Abstract
Viral production estimates show that virioplankton communities turn over rapidly in aquatic ecosystems. Thus, it is likely that the genetic identity of viral populations comprising the virioplankton also change over temporal and spatial scales, reflecting shifts in viral-host interactions. However, there are few approaches that can provide data on the genotypic identity of viral populations at low cost and with the sample throughput necessary to assess dynamic changes in the virioplankton. This study examined two of these approaches—T4-like major capsid protein (g23) gene polymorphism and randomly amplified polymorphic DNA-PCR (RAPD-PCR) fingerprinting—to ask how well each technique could track differences in virioplankton populations over time and geographic location. Seasonal changes in overall virioplankton composition were apparent from pulsed-field gel electrophoresis (PFGE) analysis. T4-like phages containing similar g23 proteins were found within both small- and large-genome populations, including populations from different geographic locations and times. The surprising occurrence of T4-like g23 within small genomic groups (23 to 64 kb) indicated that the genome size range of T4-like phages may be broader than previously believed. In contrast, RAPD-PCR fingerprinting detected high genotypic similarity within PFGE bands from the same location, time, and genome size class without the requirement for DNA sequencing. Unlike g23 polymorphism, RAPD-PCR fingerprints showed a greater temporal than geographic variation. Thus, while polymorphism in a viral signature gene, such as g23, can be a powerful tool for inferring evolutionary relationships, the degree to which this approach can capture fine-scale variability within virioplankton populations is less clear.
INTRODUCTION
Viruses that infect heterotrophic bacteria, cyanobacteria, and eukaryotic phytoplankton dominate the world's oceans and play an important role in structuring the marine microbial food web. Phage research, particularly in aquatic environments, has uncovered the extraordinary abundance of viruses as well as their impact on the ecology of our planet (3, 44). They are estimated to be the most abundant biological entities in both marine and freshwater environments (with abundances typically in the range of 107 to 109 particles/ml) and likely contain the largest pool of unknown genetic diversity on Earth (7, 22, 45, 63). It is now believed that through infection and lysis processes, viruses play an important role in shaping marine ecosystems by influencing the cycling of nutrient elements such as Fe (43) or enhancing the flow of carbon between living biomass and the dissolved pool (5, 52). As agents of mortality, viruses also play a role in structuring co-occurring host populations and communities (24, 55). Viruses, and bacteriophages in particular, are critical vectors for rampant promiscuous horizontal gene transfer between microbial populations, nonorthologous gene displacement in host cells, and lysogenic conversion (18, 40, 57).
The T4-like phages represent a large and diverse group of closely related phages that bear morphological and genetic resemblances to the tailed double-stranded DNA (dsDNA) bacteriophage T4 (1, 2). Due to their abundance, ubiquity, and high genetic diversity, the T4-like phages have provided a valuable model for examining the diversity and ecosystem dynamics of phage populations within aquatic environments. Because no single gene is shared among all viruses, the diversity of entire viral communities cannot be examined by using approaches analogous to the rRNA gene methods used for examining microbial communities. However, the burgeoning collection of genomic information from viruses is beginning to reveal signature genes which are preserved among viral subgroups and can be used for phylogenetic classification of viruses and for evaluating viral diversity in natural samples (46).
Comparative genomics and molecular phylogeny of T4-like signature genes have yielded deep insights on evolutionary history and ecology of T4-like phages (2, 7, 14, 26, 33, 38). In particular, the T4 major capsid protein, g23, a virion structural component, has proven to be among the most conserved (54) and ubiquitously distributed genes among T4-like phages. Hence, it has been widely used as a major phylogenetic marker for studying T4 phage diversity (14, 36, 54). A number of recent surveys employing degenerate g23-specific primers have resulted in the identification of uncharacterized subgroups of T4-like phages in the environment (19). For example, thousands of unique g23 sequences were found within the microbial shotgun metagenomic sequence data from the Global Ocean Sampling (GOS) expedition, indicating that infective T4-like phages are common within oceanic ecosystems (47). A large diversity of T4-like major capsid protein sequences have also been reported across a wide variety of aquatic environments, including rice paddy fields and freshwaters (10, 19, 25).
While signature gene analyses have been critical to investigations of virioplankton population ecology, other molecular genetic tools, such as pulsed-field gel electrophoresis (65), randomly amplified polymorphic DNA PCR (RAPD-PCR) fingerprinting (61), and most recently shotgun metagenomics (34), have also provided important advances in our understanding of the virioplankton. The critical distinction between these fingerprinting or random sequencing approaches and signature gene analyses is that they do not require a priori sequence information necessary for the design of a targeted PCR assay. In this study, we evaluated two low-cost, high-throughput approaches for examining the genetic similarity of viral populations—sequence polymorphism within the T4-like major capsid protein and RAPD-PCR fingerprinting—and asked how well genetic differences detected with these techniques connected with changes in time and geographic location. In addition to evaluating the utility of these approaches for observing virioplankton dynamics, the study also provided some initial insights into the genomic diversity of T4-like phages and the degree to which the dynamics of T4-like phages is reflective of larger dynamics within virioplankton populations not containing targeted signature genes. These investigations were carried out on virioplankton communities within the estuarine environments of the Chesapeake and Delaware Bays.
MATERIALS AND METHODS
Sampling sites.
Water samples were collected on six different cruise dates over a 1-year period along a transect of the Chesapeake Bay (CB) and Delaware Bay (DB). Viruses in surface water samples collected at stations CB858, CB818, CB804, CB707, and DB21 from March 2006 to July 2007 were immediately concentrated by tangential-flow filtration (TFF) (67). Locations for Chesapeake sampling stations are reported by Winget et al. (60). In July 2007, water samples from the pycnocline (designated midwater samples) and one meter above the sediment-water interface (designated bottom-water samples) were also collected.
Concentration and enumeration of viruses.
Each 50-liter seawater sample was prefiltered through a 25-μm wound cartridge filter using a diaphragm pump. Subsequently, viruses were concentrated using a multistep TFF process involving a 0.22-μm TF filter (Pellicon, Milipore Corp.), a 50-kd MWCO TF filter (Helicon, Millipore, Corp.), and a smaller 50-kDa TF filter (PrepScale, Millipore, Corp.), resulting in an approximately 250- to 300-ml viral concentrate (VC) which was postfiltered through a 0.22-μm syringe filter (Sterivex, Millipore, Corp.). The VC was dispensed into 50-ml conical tubes, snap-frozen in LN2, and stored at −80°C. In the laboratory, VCs from March and June 2006 were thawed slowly and concentrated by centrifugation using 30,000-molecular-weight-cutoff Centricon Plus-80 spin filters (Millipore Corp.) to a final volume of about 2 ml. Samples from cruises in July and November 2006 as well as February and July 2007 were concentrated by ultracentrifugation using about 100 ml of VCs. Ultracentrifugation was used, as we determined that it increased the efficiency of concentration (67). The virus pellet obtained was resuspended in a small volume of SM buffer (0.1 M NaCl, 8 mM MgSO4, 50 mM Tris-HCl, 0.005% glycerol [pH 8]). After postconcentration, viral concentrates were snap-frozen in LN2 and stored at −80°C; a subsample of each viral concentrate was stored at 4°C for pulsed-field gel electrophoresis (PFGE) and RAPD-PCR analysis. Viral abundance within concentrates was determined by epifluorescence microscopy (6).
PFGE.
Preparation of agarose plugs containing viral concentrate and subsequent analysis of viral concentrates by PFGE were carried out as described previously (65, 66). Briefly, equal volumes of viral concentrate, containing approximately 109 viruses and molten (50°C) 1.5% InCert agarose (Lonza, Rockland, ME), were mixed, vortexed, and immobilized into plug molds. The DNA was liberated within the plugs via overnight lysis (30°C in the dark with a lysis buffer [250 mM EDTA, pH 8], 1% SDS, and 1 mg/ml proteinase K solution) (65). After the plugs were washed three times with Tris-EDTA (TE) buffer, the viral genomic DNA within the plug was separated on a PFGE gel prepared from 1% (wt/vol) agarose (Bio-Rad PFGE grade). PFGE running conditions were as follows: 1× Tris-borate acetate (TBE) gel buffer, 0.5× TBE tank buffer, 14°C buffer temperature, 6.0 V/cm voltage gradient, 120° reorientation angle. The size of viral genomic DNA bands was determined on the basis of three different electrophoresis runs for each viral concentrate. Pulse ramp conditions were slightly changed for each of the three PFGE runs in order to separate a wide range of viral genomic DNA: (i) 1- to 15-s pulse ramp for 22 h for separating typical phage genome size fragments (25 to 300 kb); (ii) 0.1- to 4-s pulse ramp for 28 h for separating smaller genome fragments (25 to 100 kb); (iii) 9.65- to 44.69-s pulse ramps for 29 h for separating large genome fragments (200 to 500 kb). Concatemers of phage lambda genomic DNA (Bio-Rad), HindIII digest of phage lambda genomic DNA (Fisher Scientific), and MidRange PFG marker (New England BioLabs) were used as molecular weight markers. Gels were stained in a 1:10,000 dilution of SYBR Gold (Invitrogen) for 30 min and imaged on a Typhoon 9400 variable mode imager (GE Healthcare).
PCR amplification of g23 and DNA sequencing.
Single PFGE bands representing virioplankton genomic DNA were cut out of a preparative, low-melting-point SeaPlaque GTG agarose (Lonza) gel, run under the same conditions as the documenting gel (see above). As a precaution to prevent cross contamination, molecular weight markers were not loaded on preparative PFGE gels, and sample lanes were separated by two or more empty lanes. Viral genomic DNA bands were purified via beta-agarase (Lonza) digestion of the low-melting-temperature agarose. Purified viral genomic DNA was directly used for g23 amplification using degenerate g23 primers MZIA1bis (5′-GATATTTGNGGIGTTCAGCCIATGA-3′) and MZIA6 (5′-CGCGGTTGATTTCCAGCATGATTTC-3′), which have been previously shown (19) to amplify g23 fragments for all of the known subgroups of T4-like phages, within a range of marine environments. In a 25-μl PCR mix, the following reagents were added sequentially: 0.2 mM (each) deoxynucleoside triphosphates (dNTPs) (TaKaRa Bio Inc.), 0.8 μM each primer, a 1× final concentration of 10× Taq buffer, and 2.5 U of TaKaRa Ex Taq Hot Start version (TaKaRa Bio Inc.). Virioplankton genomic DNA from a single PFGE band was used as the template in each PCR. Purified T4 phage genomic DNA served as the positive-control template, and a reaction mix with no added DNA template served as a negative control. PCR amplification conditions were as follows: one cycle of denaturation at 94°C for 10 min, 30 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 1 min, extension at 72°C for 45 s, and a final extension at 72°C for 5 min. PCR products were separated by agarose gel electrophoresis, using 1.8% SeaPlaque GTG agarose (Lonza) in 1× Tris-acetate-EDTA (TAE) buffer at 4 V/cm for 3 h. Gels were stained with SYBR Gold (Invitrogen) nucleic acid gel stain (1:10,000 dilution) for 30 min and visualized under a transluminator. A slice of agarose containing DNA between 400 and 600 bp from all the samples showing positive g23 amplification was extracted and purified using the QIAquick gel extraction kit (Qiagen). PCR products were cloned using the pCR8/GW/TOPO TA cloning kit (Invitrogen) in accordance with the manufacturer's protocol. Ligation products were transformed into TOP10 Escherichia coli-competent cells (Invitrogen). Three to five random clones (colonies) containing g23 amplicon insert DNA were picked and sequenced after either plasmid purification using the QIAprep spin miniprep kit (Qiagen) or PCR amplification with vector-specific primers M13F and M13R directly from the clones. Each clone was sequenced bidirectionally using GWI and GW2 primers located on the TOPO vector.
Phylogenetic analysis of g23 amplicon sequences.
Sequences were edited in Sequencher 4.6 software (Gene Codes Corp.) to remove the vector and the primer sequence, and contigs were formed using both forward and reverse sequences. New g23 sequences were translated to amino acid sequences and aligned using the CLUSTALW (56) algorithm in MEGA 4.0 (53). Alignments were inspected and manually refined. Neighbor-joining analysis with the translated protein sequences was performed by using the p-distance correction model in MEGA 4.0. Clade support was identified by means of bootstrap analysis with 500 replicates, and bootstrap values above 50% were displayed on the phylograms. The phylogenetic tree was rooted using the phage T4 g23 sequence. A BLAST homology search against the GenBank non-redundant database at a maximum E score of 10−3 was performed to check the identity of each amplicon sequence as g23.
RAPD-PCR of PFGE bands.
Randomly amplified polymorphic DNA PCR (RAPD-PCR) analysis was applied to virioplankton genomic DNA from PFGE bands. In each RAPD-PCR assay, a single RAPD 10-mer, SOIL1000XC (5′-GGCGCCGGCG-3′), was used to amplify viral genomic DNA purified from the PFGE bands. Reactions were performed in a total volume of 25 μl containing 0.16 mM each dNTP, 4 μm of primer, 2.5 μl of 10× Taq buffer, and 2.5 U of TaKaRa Ex Taq HotStart version (TaKaRa Bio Inc.). Purified PFGE genomic DNA was quantified by an absorbance reading at 260 nm, and approximately 10 to 15 ng of virioplankton genomic DNA was added to each RAPD-PCR assay. PCR conditions were as follows: 94°C for 10 min; 30 cycles of 35°C for 3 min, 72°C for 1 min, 94°C for 30 s; 35°C for 3 min; with a final extension at 72°C for 10 min. A negative control without any template DNA was included with each RAPD-PCR analytical run. RAPD-PCR products were separated via agarose gel electrophoresis using 1.2% HiRes agarose (ISC Bioexpress) in 0.5× Tris-borate EDTA (TBE) buffer run at 4 V/cm for 3 h. Gels were stained in a 1:10,000 dilution of SYBR Gold (Invitrogen) and scanned using a Typhoon 9400 variable mode imager (GE Healthcare).
Image processing and RAPD-PCR fingerprints.
Images of RAPD-PCR gels were imported into GelCompar II (version 4.0; Applied Maths) and converted to binary images based on the presence and absence of bands (11, 16). The region between 350 to 2,645 bp was selected for further analysis. Once the lanes were defined, the images were processed and normalized using the molecular size marker lanes that were run at multiple locations in each gel. The process of normalization corrected gel distortion resulting from slight variability in band migration across the gel. Positive bands were identified by setting the minimum profiling value to 5%, which dictates the minimum peak height a band must have relative to the highest peak (i.e., darkest band) in that sample lane. The band calls and fragment sizes given by the program were manually reviewed and edited. Similarity between RAPD banding patterns was evaluated using the cluster analysis program in GelCompar II. Band matching (where each band is associated with a class) was also manually reviewed. Percent similarity of RAPD banding profiles was deduced using Dice's coefficient, and a dendrogram was generated using the unweighted pair group method with arithmetic mean (UPGMA) algorithm.
Nucleotide sequence accession numbers.
T4-like major capsid protein gene PCR amplicon sequences have been deposited in GenBank with accession numbers JX028427 to JX028509.
RESULTS
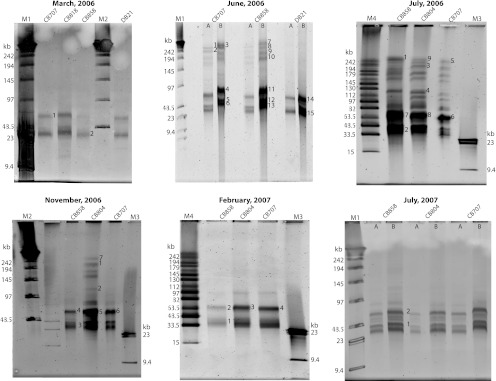
Changes in virioplankton assemblages were examined both within and between cruises across four stations (CB858, CB804, CB707, and DB21) along transects of the Chesapeake and Delaware Bays. Differences in virioplankton PFGE fingerprints in terms of presence and absence of bands was observed across the data set (Fig. 1). Designations of virioplankton genome sizes are as follows: small, 23 to 63.5 kb; medium, 64 to 130 kb; and large, 131 to 242 kb. Banding patterns were similar for samples collected at different locations in the bay in a single cruise, whereas PFGE banding patterns from a single station across different cruises (times of year) were markedly different. For example, viruses with large genome sizes were present in June and July 2006 samples but were not detected during March 2006 and the cruises following July 2006 (November 2006 to July 2007). One exception was from the station CB804 during the November 2006 cruise, where some bands appeared between 131 to 242 kb. Temporal differences in PFGE banding patterns indicate that seasonal changes in the structure of Chesapeake Bay virioplankton assemblages were greater than changes according to geographic location. It was also observed that viruses with genome sizes between 33.5 and 63.5 kb dominated the viral populations and were present in all samples. The signal intensity of these smaller genome bands was consistently higher than those of the large-genome-size viruses, indicating that viruses with smaller genomes were numerically more abundant in viral concentrates than viruses with large genomes.
Fig 1.
Pulsed-field gels of virioplankton concentrates. Gels are labeled according to the month and year of sample collection. Lanes are labeled by station designation in the Chesapeake Bay (CB) and Delaware Bay (DB). Samples from the June 2006 and July 2007 cruises were loaded at two different concentrations of virus particles: 109 (A) and ∼1010 (B). DNA was purified from bands marked with numbers. Marker lanes (in kilobases) are as follows: M1, concatemers of phage λ genome mixed with HindIII digest of λ genomic DNA; M2, concatemers of phage λ genome; M3, λ DNA digested with HindIII; M4, midrange PFG marker.
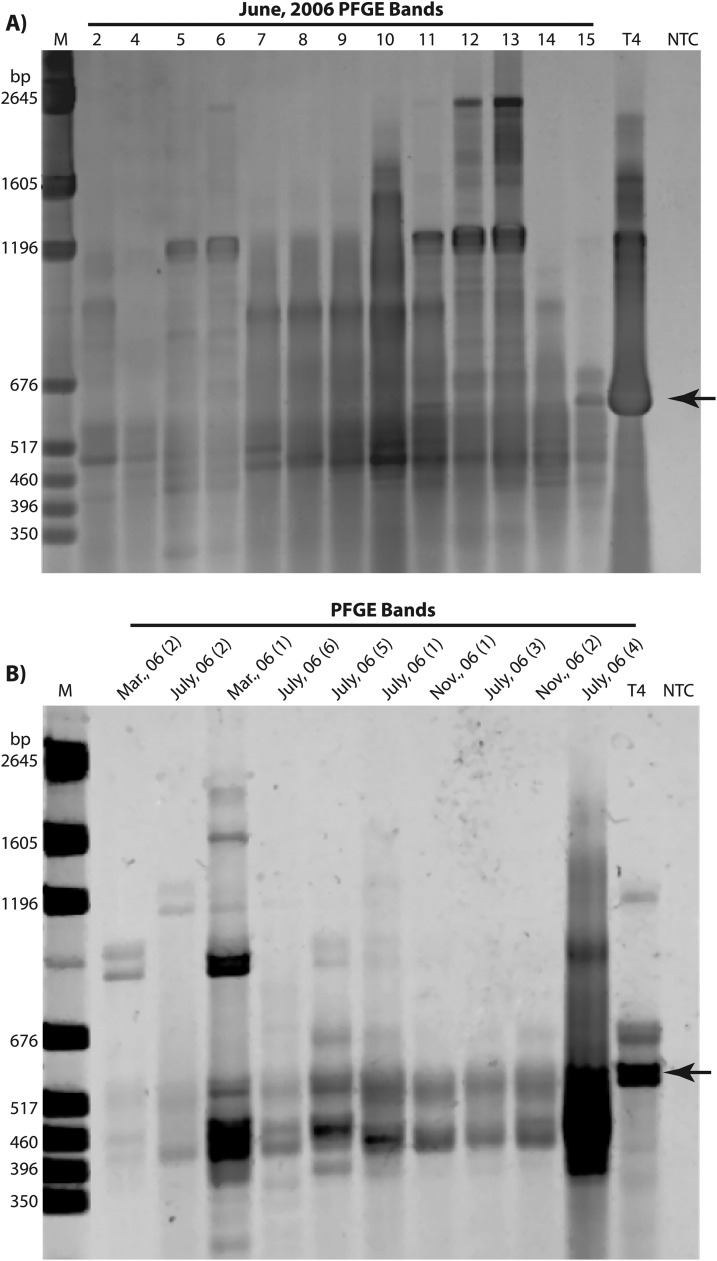
To examine the fine-scale genetic changes within viral populations (as defined by PFGE bands) the T4 major capsid protein gene (g23) was PCR amplified directly from PFGE bands (Fig. 2). The degenerate primers previously defined by Filée et al. (19) targeted a highly conserved region in all known g23 homologs. Due to insertions/deletions within the conserved region, amplicons produced by the primers varied in size between 400 and 600 bp. Amplification from small genome (23- to 63.5-kb) PFGE bands from three out of six cruise samples (November 2006, February 2007, and July 2007) did not result in g23 amplicons (see Table S1 in the supplemental material). However, bands within this size range from March, June, and July 2006 cruises resulted in a positive product when amplified with the g23 primers. All of the genome bands ranging from ca. 64 to 242 kb that were tested gave a positive g23 amplification signal.
Fig 2.
Amplification of g23 T4-like major capsid protein from single PFGE bands. (A) g23 amplicons from June 2006 PFGE bands. Numbers above lanes correspond to numbered PFGE bands in Fig. 1. (B) g23 amplicons from various PFGE bands. Lane designations are the month and year of sample collection, with numbers in parentheses corresponding to numbered PFGE bands in Fig. 1. Lanes labeled T4 were g23 amplicons from phage T4 genomic DNA. Marker lane M (in base pairs) indicates pGEM DNA. NTC, no template control. Arrow indicates the g23 amplicon from T4 genomic DNA. Amplification products from 480 bp to 600 bp were cut out and sequenced. A detailed description of bands purified and sequenced is summarized in Table S1 in the supplemental material.
Multiple sequence alignment of the 70 amino acid sequences from this study along with the T4 g23 sequence revealed two highly conserved blocks between amino acid residues 119 to 131 and 250 to 296 (see Fig. S1 in the supplemental material). Such conserved motifs have previously been established (10, 19, 30). Between these two conserved segments was a variable region, where sequences showed divergence. For all of the Chesapeake and Delaware Bay g23 sequences, divergence was limited to amino acid positions 132 to 249.
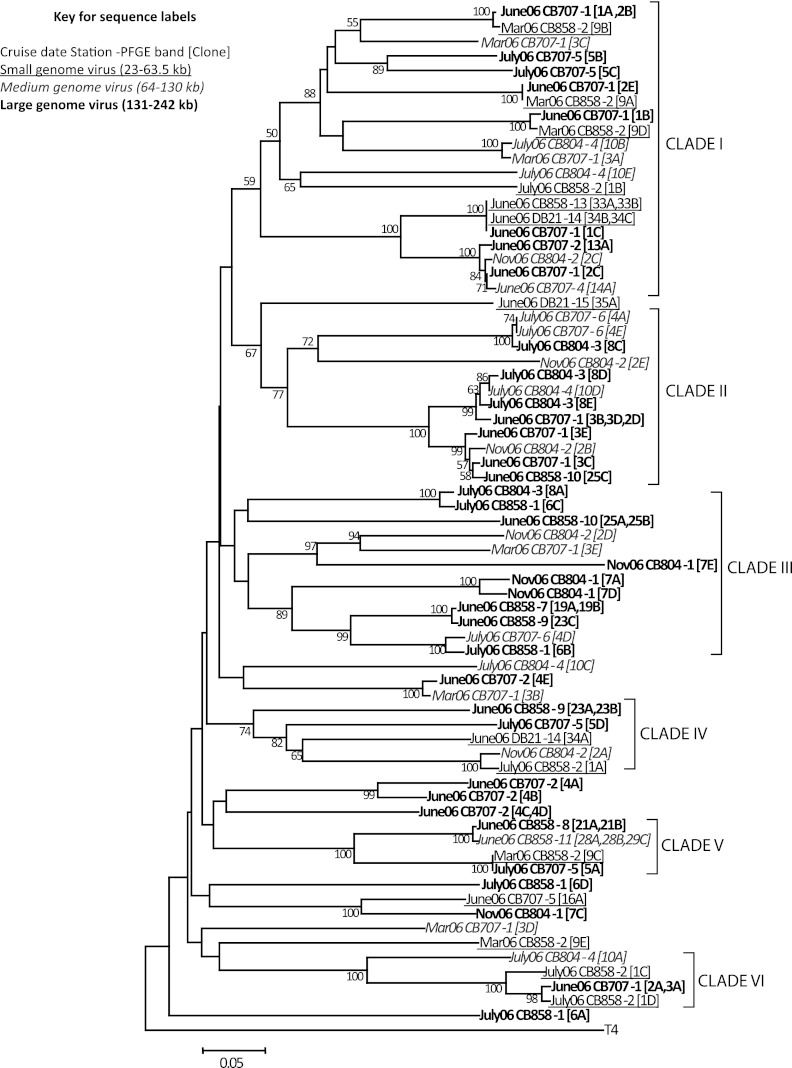
Ecological patterns of the g23 sequences obtained from this study were examined in regard to location in the Chesapeake and Delaware Bays, season, and putative phage genome size. Phylogenetic reconstruction showed that g23 sequences did not cluster based on genome size of the template phage genomic DNA or on the location and time of sample collection (Fig. 3). Evaluation of the clades marked on the phylogenetic tree shows, for example, that clade I contains sequences from four different dates (March, June, July, and November 2006), isolated from three different genome size classes (23 to 63.5 kb, 64 to 130 kb, and 131 to 242 kb). Interestingly, the sequences in this clade are also from four distinctly variable locations within the Chesapeake Bay and even from a location in another estuary—CB858, CB804, CB707, and DB21. It is evident from this clade that identical or highly similar sequences were obtained from large- and small genome viral PFGE bands that clustered together irrespective of their location or seasonal occurrence. For example, sequence June06CB707-1[1A,2B], derived from a PFGE band sized at ∼242 kb, is similar to sequence Mar06CB858-2[9B], derived from a small genome (33.5-kb) PFGE band. Sequences June06CB858-13[33A,33B], June06DB21-14[34B,34C], and June06CB707-1[1C] also reveal the same clustering pattern. A similar lack of differentiation by seasonal, spatial, or genome size characteristics was seen in g23 sequences from the clades IV, V, and VI (with bootstrap support of 74% and 100%). In contrast, clades II and III (with 77% and 97 to 89% bootstrap support, respectively), contained sequences from only medium- and large-genome-size viruses. Although this reflects a possible clustering based on the genome size, the overall sample size is small, and more sequences are needed to provide a clearer picture of associations between genome size and g23 phylogeny. In general, our findings suggest that phages carrying similar g23 genes are widespread in the Chesapeake Bay, and their distribution may not necessarily connect with location, time, or genome size.
Fig 3.
Neighbor-joining tree of g23 amino acid sequences from Chesapeake and Delaware Bay water samples. The tree is based on alignment of 178 homologous amino acid positions. The scale bar represents the number of amino acid substitutions per residue. The phage T4 g23 sequence was used as an outgroup. The values at each node indicate the bootstrap values above 50 using 500 replicates.
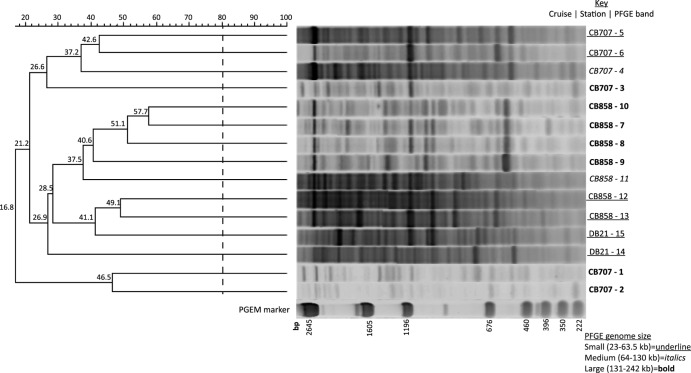
The RAPD-PCR assay produced an array of distinct and clearly reproducible bands of PCR amplicons. DNA within PFGE virioplankton genome bands from June 2006 samples was isolated and subjected to RAPD-PCR (Fig. 4). Resulting RAPD banding patterns were used to assess the genotypic diversity of virioplankton within specific viral genome size classes, as well as the genetic similarity of viruses having a particular genome size at different locations in the Chesapeake Bay. The highest similarity in RAPD profiles between two PFGE bands was 58% for PFGE bands 10 and 7 from station 858 (CB858-10 and CB858-7, respectively). For the RAPD-PCR assay, banding similarities of 80% are considered identical, as this is the established limit of sensitivity for the assay (61). Thus, no two virioplankton genome size class groups sampled in June 2006 were genotypically identical according to the assay. The lowest similarity seen in the analysis, 17%, was the clear distinction between two large-genome-class viruses at the lower Chesapeake Bay 707 station and all other June PFGE bands. In all cases, the RAPD fingerprints of PFGE bands from the higher-salinity Chesapeake station CB707 were clearly distinct from those of the lower-salinity stations CB858 and DB21 of the Chesapeake and Delaware Bays, respectively. Considering both genome size class and salinity, the RAPD fingerprint patterns of a given size class at a single station tended to clade together at ∼40% similarity. For example, large genome bands from station 858 occurred in a single clade at 41% similarity, and small genome bands from the lower salinity stations CB858 and DB21 also grouped at 41% similarity. The RAPD fingerprints from intermediate-sized-genome viruses were associated with their salinity group but were more distant. For example, PFGE bands CB858-11 and DB21-14 were outside the small-genome clades at 37.5% and 27%, similarity, respectively.
Fig 4.
Dendrogram of RAPD-PCR profiles from June 2006 PFGE band DNA. Viral concentrates were collected across three stations (CB858, CB707, DB21) in the Chesapeake and Delaware Bays. Numbers adjacent to sample names denote the PFGE band number shown in the June 2006 gel in Fig. 1. Percentage similarities of banding patterns are shown on the scale above the dendrogram and at the nodes. Size of marker band (pGEM) is given in base pairs of DNA. Fingerprints with greater than 80% similarity in banding pattern were considered identical.
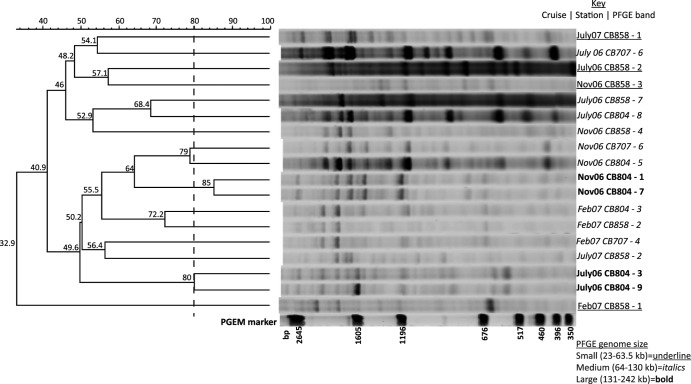
To examine the effect of seasonality in addition to location and genome size, RAPD fingerprints from PFGE bands were compared across four sampling dates (July 2006, November 2006, February 2007, and July 2007) and three sampling locations (CB858, CB804, and CB707) (Fig. 5). Unlike the June 2006 PFGE bands, there were two instances where two PFGE bands were considered identical within the limit of resolution for the RAPD-PCR fingerprinting assay. In both cases (Nov06CB804-1 and -7 and July06CB804-3 and -9), the PFGE bands were from the same location, time, and genome size class, supporting the finding of high genotypic similarity by RAPD-PCR, regardless of sampling date. There were other notable pairs of moderately similar (>60% similarity) RAPD fingerprint patterns that were within the same genome size class and time but collected at different stations (for example, July06CB858-7 and July06CB804-8, 68% similarity). The similarity of RAPD fingerprints all fell below 60% when examining relationships across time or genome size, indicating greater genotypic differences among virioplankton genome size groups sampled over time within the Chesapeake Bay.
Fig 5.
Dendrogram of RAPD-PCR profiles from PFGE bands within viral concentrates collected at four times and in three locations within the Chesapeake Bay. Numbers adjacent to sample names denote the PFGE band number shown in Fig. 1. Percentage similarities of banding patterns are shown above the dendrogram and at the nodes. The size of the marker band (pGEM) is given in base pairs of DNA. Fingerprints with greater than 80% similarity in banding pattern were considered identical.
DISCUSSION
The PFGE results indicate that the Chesapeake Bay contains both seasonal viral populations as well as those that are resident throughout the year. Previous studies of Chesapeake Bay virioplankton communities also demonstrated that Chesapeake Bay virioplankton assemblages vary with time and geographic location and are composed largely of viruses in the genome size range of 23 to 97 kb (65, 66). The predominance of dsDNA viruses with genome sizes of 23 to 97 kb is indicative of the fact that most of these viruses are likely bacteriophages. Several other studies have shown that marine viral assemblages are comprised predominantly of tailed phages in the Myoviridae, Podoviridae, and Siphoviridae families (4, 63), and the typical genome size for most phages ranges from 30 to 60 kb (50, 63).
While virioplankton PFGE patterns can vary seasonally in marine environments, such as the Chesapeake Bay (63–65) and Norwegian coastal waters (48), this study clearly demonstrates that in comparison to genotypic approaches, PFGE lacks the sensitivity necessary to examine the dynamic characteristics of virioplankton populations. Nevertheless, PFGE can be useful as a means for separating virioplankton communities into subpopulations based on genome size. Subsequent assays applied to these viral subpopulations can be used to examine finer-scale genetic changes within the virioplankton. The degree to which particular subgroups of virioplankton sharing a common PFGE genome size vary according to a genotypic criterion remains an open question. We addressed this question using two genotyping approaches targeted at individual PFGE bands: (i) sequence polymorphism of the T4-like major capsid protein (g23) gene and (ii) randomly amplified polymorphic DNA PCR. These two genotyping approaches are fundamentally different in that the g23 gene is believed to be restricted to a single group of bacteriophages, the T4-like myoviruses (14, 36, 54), while the RAPD-PCR assay is blind to gene content and instead randomly samples viral genotypic diversity across populations according to the presence of a specific 10-mer sequence within virioplankton genomic DNA (61).
One of the more surprising outcomes of the g23 survey was identification of g23 sequences from a broad range of viral genome size classes (∼23 to 242 kb). The genome size of most cultured T4-like phages (http://phage.bioc.tulane.edu) is known to be large (∼160 to 250 kb) (26, 35, 36, 42, 51), and the average genome size of phages having g23 proteins within the PHA2541 major capsid protein cluster (NCBI Protein Clusters database) is 180.6 kb (n = 24; standard deviation [SD], ±24.7 kb). Thus, the presence of g23 sequences within PFGE bands smaller than 100 kb suggests that the T4-like major capsid protein is carried by a broad diversity of phages, including those with genomes significantly smaller than the T4-like myoviruses. Furthermore, the fact that no g23 amplification occurred in smaller viral genomes (33.5 to 63.5) from November 2006 and February 2007 samples indicates a possible seasonality in the abundance of small-genome-size g23-carrying phages. This finding agrees with previous work examining the genome size distributions of cyanophages carrying the g20 vertex portal protein (48) and the two subunit genes of photosystem II (49). In both cases, these genes were detected within a broader range of genome size classes than would be indicated from known phages carrying these genes.
Seasonal variation in virioplankton composition and diversity has been observed in previous studies using single-gene approaches (15, 48, 58, 65) as well as PFGE whole-community fingerprinting (39, 50, 65). The seasonality of T4-like myoviral populations was recently established through monthly interannual observations of g23 genotypes at the San Pedro Ocean Time series (SPOTs) (8). Statistical analysis found that T4-like myoviral communities separated by a year were more alike than ones separated by three to six months. These oscillations in T4 communities were reminiscent of similar oscillations in bacterioplankton according to 16S genotyping (23). Our documentation of seasonal changes in the occurrence of viral populations based on g23 genotypes adds to the growing evidence of seasonality in the virioplankton. The fact that we have observed g23 in smaller-genome viruses supports previous observations of the wide distribution and high polymorphism among environmental g23 sequences (19, 25, 30) and argues for the ancient nature of the g23 gene (31).
Ancestral g23 sequences seem to have evolved and spread in different marine and terrestrial environments, such as rice paddy soils (19, 25, 30), possibly through horizontal gene transfer and insertion and deletion events at specific regions in the gene. The clustering of g23 sequences obtained from PFGE bands largely supports the view that g23 is a phage gene that is widespread in nature with a phylogeny that does not seem to conform to environmental context or the biological characteristics (i.e., genome size) of the phage that carry the gene. Phylogenetic analyses of g20 (48) and the psbA (49) gene sequences collected from PFGE bands showed little relationship to genome size, whereas the psbD gene showed a closer connection, possibly reflecting a longer period of purifying selection or less selective constraint on this gene within cyanophages (49).
Within the dynamic estuarine environments of the Chesapeake and Delaware Bays, the g23 gene shows a high degree of polymorphism. Nearly all of the major clades within the dendrogram of only Chesapeake and Delaware Bay g23 sequences contained sequences from each of the genome size bins and three or more sampling times and locations (Fig. 3). We can only speculate as to the mechanisms generating the observed polymorphism in the T4-like major capsid protein; however, there is ample evidence of horizontal gene transfer among tailed bacteriophages through the highly mosaic nature of their genome structures (17, 28). Since g23 is a core gene among T4-like myoviruses (9, 13, 26, 55), we speculate that g23 could be a frequent target of homologous recombination events, contributing to its ubiquitous dispersion and polymorphic nature.
Membership within the Myoviridae morphological family is the only consistent biological feature believed to be connected to the presence of the g23 gene in known phage genomes. All member phages of the UniRef 50 cluster of the T4-like major capsid protein (UniRef50_P04535) are myoviruses. Many myoviruses have genomes larger than 100 kb, although smaller-genome phages also fall in this group, such as the P2-like phages (NCBI taxid 140410) and the phage mu-like group (NCBI taxid 186777). Within this study, g23 sequences were less frequent within small genome viral PFGE bands and did not occur at all in the November 2006, February 2007, or July 2007 samples. While it is possible the small genome g23-positive viral PFGE bands we detected were myoviruses, reports of g23 amplicons from siphoviral isolates demonstrate that the linkage between g23 and myoviruses may not be absolute (29, 37). Finally, structural biological investigations support the idea that T4-like phages may have a broader genome size range than previously described. Mutations in g23 can affect the head length (32), which would have direct effects on the amount of DNA packaged in the phage head (i.e., phage genome size), and substitutions of g23 for g24, the vertex protein, have been shown to produce small phage capsids (20).
Overall, phylogenetic analysis did not reveal underlying connections between g23 sequence polymorphism and phage genome size, sampling location, or collection time. Thus, an alternative approach, RAPD-PCR, was used to examine the genotypic dynamics of virioplankton genome size groups over time and space. Several studies have used RAPD-PCR to contrast genotypic changes in whole viral assemblages (27, 61, 62). These investigations have found RAPD-PCR to be a more robust and sensitive means than PFGE for examining dynamic changes in viral assemblages. In this investigation, the number of RAPD-PCR bands obtained from viral genomic DNA in a single PFGE band equaled or exceeded that seen in RAPD-PCR assays of entire viral assemblages (61, 62). While this result seems counterintuitive, it likely reflects the problem of template dilution in PCR, where a high ratio of priming sites to primer molecules causes inefficiencies in PCR amplification (61). Because the genomic DNA template from an entire virioplankton community contains more sequence diversity (and thus more priming sites) than the DNA from a single PFGE band, fewer amplicons reach the critical threshold for exponential amplification within the 30 cycles of the RAPD-PCR assay. As a consequence, the sensitivity of RAPD-PCR for detecting genotypic changes in a single virioplankton genome size class may be greater than for entire virioplankton assemblages. Unlike g23 polymorphism, the similarities of RAPD-PCR fingerprinting patterns loosely reflected viral genome size, location, and time. Thus, RAPD fingerprints of two PFGE bands of similar size collected at the same time and location had higher similarity than those of two bands that differed by any one of these parameters.
The observation that environmental factors such as salinity appeared to influence the composition of virioplankton populations within genome size groups according to RAPD PCR was an encouraging outcome of the study. The significant influence of salinity and depth as factors structuring the composition of bacterioplankton communities has been firmly established (21). While the PFGE data from this study and a previous one show slight changes in virioplankton communities along the Chesapeake salinity gradient (65), the RAPD data more clearly resolved spatial differences in virioplankton communities for the June 2006 samples. Other work examining microbial communities across a broad transect of the subtropical Atlantic found that virioplankton communities partitioned clearly by depth according to RAPD-PCR fingerprints (12). Thus, future application of RAPD-PCR fingerprinting across environmental gradients may be sensitive enough to detect those environmental and biological variables that most influence changes in the genotypic composition of the virioplankton. Moreover, RAPD fingerprinting can be applied to environmental samples with high throughput and low per-sample cost, distinct advantages over shotgun metagenomic approaches.
In a few cases, nearly identical RAPD fingerprints were obtained from viral genomes within different genome size classes. These fingerprints were within the percent similarity threshold for true replicates in the RAPD-PCR assay (59, 61). Although the underlying sequence identity between bands in the fingerprint is unknown, the likelihood that the agreement in banding patterns occurred through coincidence is quite low when considering the rarity of 10-mer binding sites within random DNA sequence. In total, there exist 1,047,576 possible 10-mers (410 decamers), with ∼800,000 containing at least seven Gs and/or Cs (the design constraint of primer 5′-GGCGCCGGCG-3′ used in this study). The probability of two 10-mer sites facing one another and producing bands of identical sizes in two different DNA samples is very small. The probability is lower when considering entire RAPD-PCR fingerprints. Alternate and more likely explanations for the high agreement between RAPD-PCR fingerprints of two different-sized genome groups is either the occurrence of genetically identical regions within viruses having different-sized genomes or the existence of gene deletions within the amplified region from the larger genome or gene insertion in the smaller genome. The fact that viruses demonstrate a high degree of genetic plasticity, i.e., deletions, insertions, and genetic reassortment are very common in viral genomes (28, 41), also supports this argument. One means to improve the resolution of RAPD-PCR for detecting genotypic differences among closely separated viral genome size groups would be to run independent RAPD-PCR assays of each band using different decamer primers.
For cellular microbial life, sequence polymorphism within the 16S rRNA gene has become the de facto standard for examining the dynamic behavior of communities. The lack of such a universal marker gene among viruses has required researchers to explore other means for examining the diversity and compositional dynamics of viral communities. These investigations demonstrate the advantages and limitations of using single gene marker approaches for examining a virioplankton population as defined by the property of genome size and environmental context. Polymorphism within the g23 protein provides high sensitivity and the advantage of a growing phylogenetic framework of genes amplified from viral isolates and environmental samples; however, the degree to which g23 sequences connect with other genotypic or phenotypic properties of phages is an open question. As shown here, the presence of the gene may not be restricted to large-genome virulent phages. Unlike g23 polymorphism approaches, RAPD-PCR requires no a priori sequence information or assumptions regarding the universality of degenerate primers used in PCR amplification. These investigations indicate some connection between the similarity of RAPD-PCR fingerprints and exogenous environmental factors or endogenous biological properties, such as viral genome size. However, without sequencing, RAPD-PCR will not detect genotypic variation in viral populations. Nevertheless, RAPD-PCR performed well as a means of quickly and inexpensively examining the dynamics of virioplankton subpopulations at high sensitivity.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the support of National Science Foundation grants (EF-0626826, MCB-0132070, and MCB-0731916) to K.E.W.
We are indebted to the crew of the R/V Cape Henlopen and Hugh R. Sharp for their tireless assistance in the field.
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 [DOI] [PubMed] [Google Scholar]
- 2. Ackermann HW, Krisch HM. 1997. A catalogue of T4-type bacteriophages. Arch. Virol. 142:2329–2345 [DOI] [PubMed] [Google Scholar]
- 3. Bergh O, Borsheim KY, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340:467–468 [DOI] [PubMed] [Google Scholar]
- 4. Breitbart M, et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brussaard CP, et al. 2008. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2:575–578 [DOI] [PubMed] [Google Scholar]
- 6. Chen F, Lu JR, Binder BJ, Liu YC, Hodson RE. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 67:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chibani-Chennoufi S, Bruttin A, Dillmann M-L, Brussow H. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow C-ET, Fuhrman JA. 2012. Seasonality and monthly dynamics of marine myovirus communities. Environ. Microbiol. 14:2171–2183 [DOI] [PubMed] [Google Scholar]
- 9. Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
- 10. Comeau AM, Krisch HM. 2008. The capsid of the T4 phage superfamily: the evolution, diversity, and structure of some of the most prevalent proteins in the biosphere. Mol. Biol. Evol. 25:1321–1332 [DOI] [PubMed] [Google Scholar]
- 11. Comeau AM, Short S, Suttle CA. 2004. The use of degenerate-primed random amplification of polymorphic DNA (DP-RAPD) for strain-typing and inferring the genetic similarity among closely related viruses. J. Virol. Methods 118:95–100 [DOI] [PubMed] [Google Scholar]
- 12. De Corte D, Sintes E, Yokokawa T, Reinthaler T, Herndl GJ. 2010. Links between viruses and prokaryotes throughout the water column along a North Atlantic latitudinal transect. ISME J. 4:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desplats C, Dez C, Tetart F, Eleaume H, Krisch HM. 2002. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. J. Bacteriol. 184:2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desplats C, Krisch HM. 2003. The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154:259–267 [DOI] [PubMed] [Google Scholar]
- 15. Dorigo U, Jacquet S, Humbert J-F. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol. 70:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrari VC, Hollibaugh JT. 1999. Distribution of microbial assemblages in the Central Arctic Ocean Basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia 401:55–68 [Google Scholar]
- 17. Filée J, Bapteste E, Susko E, Krisch HM. 2006. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol. Biol. Evol. 23:1688–1696 [DOI] [PubMed] [Google Scholar]
- 18. Filée J, Forterre P, Sen-Lin T, Laurent J. 2002. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54:763–773 [DOI] [PubMed] [Google Scholar]
- 19. Filée J, Tétart F, Suttle CA, Krisch HM. 2005. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. U. S. A. 102:12471–12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fokine A, et al. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 102:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. 2012. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 6:554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548 [DOI] [PubMed] [Google Scholar]
- 23. Fuhrman JA, et al. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. U. S. A. 103:13104–13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuhrman JA, Suttle CA. 1993. Viruses in marine planktonic systems. Oceanography 6:51–63 [Google Scholar]
- 25. Fujii T, et al. 2008. Novel capsid genes (g23) of T4-type bacteriophages in a Japanese paddy field. Soil Biol. Biochem. 40:1049–1058 [Google Scholar]
- 26. Hambly E, et al. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. U. S. A. 98:11411–11416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helton RR, Wommack KE. 2009. Seasonal dynamics and metagenomic characterization of estuarine viriobenthos assemblages by randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 75:2259–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hendrix RW, Smith MCM, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenkins CA, Hayes PK. 2006. Diversity of cyanophages infecting the heterocystous filamentous cyanobacterium Nodularia isolated from the brackish Baltic Sea. J. Mar. Biol. Assoc. U. K. 86:529–536 [Google Scholar]
- 30. Jia Z, Ishihara R, Nakajima Y, Asakawa S, Kimura M. 2007. Molecular characterization of T4-type bacteriophages in a rice field. Environ. Microbiol. 9:1091–1096 [DOI] [PubMed] [Google Scholar]
- 31. Koonin E, Senkevich T, Dolja V. 2006. The ancient Virus World and evolution of cells. Biol. Direct 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60:2356–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mann NH, et al. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marine R, et al. 2011. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl. Environ. Microbiol. 77:8071–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller ES, et al. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monod C, Repoila F, Kutateladze M, Tétart F, Krisch HM. 1997. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J. Mol. Biol. 267:237–249 [DOI] [PubMed] [Google Scholar]
- 37. Nakayama N, Tsuge T, Asakawa S, Kimura M. 2009. Morphology, host range and phylogenetic diversity of Sphingomonas phages in the floodwater of a Japanese paddy field. Soil Sci. Plant Nutr. 55:53–64 [Google Scholar]
- 38. Nolan J, Petrov V, Bertrand C, Krisch H, Karam J. 2006. Genetic diversity among five T4-like bacteriophages. Virol. J. 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parada V, Baudoux A-C, Sintes E, Weinbauer MG, Herndl GJ. 2008. Dynamics and diversity of newly produced virioplankton in the North Sea. ISME J. 2:924–936 [DOI] [PubMed] [Google Scholar]
- 40. Patrick F. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33:457–465 [DOI] [PubMed] [Google Scholar]
- 41. Pedulla ML, et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182 [DOI] [PubMed] [Google Scholar]
- 42. Petrov V, Ratnayaka S, Nolan J, Miller E, Karam J. 2010. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. 2004. Viral release of iron and its bioavailability to marine plankton. Limnol. Oceanogr. 49:1734–1741 [Google Scholar]
- 44. Proctor LM, Fuhrman JA. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62 [Google Scholar]
- 45. Rohwer F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 46. Rohwer F, Edwards R. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rusch DB, et al. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sandaa R-A, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl. Environ. Microbiol. 72:4610–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sandaa RA, Clokie M, Mann NH. 2008. Photosynthetic genes in viral populations with a large genomic size range from Norwegian coastal waters. FEMS Microbiol. Ecol. 63:2–11 [DOI] [PubMed] [Google Scholar]
- 50. Steward GF, Montiel JL, Azam F. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697–1706 [Google Scholar]
- 51. Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:790–806 doi:10.1371/journal.pbio.0030144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suttle CA. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 53. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 54. Tetart F, et al. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thingstad TF, Lignell R. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19–27 [Google Scholar]
- 56. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914 [DOI] [PubMed] [Google Scholar]
- 58. Wang K, Chen F. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105–116 [Google Scholar]
- 59. Williamson SJ, et al. 2008. Lysogenic virus-host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2:1112–1121 [DOI] [PubMed] [Google Scholar]
- 60. Winget DM, et al. 2011. Repeating patterns of virioplankton production within an estuarine ecosystem. Proc. Nat. Acad. Sci. 108:11506–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Winget DM, Wommack KE. 2008. Randomly amplified polymorphic DNA (RAPD)-PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 76:6724–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winter C, Weinbauer MG. 2010. Randomly amplified polymorphic DNA reveals tight links between viruses and microbes in the bathypelagic zone of the Northwestern Mediterranean Sea. Appl. Environ. Microbiol. 76:6724–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wommack KE, Hill RT, Kessel M, Russek-Cohen E, Colwell RR. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wommack KE, Ravel J, Hill RT, Chun J, Colwell RR. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wommack KE, Ravel J, Hill RT, Colwell RR. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wommack KE, Sime-Ngando T, Winget DM, Jamindar S, Helton RR. 2010. Filtration-based methods for the collection of viral concentrates from large water samples, p 110–117 In Wilhelm SW, Weinbauer MG, Suttle CA. (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.