity.
In order to investigate the effects of pH and temperature, enzyme activities of reductive amination with pyruvic acid as the amino acceptor and oxidative deamination with meso-DAP as the substrate were investigated under different conditions according to the standard enzyme activity assay. While investigating the effects of pH on activity, Na2CO3-NaHCO3 buffer (pH 8.5 to 10.7) and Na2HPO4-NaOH buffer (pH 10.9 to 12.0) were chosen to prepare reaction mixtures with different pH values. For temperature stability, the enzyme was incubated at different temperatures between 30°C and 80°C for 1 h. The remaining enzyme activities were measured at 30°C after being cooled.
Substrate specificity and biotransformation of 2-keto acids to d-amino acids.
2-Keto acids and derivatives were screened with the standard enzyme activity protocol for reductive amination. All l- and d-amino acids were screened by following the protocol for oxidative deamination, except that the concentrations of substrate were increased to 20 mM.
In order to determine the enantiomeric excess (ee) values of products from reductive amination, biotransformation reactions were carried out as follows: 1 mmol 2-keto acid, 3 mmol NH4Cl, 4 mmol glucose, 10 μmol NADP+, 6 U meso-DAPDH, and 4.8 U glucose dehydrogenase (GDH) were added to 10 ml Na2CO3-NaHCO3 buffer (200 mM, pH 8.5). The mixture was shaken at 37°C and 200 rpm for 24 h. Then, 1.3 mmol N-(9-fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu) in tetrahydrofuran solution was added to the mixture, while Na2CO3 was added to maintain the solution pH at 8.0 to 9.0. After 10 h, the mixture was washed with methyl tert-butyl ether three times and precipitated at pH 2 to 3 by addition of 6 N HCl. The aqueous phase was then extracted by ethyl acetate for three times. The extracts were analyzed by high-pressure liquid chromatography (HPLC) with a chiral column (S,S; HELK-O1; 25 cm by 4.6 mm; Regis) at 254 nm to measure the ee value of the product. The mobile phase was hexane-isopropanol-acetic acid (2:8:0.1), and the flow rate was 0.6 ml · min−1.
Nucleotide sequence accession numbers.
The amino acid and nucleotide sequences of meso-DAPDH from S. thermophilum have been deposited in the GenBank database under accession numbers BAD40410.1 and AP006840.1, respectively.
RESULTS AND DISCUSSION
Sequence analysis.
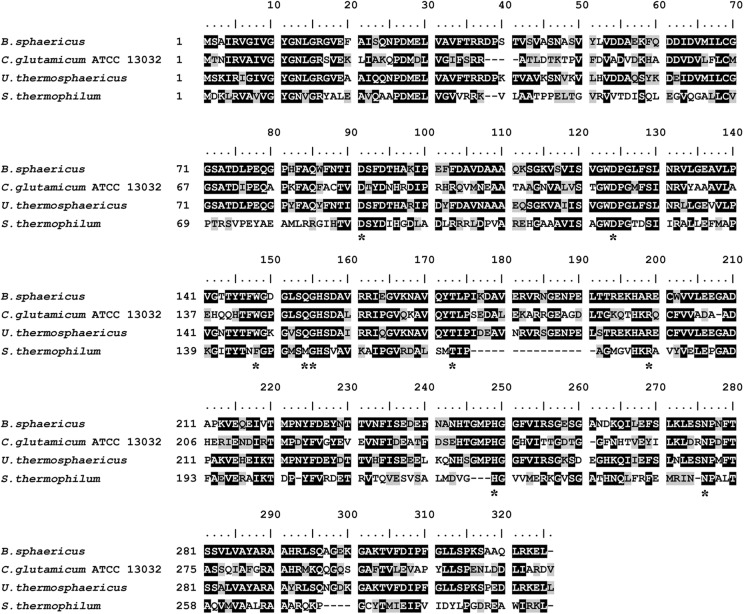
The amino acid sequence (GenBank accession number BAD40410.1) of meso-DAPDH from S. thermophilum was aligned with the amino acid sequences of meso-DAPDH from C. glutamicum ATCC 13032 (8), B. sphaericus (18), and Ureibacillus thermosphaericus (2) using the BioEdit software package (version 7.0.1) (Fig. 1). The identities of the S. thermophilum meso-DAPDH sequence with the sequences of three known meso-DAPDHs were 28.4, 31.2, and 29.9%, respectively. Of the nine amino acid residues in the substrate binding site, Phe146 and Met152 were different from the sequences in other meso-DAPDHs.
Fig 1.
Multiple-amino-acid-sequence alignment between meso-DAPDHs from S. thermophilum, C. glutamicum ATCC 13032 (8), B. sphaericus (18), and Ureibacillus thermosphaericus (2). Substrate binding sites reported previously (3) are indicated (*).
Cloning, expression, and purification.
In order to facilitate the purification of recombinant protein, the meso-DAPDH gene (GenBank accession number AP006840.1) from S. thermophilum was subcloned into the pET-32a (+) vector with a 6× His tag at the N terminus of the protein. It was expressed in E. coli BL21(DE3) host cells as a soluble protein and inclusion body at a ratio of 1:1. The protein was purified by affinity chromatography with an Ni column, and the results of SDS-PAGE analysis are shown in Fig. 2A. The specific activity of the enzyme toward meso-DAP was increased by 3.4-fold with 90.7% yield. Three mutant enzymes, F146W, M152Q, and F146W M152Q, were also prepared by the same procedure (Fig. 2B).
Fig 2.
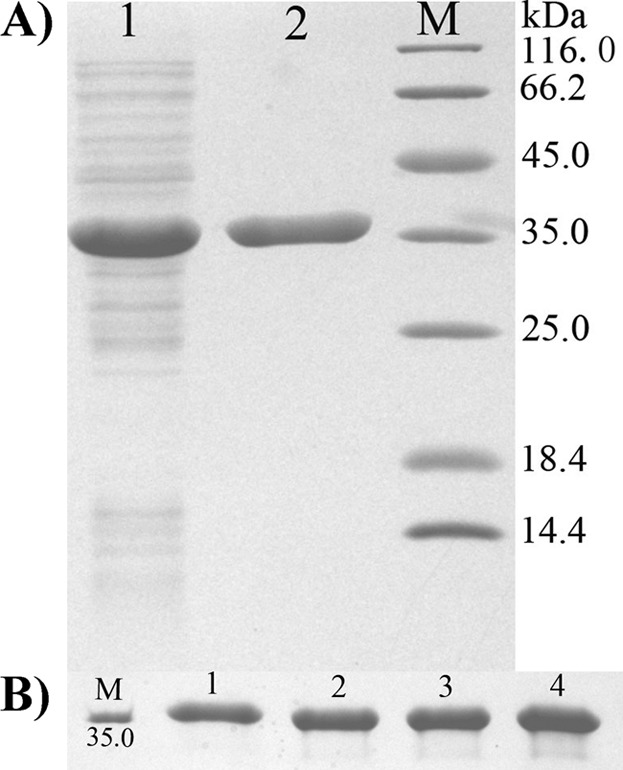
SDS-PAGE analysis of purified wild-type and mutant meso-DAPDHs from S. thermophilum. (A) Purification of wild-type meso-DAPDH. Lane M, protein ruler; lane 1, crude enzyme extract; lane 2, enzyme purified by affinity chromatography. (B) Purified wild-type and mutant meso-DAPDHs by affinity chromatography. Lane M, protein ruler (35.0 kDa); lane 1, wild type; lane 2, F146W; lane 3, M152Q; lane 4, F146W M152Q.
By SDS-PAGE analysis, the molecular mass of the subunit was about 36 kDa (Fig. 2A), which was close to the calculated value of 32.3 kDa. The native molecular mass was determined to be 143.7 kDa by gel filtration, implying that the enzyme exists as a tetramer.
Effects of pH and temperature on enzyme activity.
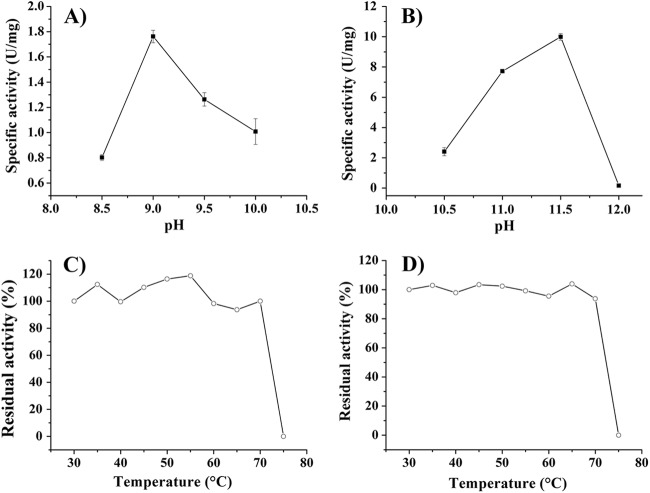
The purified enzyme exhibited an optimum activity at pH 9.0 for reductive amination (Fig. 3A) and pH 11.5 for oxidative deamination (Fig. 3B), which were both a little more alkaline than those reported for meso-DAPDHs from C. glutamicum (14), B. sphaericus (16), and a Brevibacterium sp. (15). After being incubated at 70°C for 1 h, there was still 94% activity remaining for deamination, and almost no activity loss was observed for amination by the meso-DAPDH from S. thermophilum (Fig. 3C and D). The activity was quickly lost when the enzyme was placed at 75°C for 1 h. meso-DAPDH from U. thermosphaericus is the most thermostable meso-DAPDH reported in the literature and showed almost no loss of activity for deamination when it was incubated at 60°C for 30 min, but it lost 50% of its activity when it was incubated at 65°C for 30 min (2). As such, the enzyme from S. thermophilum was more thermostable than all known meso-DAPDHs reported so far.
Fig 3.
Effects of pH and temperature on enzyme activity. meso-DAP and pyruvic acid were used as the substrates for deamination and amination, respectively. (A) pH profile for amination. Na2CO3-NaHCO3 buffer (pH 8.5 to 10.7) was used. (B) pH profile for deamination. Na2HPO4-NaOH buffer (pH 10.9 to 12) was used. (C) Temperature stability for amination in Na2CO3-NaHCO3 buffer (pH 9.0). (D) Temperature stability for deamination in Na2CO3-NaHCO3 buffer (pH 10.0).
Kinetic parameter determination of wild-type S. thermophilum meso-DAPDH.
The kinetic parameters of the meso-DAPDH from S. thermophilum for oxidative deamination of meso-DAP and reductive amination of pyruvic acid were measured (Table 1). In the reductive amination of pyruvic acid using NH4Cl as the amino donor, the Km and kcat toward NH4Cl were 120.5 ± 26.6 mM and (1.29 ± 0.09) × 104 s−1, respectively. The Km toward meso-DAP of the meso-DAPDHs from S. thermophilum was in the range of values for other meso-DAPDHs from U. thermosphaeticus, B. sphaericus, C. glutamicum, and a Brevibacterium sp., which were 1.6 mM (2), 2.5 mM (16), 3.1 mM (14), and 6.25 mM (15), respectively. In other words, the affinity of the enzyme from S. thermophilum to meso-DAP was similar to that of other meso-DAPDHs. By comparing Km and kcat values between amination and deamination, it can be seen that the affinity to meso-DAP in deamination was much larger than that to pyruvic acid in amination, and the catalytic rate of deamination was higher than that of amination. Therefore, the meso-DAPDH from S. thermophilum catalyzes the oxidative deamination of meso-DAP with a higher efficiency than it catalyzes the reductive amination of pyruvic acid.
Table 1.
Kinetic parameters for wild-type and mutant enzymesa
| Strain |
meso-DAP |
Pyruvic acid |
||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | Km (mM) | kcat (s−1) | |
| Wild type | 1.9 ± 0.2 | (1.27 ± 0.04) × 105 | 13.5 ± 2.8 | (7.85 ± 0.43) × 103 |
| F146W | 2.5 ± 0.2 | (1.51 ± 0.03) × 104 | 12.2 ± 3.2 | (1.01 ± 0.08) × 104 |
| M152Q | 16.7 ± 1.2 | (3.57 ± 0.09) × 104 | 24.8 ± 3.5 | (1.01 ± 0.06) × 103 |
| F146W M152Q | 15.7 ± 1.1 | (6.78 ± 0.17) × 103 | 23.6 ± 3.5 | (6.64 ± 0.23) × 102 |
Amination was performed in 100 mM Na2CO3-NaHCO3 buffer (pH 8.5), while deamination was carried out in 100 mM Na2CO3-NaHCO3 buffer (pH 10.0). The temperature was controlled at 30°C.
Substrate profiles and enantiomeric purity determination.
For oxidative deamination, among the tested amino acids, both d- and l-enantiomers of leucine, lysine, valine, serine, tyrosine, aspartic acid, phenylalanine, glutamic acid, threonine, cysteine, and tryptophan, the meso-DAPDH from S. thermophilum showed activities toward d-lysine, d-alanine, and d-valine which were much lower than those of the native substrate meso-DAP (Table 2). The enzyme from S. thermophilum also catalyzed the reductive amination of some 2-keto acids and derivatives using NH4Cl as the amino donor to afford d-amino acids, and the results are summarized in Table 2. The enzyme showed the highest specific activity toward pyruvic acid and lower activities toward other 2-keto acids, such as 4-methyl-2-oxopentanoic acid, 3-methyl-2-oxobutanoic acid, 2-oxosuccinic acid, and 2-oxo-2-phenylpropanoic acid.
Table 2.
Specific activities of meso-DAPDH from S. thermophilum and C. glutamicuma for both amination and deamination
| Substrate | Product | Sp act (U · mg−1) |
||||
|---|---|---|---|---|---|---|
|
meso-DAPDH from S. thermophilum |
meso-DAPDH from C. glutamicum | |||||
| Wild type | F146W | M152Q | F146W M152Q | |||
| Pyruvic acid | d-Alanine | 2.87 ± 0.16 | 2.52 ± 0.02 | 0.10 ± 0.03 | 0.18 ± 0.01 | 0.009 |
| Ethyl pyruvate | d-Alanine | 0.32 ± 0.04 | 0.36 ± 0.03 | 0.085 ± 0.001 | 0.060 ± 0.002 | |
| 4-Methyl-2-oxopentanoic acid | d-Leucine | 0.11 ± 0.03 | 0.12 ± 0.02 | 0.009 ± 0.002 | 0.022 ± 0.001 | 0.004 |
| Sodium 3-methyl-2-oxobutanoate | d-Valine | 0.13 ± 0.01 | 0.24 ± 0.01 | 0.066 ± 0.004 | 0.04 ± 0 | 0.005 |
| Ethyl 3-methyl-2-oxobutanoate | d-Valine | 0.097 ± 0.001 | 0.11 ± 0.01 | 0.029 ± 0.002 | 0.015 ± 0 | |
| 2-Oxosuccinic acid | d-Asparagine | 0.26 ± 0.01 | 0.096 ± 0.004 | 0.026 ± 0.004 | 0.015 ± 0.001 | NAb |
| 2-Oxo-3-phenylpropanoic acid | d-Phenylalanine | 0.051 ± 0.001 | 0.092 ± 0.005 | 0.018 ± 0.001 | 0.018 ± 0.002 | NA |
| meso-DAP | l-2-Amino-6-oxopimelate | 11.33 ± 0.46 | NTc | NT | NT | 131.6d |
| d-Lysine | 6-Amino-2-oxohexanoic acid | 0.004 ± 0 | NT | NT | NT | NA |
| d-Alanine | Pyruvic acid | 0.004 ± 0 | NT | NT | NT | NA |
| d-Valine | 3-Methyl-2-oxobutanoate | 0.005 ± 0 | NT | NT | NT | NA |
The reductive aminations of several 2-keto acids and derivatives were studied by using d-glucose dehydrogenase/d-glucose as an NADPH recycling system. The enantiomeric purity of the products was determined by chiral HPLC analysis after being derivatized with Fmoc-OSu (Table 3; see Fig. S1 to S3 in the supplemental material). The S. thermophilum meso-DAPDH catalyzed the reductive amination of pyruvic acid to yield d-alanine with excellent conversion (99%) and an excellent ee value, and the 9-fluorenylmethoxy carbonyl (Fmoc)-derivatized d-alanine was isolated in 68% yield. The enantioselectivity was affected by the structure of the substrates, because for other 2-keto acids, the products were obtained with a lower optical purity. In addition to 2-keto acids, the ethyl esters of pyruvic acid and 3-methyl-2-oxobutanoic acid were also active but had lower specific activities. Since no hydrolysis products of ethyl pyruvate and ethyl 3-methyl-2-oxobutanoate were observed and ethyl esters of d-alanine and d-valine were not observed as products from reactions of these two substrates, the esters may first be hydrolyzed to the acids, followed by the enzyme-catalyzed reductive amination, and the ester hydrolysis may be the rate-determining step. Recently, a FAD-dependent d-amino acid dehydrogenase, DauA, was identified to be one of the two components which perform d-to-l racemization of arginine. DauA catalyzes oxidative deamination of several d-amino acids into the corresponding 2-keto acids and ammonia, with d-Arg and d-Lys being the two most effective substrates (10, 11). The wild-type meso-DAPDH from C. glutamicum has been reported to have barely measurable activity (0.004 to 0.009 U/mg) toward pyruvic acid, 4-methyl-2-oxopentanoic acid, and 3-methyl-2-oxobutanoic acid (Table 2) (23). So, to the best of our knowledge, the meso-DAPDH from S. thermophilum was the first wild-type NAD(P)H-dependent meso-DAPDH with high activity for the reductive amination of 2-keto acids and potential for application to d-amino acid synthesis.
Table 3.
Enantiomeric excess and yield of reductive amination by the meso-DAPDH from S. thermophilum
| Substrate | ee (%) | Yield (%) |
|---|---|---|
| Pyruvic acid | 99 | 68 |
| Ethyl pyruvate | 99 | 36 |
| 4-Methyl-2-oxopentanoic acid | 81 | 32 |
| Sodium 3-methyl-2-oxobutanoate | 73 | 60 |
| Ethyl 3-methyl-2-oxobutanoate | 76 | 58 |
Site-directed mutations of meso-DAPDH from S. thermophilum.
From multiple-sequence alignment (Fig. 1), we found that Phe146 and Met152 in the substrate binding sites of the meso-DAPDH from S. thermophilum were different from the sequences of other known meso-DAPDHs with strict substrate specificity. Therefore, three mutants, F146W, M152Q, and F146W M152Q, were created by mutating the two residues into the conserved amino acids in other known meso-DAPDHs by site-directed mutagenesis. The three mutant enzymes were purified with an Ni column (Fig. 2B), and their activities toward the 2-keto acids were assayed. As shown in Table 2, all of the three mutant enzymes showed the same substrate profile as the wild type, although they had different specific activities. The mutation at M152 decreased the enzyme activity, but nearly no effect was observed for the enzyme with the mutation at F146. This is consistent with the kcat for the reductive amination of pyruvic acid by wild-type and mutant meso-DAPDHs (Table 1). The Km value of the mutant with the F146W mutation was similar to that of the wild-type enzyme, indicating that the mutation exerted little effect on the binding ability for the tested substrate. The Km values of the mutant enzyme with the mutation at M152 were almost 2- and 8-fold of those for the wild type for pyruvic acid and meso-DAP, respectively, indicating that the amino acid residue at M152 has a strong effect on substrate binding. F146 was the residue close to the l-center, while M152 interacted with the d-center, determined by sequence alignment with the enzyme from C. glutamicum and comparison with its crystal structure (Protein Data Bank accession number 1f06). This might be the reason for the discrepancies between these two residues. In addition, mutation of the two amino acid residues had no effect on the enantiomeric excess of the product. Although F146 and M152 exerted some effects on enzyme activity, they were not critical in determining the enzyme's substrate specificity toward the tested substrates.
In conclusion, the S. thermophilum meso-DAPDH is the most thermostable and a novel native NAD(P)H-dependent meso-diaminopimelate dehydrogenase with a broad substrate profile. Although it is not clear what has caused such unusual substrate specificity, the S. thermophilum enzyme could serve as an excellent starting enzyme for creating effective d-amino acid dehydrogenases by protein engineering, which is under way in the national engineering laboratory.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (grant no. 21072151) and the National Basic Research Program of China (973 Program, grant no. 2011CB710801).
Footnotes
Published ahead of print 28 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akita H, Doi K, Kawarabayasi Y, Ohshima T. 2012. Creation of a thermostable NADP+-dependent d-amino acid dehydrogenase from Ureibacillus thermosphaericus strain A1 meso-diaminopimelate dehydrogenase by site-directed mutagenesis. Biotechnol. Lett. 34:1693–1699 [DOI] [PubMed] [Google Scholar]
- 2. Akita H, Fujino Y, Doi K, Ohshima T. 2011. Highly stable meso-diaminopimelate dehydrogenase from an Ureibacillus thermosphaericus strain A1 isolated from a Japanese compost: purification, characterization and sequencing. AMB Express. 1:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cirilli M, Scapin G, Sutherland A, Vederas JC, Blanchard JS. 2000. The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-l-2-amino-6-methylene-pimelate. Protein Sci. 9:2034–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedman M. 2010. Origin, microbiology, nutrition, and pharmacology of d-amino acids. Chem. Biodivers. 7:1491–1530 [DOI] [PubMed] [Google Scholar]
- 5. Friedman M, Levin C. 2012. Nutritional and medicinal aspects of d-amino acids. Amino Acids 42:1553–1582 [DOI] [PubMed] [Google Scholar]
- 6. Groeger H, May O, Werner H, Menzel A, Altenbuchner J. 2006. A “second-generation process” for the synthesis of l-neopentylglycine: asymmetric reductive amination using a recombinant whole cell catalyst. Org. Proc. Res. Dev. 10:666–669 [Google Scholar]
- 7. Hanson RL, et al. 2007. Preparation of an amino acid intermediate for the dipeptidyl peptidase IV inhibitor, saxagliptin, using a modified phenylalanine dehydrogenase. Adv. Synth. Catal. 349:1369–1378 [Google Scholar]
- 8. Ishino S, Mizukami T, Yamaguchi K, Katsumata R, Araki K. 1987. Nucleotide sequence of the meso-diaminopimelate d-dehydrogenase gene from Corynebacterium glutamicum. Nucleic Acids Res. 15:3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 10. Li C, Lu C-D. 2009. Arginine racemization by coupled catabolic and anabolic dehydrogenases. Proc. Natl. Acad. Sci. U. S. A. 106:906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li C, Yao X, Lu C-D. 2010. Regulation of the dauBAR operon and characterization of d-amino acid dehydrogenase DauA in arginine and lysine catabolism of Pseudomonas aeruginosa PAO1. Microbiology 156:60–71 [DOI] [PubMed] [Google Scholar]
- 12. Ma JS. 2003. Unnatural amino acids in drug discovery. Chim. Oggi 21:65–68 [Google Scholar]
- 13. Menzel A, Werner H, Altenbuchner J, Groeger H. 2004. From enzymes to “designer bugs” in reductive amination: a new process for the synthesis of l-tert-leucine using a whole cell-catalyst. Eng. Life Sci. 4:573–576 [Google Scholar]
- 14. Misono H, Ogasawara M, Nagasaki S. 1986. Characterization of meso-diaminopimelate dehydrogenase from Corynebacterium glutamicum and its distribution in bacteria (microbiology & fermentation industry). Agric. Biol. Chem. 50:2729–2734 [Google Scholar]
- 15. Misono H, Ogasawara M, Nagasaki S. 1986. Purification and properties of meso-diaminopimelate dehydrogenase from Brevibacterium sp. Agric. Biol. Chem. 50:1329–1330 [Google Scholar]
- 16. Misono H, Soda K. 1980. Properties of meso-alpha, epsilon-diaminopimelate d-dehydrogenase from Bacillus sphaericus. J. Biol. Chem. 255:10599–10605 [PubMed] [Google Scholar]
- 17. Misono H, Togawa H, Yamamoto T, Soda K. 1979. meso-alpha, epsilon-Diaminopimelate d-dehydrogenase: distribution and the reaction product. J. Bacteriol. 137:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakamoto S, Seki M, Nagata S, Misono H. 2001. Cloning, sequencing, and expression of the meso-diaminopimelate dehydrogenase gene from Bacillus sphaericus. J. Mol. Catal. B Enzymatic 12:85–92 [Google Scholar]
- 19. Scheffers D-J, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69:585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snyder SH, Kim PM. 2000. d-Amino acids as putative neurotransmitters: focus on d-serine. Neurochem. Res. 25:553–560 [DOI] [PubMed] [Google Scholar]
- 21. Stewart JD. 2001. Dehydrogenases and transaminases in asymmetric synthesis. Curr. Opin. Chem. Biol. 5:120–129 [DOI] [PubMed] [Google Scholar]
- 22. Ueda K, et al. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vedha-Peters K, Gunawardana M, Rozzell JD, Novick SJ. 2006. Creation of a broad-range and highly stereoselective d-amino acid dehydrogenase for the one-step synthesis of d-amino acids. J. Am. Chem. Soc. 128:10923–10929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wenko LK, Treick RW, Wilson KG. 1985. Isolation and characterization of a gene encoding meso-diaminopimelate dehydrogenase from Glycine max. Plant Mol. Biol. 4:197–204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.