Abstract
Here we report the cloning of the Pa_3_10940 gene from the coprophilic fungus Podospora anserina, which encodes a C-terminal family 1 carbohydrate binding module (CBM1) linked to a domain of unknown function. The function of the gene was investigated by expression of the full-length protein and a truncated derivative without the CBM1 domain in the yeast Pichia pastoris. Using a library of polysaccharides of different origins, we demonstrated that the full-length enzyme displays activity toward a broad range of β-glucan polysaccharides, including laminarin, curdlan, pachyman, lichenan, pustulan, and cellulosic derivatives. Analysis of the products released from polysaccharides revealed that this β-glucanase is an exo-acting enzyme on β-(1,3)- and β-(1,6)-linked glucan substrates and an endo-acting enzyme on β-(1,4)-linked glucan substrates. Hydrolysis of short β-(1,3), β-(1,4), and β-(1,3)/β-(1,4) gluco-oligosaccharides confirmed this striking feature and revealed that the enzyme performs in an exo-type mode on the nonreducing end of gluco-oligosaccharides. Excision of the CBM1 domain resulted in an inactive enzyme on all substrates tested. To our knowledge, this is the first report of an enzyme that displays bifunctional exo-β-(1,3)/(1,6) and endo-β-(1,4) activities toward beta-glucans and therefore cannot readily be assigned to existing Enzyme Commission groups. The amino acid sequence has high sequence identity to hypothetical proteins within the fungal taxa and thus defines a new family of glycoside hydrolases, the GH131 family.
INTRODUCTION
Podospora anserina is a late grower in the droppings of grass herbivores. It has been suggested that P. anserina is likely to target lignocellulose, since most hemicellulose and pectin would be consumed by zygomycetes and early ascomycetes. Genomic analysis of this coprophilic ascomycete has revealed a large and highly specialized set of genes involved in utilization of natural carbon sources commonly found in its natural biotope (7, 10). Despite this organism and other fungi possessing similar numbers of putative enzymes, the distribution of the possible enzyme functions related to plant cell wall degradation (i.e., carbohydrate-active enzymes [CAZymes]) (5; www.cazy.org) in P. anserina is significantly different from that in other fungi. P. anserina has one of the largest fungal sets of candidate enzymes for cellulose degradation described to date, with one of the largest numbers of carbohydrate-binding modules (CBMs) of all the fungal genomes. This is particularly remarkable for the numbers of GH61 (copper-dependent monooxygenases acting on cellulose) and CBM1 (targeting cellulose) modules encoded in the genome of P. anserina, which are larger than the sets in the white rot fungus Phanerochaete chrysosporium and the phytopathogen Magnaporthe grisea. Furthermore, the large number of GH18 and CBM18 modules could indicate that P. anserina has the ability to degrade exogenous chitin and possibly to degrade available fungal cell material from the set of fungi that grow earlier on dung of herbivores.
Recently, we cloned and characterized five hemicellulases from P. anserina, belonging to different glycoside hydrolase (GH) families, some of which efficiently supplemented a Trichoderma reesei enzymatic cocktail for the saccharification of lignocellulosic biomass (8). Among the components of this unexpected unique enzymatic equipment, P. anserina's genome encodes a number of other interesting enzymes potentially acting on the recalcitrant components of the plant cell wall. However, the presence of hypothetical proteins hampers our understanding of the mechanisms by which this carbon cycle is carried out. Here we report the cloning of a gene from P. anserina that encodes a CBM1 fused to a catalytic domain of unknown function. We present the detailed enzymatic characterization of the corresponding enzyme, which belongs to a new GH family.
MATERIALS AND METHODS
Bioinformatic analysis.
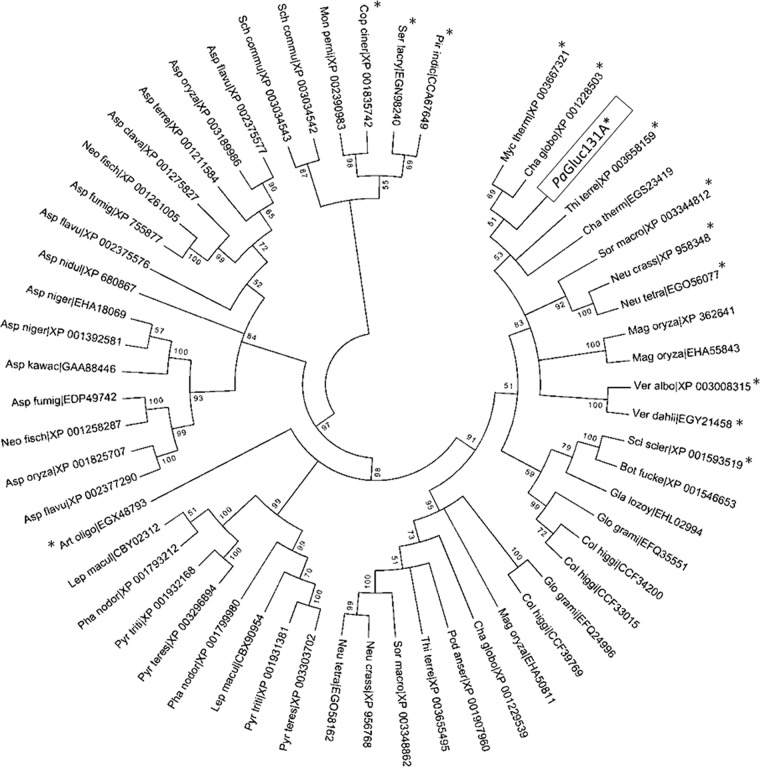
The P. anserina genome was searched for genes encoding enzymes containing a CBM1 module. Among the genes for 30 proteins bearing a CBM1, we selected the Pa_3_10940 gene because the encoded catalytic module was of unknown function. As a complement, the signal peptide cleavage site was predicted using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). Phylogenetic analysis involved 52 amino acid sequences which displayed high sequence identity to PaGluc131A (catalytic domain only), encoded by the Pa_3_10940 gene. The evolutionary history was inferred using the minimum evolution method (24). The evolutionary distances were computed using the P-distance method (19) and are reported as the number of amino acid differences per site. The minimum evolution tree was searched using the close neighbor interchange algorithm (19) at a search level of 0. The neighbor-joining algorithm (25) was used to generate the initial tree. All ambiguous positions were removed for each sequence pair. There were a total of 288 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (29).
Cloning of the Pa_3_10940 gene.
The homokaryotic Podospora anserina strain S mat+ used in this study was kindly provided by P. Silar (CNRS, Paris, France). Currently used methods and culture media can be accessed at the Podospora anserina Genome Project website (http://podospora.igmors.u-psud.fr). P. anserina was grown on M2 medium supplemented with either sugar beet pulp at 15 g liter−1, autoclaved fraction of maize bran at 15 g liter−1, or a mixture of pectin, birchwood xylan, and larchwood xylan (Sigma-Aldrich, Lyon, France) at 5 g of each per liter. The autoclaved fraction of maize bran was prepared as described previously (3), and its sugar composition has also been described previously (9). Growth was carried out in baffled flasks at 120 rpm and 27°C. The mycelium was recovered by sterile filtration using Miracloth after 3, 4, and 5 days of culture and directly frozen at −80°C. Total RNA extraction and first-strand cDNA synthesis were performed as described previously (8). DNA was amplified by a PCR using Pwo Super Yield DNA polymerase (GE Healthcare, Buc, France) and the following primers, designed based on the Pa_3_10940 gene sequence: EcoRIFor, TTTGAATTCGGCACCATCCTGTGGGATGGCC; and XbaIRev, TTTTCTAGACCCAAGCACTGGGAGTACCAG (restriction sites are underlined). The amplified fragment was subcloned into the pCRII-TOPO vector (Invitrogen) and subjected to sequencing. In order to delete the CBM1, amplification was performed by a PCR using primers EcoRIFor and dCBMXbaR2 (TTTTCTAGACCGCCACCGTCACCGGGGAT). The full-length open reading frame (ORF) (PaGluc131A) and that for the catalytic module only (PaGluc131AΔCBM) were inserted at the corresponding sites (EcoRI and XbaI) into the pPICZαA vector in frame with both the yeast α-secretion factor- and C-terminal His6 tag-encoding sequences. Each recombinant plasmid was sequenced to check the integrity of the constructs. The resulting recombinant expression plasmid was linearized with PmeI and transformed into P. pastoris X-33 (Invitrogen) by electroporation. P. pastoris transformants were isolated on plates containing increasing concentrations of zeocin ranging from 100 μg/ml to 1,000 μg/ml. Zeocin-resistant P. pastoris transformants were then screened for protein expression by growth in 10 ml of buffered minimal glycerol medium (BMGY) (in 50-ml tubes) at 30°C in an orbital shaker (200 rpm) for 16 h, to an optical density at 600 nm (OD600) of 2 to 6, with expression induced by transferring cells into 2 ml buffered minimal methanol medium (BMMY) and incubating them for another 3 days. Each day, the medium was supplemented with 3% (vol/vol) methanol. The supernatant was then analyzed by SDS-PAGE to determine the transformant with the best secretion yield for each enzyme.
PaGluc131A production and purification.
For each construct, the best-producing transformant was grown in 200 ml BMGY in a 1-liter baffled flask, and then the cells were transferred to 40 ml BMMY in a 200-ml flask at 200 rpm and 30°C. The suspension was recovered by centrifugation (10 min, 4,000 × g) after 3 days of induction, and the supernatant was concentrated 10-fold at 4,000 × g using Amicon centrifugal units (10-kDa cutoff; polyether sulfone) (Millipore, Molsheim, France). Purification of PaGluc131AΔCBM was performed as described previously (8). As PaGluc131A did not bind to the nickel column, purification was performed as described previously (1). Briefly, the supernatant was concentrated using Amicon centrifugal units as described above, loaded onto a 2- by 60-cm Superdex 200 column, and eluted in 50 mM sodium acetate-150 mM sodium chloride at a flow rate of 0.5 ml/min. The fractions corresponding to the middle of the eluted peak were pooled and checked for purity by SDS-PAGE and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (not shown).
Biochemical characterization.
The protein concentration was determined using a Bio-Rad protein assay kit with bovine serum albumin as the standard (Bio-Rad, Marnes la Coquette, France). SDS-PAGE was performed on a 12% (wt/vol) polyacrylamide gel (Bio-Rad), using a Pharmacia LMW electrophoresis calibration kit (GE Healthcare). Native isoelectric focusing (IEF) was carried out at 4°C in a Bio-Rad gel system, using pI standards ranging from 4.45 to 8.2 (Bio-Rad). Proteins were visualized either by Coomassie blue staining or by use of IEF gel staining solution (0.04% [wt/vol] Coomassie blue R250, 0.05% [wt/vol] Cocreïn scarlet, 10% [vol/vol] acetic acid, 27% [vol/vol] isopropanol).
Substrates.
Birchwood xylan, carboxymethyl cellulose (CMC), Avicel PH101, hydroxyethyl cellulose (HEC), chitin, pectin, laminarin, locust bean gum, and gentiobiose were purchased from Sigma-Aldrich. Low-viscosity wheat arabinoxylan, konjak glucomannan, xyloglucan, barley-β-glucan, lichenan, curdlan, and pachyman were purchased from Megazyme International (Wicklow, Ireland). Walseth cellulose and pustulan were provided by IFPEN (Paris, France) and CERMAV-CNRS (Grenoble, France), respectively. Cello-oligosaccharides, β-(1,3;1,4)-gluco-oligosaccharides, and β-(1,3)-oligosaccharides were purchased from Megazyme International. pNP-β-d-glucopyranoside, pNP-β-d-xylopyranoside, pNP-β-d-mannopyranoside, pNP-α-l-arabinofuranoside, pNP-β-d-lactose, pNP-β-d-saccharose, and pNP-β-d-cellobioside were purchased from Sigma-Aldrich.
Enzyme assays.
Activity screening using the library of substrates was performed using the DNS assay (18) and high-performance anion-exchange chromatography (HPAEC) coupled with pulsed amperometric detection (PAD) (ICS 3000; Dionex, Sunnyvale, CA). Unless otherwise indicated, assay mixtures contained substrate and suitably diluted enzyme in 50 mM sodium acetate buffer, pH 5.5, at 40°C. Briefly, 20 μl (1.2 to 5.2 nmol of enzyme) of PaGluc131A was mixed with 100 μl of polymeric substrate (10 mg ml−1) or oligosaccharides (100 μM). The reaction was terminated by the addition of 120 μl of 18 mM NaOH. The reaction mixtures were transferred to a microfiltration plate and centrifuged for 2 min at 900 × g. For HPAEC analysis, 5 μl was injected and elution was carried out in 130 mM NaOH, using a 25-min linear gradient program from 100% phase A (130 mM NaOH) to 60% phase A and 40% phase B (500 mM NaOAc, 130 mM NaOH), as described previously (8).
The optimal pH was estimated using laminarin at 10 mg ml−1 in 50 mM sodium citrate buffer (pHs from 2.5 to 3.5), 50 mM sodium acetate buffer (pHs from 4.0 to 6.0), and 50 mM sodium phosphate buffer (pHs from 6.5 to 8.0). The optimal temperature was estimated at temperatures ranging from 4 to 80°C.
Specific activities toward the different complex polysaccharide substrates were measured using HPAEC-PAD. Monosaccharides and oligosaccharides generated after hydrolysis of the different polymeric substrates [β-(1,4)-glucan, β-(1,3)-glucan, β-(1,3;1,4)-glucan, and β-(1,6)-glucan] and oligosaccharides [cello-oligosaccharides, β-(1,3;1,4)-gluco-oligosaccharides, and β-(1,3)-gluco-oligosaccharides] by PaGluc131A were analyzed as described above, using appropriate standards as controls. Calibration curves were constructed using the standards, from which response factors were calculated (Chromeleon program; Dionex) and used to estimate the amount of product released in test incubations. All assays were carried out in triplicate. Results are expressed in μmol of sugar released per min per mg of enzyme. The specificity constants were calculated using the Matsui equation for oligosaccharides (2, 16). For determination of Michaelis-Menten constants on laminarin and pustulan, the initial velocities were measured for substrate concentrations ranging from 1.25 to 20 mg ml−1. The degrees of polymerization of pustulan and laminarin used to calculate their molar concentrations were 94 (22) and 28 (17), respectively. The kinetic parameters were estimated using weighted nonlinear least-squares regression analysis with Grafit software (Erithacus Software, Horley, United Kingdom).
Activities toward pNP substrates (1 mM) were determined by measuring the release of 4-nitrophenol in 50 mM sodium phosphate buffer, pH 5.5, at 37°C, using a 100-μl reaction volume. The reaction was stopped by the addition of 200 μl of 1 M sodium carbonate, pH 9, and the release of 4-nitrophenol was quantified at 405 nm by using the molar extinction coefficient of 4-nitrophenol (18,300 M−1 cm−1). One unit of enzyme activity was defined as the amount of protein that released 1 μmol of glucose per min.
Nucleotide sequence accession number.
The Pa_3_10940 gene sequence has been deposited in GenBank under accession number XM_001903499.
RESULTS
Cloning of the Pa_3_10940 gene and sequence analysis.
The P. anserina Pa_3_10940 gene was selected based on BLASTp searches against the P. anserina genome, since the coding sequence of the Pa_3_10940 gene encodes a CBM1 module linked to a domain of unknown function. The cloning of the Pa_3_10940 gene was carried out by reverse transcription-PCR (RT-PCR) using mRNAs purified from P. anserina grown on different natural inducers. All inducers (i.e., sugar beet pulp, maize bran, and a mixture of pectin and xylan) used were found to induce the expression of the Pa_3_10940 gene after 5 days of growth (data not shown). After sequencing of the cDNA, its comparison with the predicted coding sequence of Pa_3_10940 revealed an insertion of 66 nucleotides in the cDNA sequence that led to 22 extra amino acids without any change in the open reading frame, probably due to a missed intron in the genome annotation. The cDNA (GenBank accession no. JX310321) consisted of 1,035 bp corresponding to a protein of 345 amino acids, with a predicted molecular mass of the mature protein of 37 kDa. The catalytic domain (CD) (residues 1 to 252) and the CBM1 (residues 307 to 345) are separated by a linker carrying numerous potential O-glycosylation sites (Ser and Thr residues) (Fig. 1). No putative N-glycosylation site was identified in the full-length protein. Interestingly, the results of BLAST searches using the amino acid sequence of the PaGluc131A CD revealed that this family of proteins is encountered only in fungi. A phylogenetic analysis of 52 fungal protein sequences of unknown function that display similarity to the PaGluc131A CD is presented in Fig. 2. Fungi generally carry one or two members of this family. The phylogenetic tree shows clear branches that correspond to the fungal taxa. The PaGluc131A protein sequence clustered within the Sordariales group, i.e., it was closely related (>53% amino acid sequence identity) to GH131 proteins from Myceliophthora thermophila (GenBank accession no. XP_003667321), Chaetomium globosum (GenBank accession no. XP_001228503), Thielava terrestris (GenBank accession no. XP_003658159), Chaetomium thermophilum (GenBank accession no. EGS23419), Sordaria macrospora (GenBank accession no. XP_003344812), Neurospora crassa (GenBank accession no. XP_958348), and Neurospora tetrasperma (GenBank accession no. EGO56077), which all contain a CBM1 module (except for C. thermophilum GH131) (Fig. 2).
Fig 1.
Amino acid sequence of PaGluc131A. The catalytic domain (residues 1 to 252), linker sequence (underlined; residues 253 to 306), and CBM1 (residues 307 to 345) are shown.
Fig 2.
Phylogenetic representation of GH131 family. The analysis involved 52 fungal amino acid sequences which displayed high sequence identity to the PaGluc131A catalytic domain. The phylogenetic tree highlights the relative positions of the proteins, each labeled with the abbreviation of the species name and the reference public database accession number (GenBank). The presence of the CBM1 module is indicated with an asterisk.
Heterologous expression of PaGluc131A in Pichia pastoris.
The full-length sequence and the catalytic module without CBM1 were inserted into the P. pastoris expression vector in frame with sequences encoding the yeast α-factor secretion peptide and a His6 tag located at the C terminus. After introduction of each construct into the Pichia genome under the control of a methanol-inducible promoter, multicopy transformants were screened in small cultures to select transformants that exhibited satisfactory levels of production by the analysis of each culture supernatant by means of SDS-PAGE. Both recombinant proteins were successfully detected in the supernatant after induction, indicating correct processing of the α-factor signal sequence. SDS-PAGE analysis showed that expression yields of PaGluc131A and its truncated derivative, PaGluc131AΔCBM, reached 1.8 and 1.4 g per liter of culture, respectively (data not shown). Only traces of endogenous proteins were detected in the culture supernatants of transformants (data not shown). After purification, the two recombinant proteins were analyzed by SDS-PAGE for assessment of their apparent molecular masses (Fig. 3). They displayed apparent molecular masses that were higher than the theoretical ones, especially for PaGluc131A, which possesses several O-glycosylation sites in the linker region. PaGluc131AΔCBM appeared to have a minor band of lower molecular mass (Fig. 3), probably due to cleavage or a different pattern of O-glycosylation. The experimental pI in both cases revealed single isoforms, in good agreement with the theoretical pI, as PaGluc131A displayed a pI of 5.93 (theoretical pI, 5.46) and PaGluc131AΔCBM displayed a pI of 4.16 (theoretical pI, 5.25).
Fig 3.

SDS-PAGE analysis of PaGluc131A constructs. Purified recombinant proteins were loaded onto a 12% Tris-glycine SDS-PAGE gel and were stained with Coomassie blue. Lane 1, molecular mass markers (in kDa); lane 2, PaGluc131AΔCBM1 (6 μg); lane 3, PaGluc131A (8 μg).
Kinetic properties of PaGluc131A.
Using a library of plant-, bacterium-, and fungus-derived polysaccharides (cellulose, mannan, xylan, chitin, pectin, β-glucan, xyloglucan, galactan, laminarin, curdlan, pachyman, and pustulan), we searched for enzymatic activity of PaGluc131A and PaGluc131AΔCBM. Although PaGluc131AΔCBM displayed no enzymatic activity under all conditions tested, PaGluc131A released soluble sugars from a wide range of beta-glucan substrates, including laminarin, curdlan, pachyman, lichenan, pustulan, and cellulose derivatives (CMC, HEC, Avicel, and Walseth cellulose).
PaGluc131A displayed an apparent optimum pH of 5.5 and an apparent optimum temperature of 40°C, using laminarin as the substrate (data not shown). PaGluc131A was stable at 40°C, with a residual activity of >80% after 48 h. No effect of EDTA on enzymatic activity was observed (data not shown). At pH 5.5 and 40°C, PaGluc131A displayed similar activities toward β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans, with the highest specific activity obtained using laminarin (Table 1). No hydrolysis of xylan, mannan, arabinan, arabinogalactan, chitin, pectin, or xyloglucan was observed, even after extended incubation. To a lesser extent, PaGluc131A displayed activity toward pNP-β-d-glucopyranoside, pNP-β-d-xylopyranoside, and pNP-β-d-cellobiose. However, no activity was detected on pNP-β-d-lactose, pNP-β-d-saccharose, pNP-α-d-galactopyranoside, and pNP-α-l-arabinofuranoside.
Table 1.
Specific activities of PaGluc131A on substrates from different sources
| Substrate | Sp act (U mg−1)a | Relative activity (%) |
|---|---|---|
| β-(1,3)-Linked substrates | ||
| Laminarin [algal β-d-(1,3;1,6)-glucan] | 84.3 ± 1.4 | 100 |
| Curdlan [fungal β-d-(1,3)-glucan] | 19.1 ± 0.8 | 22.7 |
| Pachyman [bacterial β-d-(1,3)-glucan] | 8.1 ± 1.5 | 9.6 |
| β-(1,3;1,4)-Linked substrates | ||
| Barley β-glucan [ β-d-(1,3;1,4)-glucan; 1:3 ratio] | 13.3 ± 1.8 | 15.8 |
| Lichenan [ β-d-(1,3;1,4)-glucan; 1:2 ratio] | 4.0 ± 0.7 | 4.7 |
| β-(1,4)-Linked substrates | ||
| Carboxymethyl cellulose (CMC) | 15.2 ± 2.7 | 18.0 |
| Hydroxyethyl cellulose (HEC) | 6.3 ± 0.3 | 7.5 |
| Avicel PH101 (crystalline cellulose) | 2.0 ± 0.4 | 2.4 |
| Walseth cellulose (swollen cellulose) | 2.0 ± 0.1 | 2.4 |
| β-(1,6)-Linked substrate | ||
| Pustulan [plant β-d-(1,6)-glucan] | 13.2 ± 0.7 | 15.7 |
| Synthetic sugars | ||
| pNP-β-d-glucopyranoside | 4.3 ± 0.0 | 5.1 |
| pNP-β-d-xylopyranoside | 0.03 ± 0.00 | 0.04 |
| pNP-β-d-cellobiose | 0.0035 ± 0.0005 | 0.004 |
Specific activities were determined as described in Materials and Methods. Values are means for triplicate independent measures, and standard errors of the means are also given.
To evaluate the mode of action of PaGluc131A, the soluble sugars generated upon hydrolysis of either cellulose derivatives, β-(1,3)-glucans, β-(1,3;1,4)-glucans, β-(1,6)-glucans, or oligosaccharides with different types of linkages were analyzed by anion-exchange chromatography (Fig. 4; Table 2). Analysis of the products formed upon hydrolysis of polymeric substrates showed striking differences depending on the type of linkages of the β-glucan substrates (Fig. 4). Hydrolysis of β-(1,3)-glucans (laminarin, curdlan, and pachyman) and β-(1,6)-glucans (pustulan) yielded glucose as a unique end product. No oligosaccharides were detected even at the initial rate of the reaction. However, upon hydrolysis of β-(1,4)-glucans (CMC, HEC, Walseth cellulose, and Avicel), PaGluc131A yielded a mixture of oligosaccharides, i.e., G1 to G6, toward the end of the reaction (Fig. 4). Moreover, hydrolysis of β-(1,3;1,4)-glucans (barley β-glucan and lichenan) yielded a mixture of gluco-oligosaccharides and glucose.
Fig 4.
HPAEC analysis of end products released by PaGluc131A. The hydrolysis was performed with 88 μg of PaGluc131A for 20 h at 40°C. The dashed line represents a control experiment without enzyme. (A) Laminarin; (B) pustulan; (C) CMC; (D) barley-β-glucan. G1, glucose; G2, cellobiose; G3, cellotriose; G4, cellotetraose; G5, cellopentaose; G6, cellohexaose; L2, laminaribiose; G3B, Glc-β1-4-Glc-β1-3-Glc-OH; G4A, Glc-β1-3-Glc-β1-4-Glc-β1-4-Glc-OH; G4C, Glc-β1-4-Glc-β1-3-Glc-β1-4-Glc-OH.
Table 2.
Specificity and catalytic efficiencies of PaGluc131A on gluco-oligosaccharides
| Substrate | kcat/Km (s−1 M−1)a | Major product(s) | Minor product |
|---|---|---|---|
| β-(1,3)-Linked substrates | |||
| Laminarin | 12.6 | G1 | |
| Laminarihexaose (L6) | 2.42 | G1 | |
| Laminaripentaose (L5) | 2.54 | G1 | |
| Laminaritetraose (L4) | 3.28 | G1 | |
| Laminaritriose (L3) | 7.97 | G1 | |
| Laminaribiose (L2) | 0.19 | G1 | |
| β-(1,3;1,4)-Linked substrates | |||
| G3-G4-G4-G-OH (G4A) | 0.59 | G1, G3 | G2 |
| G4-G4-G3-G-OH (G4B) | ND | ||
| G4-G3-G4-G-OH (G4C) | ND | ||
| G3-G4-G-OH (G3A) | 0.72 | G1, G2 | L2 |
| G4-G3-G-OH (G3B) | ND | ||
| β-(1,4)-Linked substrates | |||
| Cellohexaose (G6) | 2.95 | G2, G4 | G3 |
| Cellopentaose (G5) | 0.53 | G3, G2 | |
| Cellotetraose (G4) | 0.055 | G2 | |
| Cellotriose (G3) | ND | ||
| Cellobiose (G2) | ND | ||
| β-(1,6)-Linked substrates | |||
| Pustulan | 45.4 | G1 | |
| Gentiobiose | ND |
ND, no activity detected.
PaGluc131A also showed different hydrolytic patterns toward β-(1,3)- and β-(1,4)-linked gluco-oligosaccharides. β-(1,3)-Gluco-oligosaccharides (L2 to L6) were all hydrolyzed by PaGluc131A, releasing glucose as the main product. Initial rate data from hydrolysis of L2 to L6 indicated that the catalytic efficiencies (kcat/Km) were similar, with the highest being obtained using laminaritriose (L3), at 7.97 s−1 M−1 (Table 2). Catalytic efficiencies measured on β-(1,3)-gluco-oligosaccharides were on the same order of magnitude as that on laminarin (i.e., kcat/Km of 12.6 s−1 M−1). Cello-oligosaccharides (G4 to G6) were also hydrolyzed efficiently by PaGluc131A. G6 was hydrolyzed to produce G4 and G2, with trace amounts of G3, and G5 was hydrolyzed to form exclusively G3 and G2. G4 was hydrolyzed at a lower rate, producing exclusively G2, while G3 was not hydrolyzed. Initial rate data from hydrolysis of G4, G5, and G6 indicated that the catalytic efficiency increased with increasing chain length, since the respective ratio of kcat/Km values was 1:10:54 (Table 2). A range of β-(1,3;1,4) oligosaccharides (i.e., G3A, G3B, G4A, G4B, and G4C) originating from barley β-glucan were also tested as substrates. Only G4A and G3A, which possess a β-(1,3) linkage at the nonreducing end, were hydrolyzed by PaGluc131A, yielding mainly glucose together with G3 and G2, respectively (Table 2). Although pustulan [β-(1,6)-glucan] was efficiently hydrolyzed by PaGluc131A, with a catalytic efficiency of 45.4 s−1 M−1, the corresponding dimeric form (gentiobiose) was not hydrolyzed even with high enzyme loading.
DISCUSSION
β-(1,3;1,4)-Glucans consist of unbranched and unsubstituted chains of β-(1,3)- and β-(1,4)-glucosyl residues, in which the ratio of β-(1,4)-glucosyl residues to β-(1,3)-glucosyl residues is quite variable (4). This ratio combined with the distribution of the linkages appears to influence the physicochemical properties of polysaccharides and their functional properties in cell walls. β-(1,3;1,4)-Glucans are widely distributed as noncellulosic matrix-phase polysaccharides in cell walls of grasses and commercially important cereal species. β-(1,3;1,4)-Glucans have also been detected in fungi, where they exhibit distinctive linkage characteristics (11). Fungal β-d-glucans are also found linked to chitin (linear polymer of β-1,4-N-acetylglucosamine), through the nonreducing end of the β-(1,3)-glucan chain (15). Some fungi and bacteria also contain β-(1,6)-glucans that are minor components of the cell wall.
Fungi produce a wide range of β-(1,3)- and/or β-(1,4)-glucanases that act in cooperation. β-(1,3)- and β-(1,4)-glucanases are differentiated according to their mode of action (endo- or exo-type glucanase) and the category of substrate hydrolyzed (23). Enzymes hydrolyzing β-(1,3)-glucans, found in the GH9 and GH16 families, have been termed laminarinases (EC 3.2.1.6), since laminarin was used as the test substrate. Glucan endo-β-1,3-glucosidases (EC 3.2.1.39) found in the GH16, GH17, GH55, GH81, and GH128 families act on β-(1,3)-glucosidic linkages in β-(1,3)-glucans (e.g., laminarin and pachyman). They have very limited action on mixed-linkage (1,3;1,4)-β-d-glucans. Glucan β-(1,3)-glucosidases or exo-β-(1,3)-glucosidases (EC 3.2.1.58) found in the GH3, GH5, GH17, and GH55 families catalyze the successive hydrolysis of β-d-glucose units from the nonreducing ends of β-(1,3)-glucans, releasing α-glucose. Licheninases (EC 3.2.1.73) found in the GH5, GH7, GH12, GH16, and GH17 families act on β-(1,4)-glucosidic linkages in β-d-glucans containing β-(1,3) and β-(1,4) bonds (e.g., lichenin and cereal β-d-glucans).
In the present study, we characterized a glucanase from P. anserina that exhibits broad specificity with respect to linkage position in β-glucan substrates (Table 1). PaGluc131A hydrolyzed noncellulosic β-(1,3)- and β-(1,6)-glucans as a glucan exo-β-glucosidase and hydrolyzed cellulosic β-(1,4)-glucans as an endo-β-glucanase. Hydrolysis of short β-(1,3), β-(1,4), and β-(1,3)/β-(1,4) gluco-oligosaccharides confirmed this striking feature and revealed that the enzyme was performing in an exo-type mode on the nonreducing end of β-(1,3)-gluco-oligosaccharides. PaGluc131A displayed comparable specific activities with characterized β-glucanases from the GH3 (27), GH5 (21), GH12 (28), GH16 (6), GH55 (14, 20), and GH128 (26) families but appeared to behave differently in terms of substrate specificity. The substrate specificity of PaGluc131A could be related to that of family GH3 plant β-glucanases that remove single glucosyl residues from the nonreducing termini of a broad range of β-d-linked glucosidic substrates (12, 13), but they hydrolyze all types of beta-glucans [including β-(1,4)-glucans] through an exohydrolytic mechanism of action.
To our knowledge, our study is the first report of an enzyme that displays bifunctional exo-β-(1,3)/(1,6) and endo-β-(1,4) activities toward beta-glucans and therefore cannot readily be assigned to existing Enzyme Commission groups. The amino acid sequence of PaGluc131A has high sequence identity to hypothetical proteins in the fungal taxa and defines a new family of GHs in the CAZy database, namely, the GH131 family. Phylogenetic analysis of the GH131 family shows that PaGluc131A belongs to a group of sequences that contain a CBM1 module (except for that of C. thermophilum), which may explain why excision of the CBM1 domain resulted in an inactive enzyme on all substrates tested.
We have shown that the gene is functionally transcribed, since we detected its expression using several natural substrates. The protein was also identified in the secretome of P. anserina by means of proteomics following induction using sugar beet pulp (L. Poidevin et al., unpublished results). Moreover, we also detected another member of this new GH family in the secretome of Aspergillus niger (GenBank accession no. AM270168) (9). The relatively broad specificity of PaGluc131A raises questions regarding its function when secreted by P. anserina. The results of the present study emphasize that P. anserina can depend on different cell wall components from recalcitrant plant materials and from bacteria, yeast, and fungi. This highlights the high environmental pressure on evolution as well as the high level of specialization that occurs in the fungal kingdom. Together with our previous study (8), the present findings give more insights into P. anserina's enzymatic machinery for the deconstruction of plant cell wall polysaccharides.
ACKNOWLEDGMENTS
This work was funded by the French National Research Agency (ANR) (E-TRICEL grant ANR-07-BIOE-006).
We are grateful to P. Silar for advice on the P. anserina culture method, J. Féliu for technical assistance, N. Lopes-Ferreira for providing Walseth cellulose, and W. Helbert for providing pustulan. We thank P. M. Coutinho and B. Henrissat for fruitful discussions.
Footnotes
Published ahead of print 28 September 2012
REFERENCES
- 1. Berrin JG, et al. 2000. High-level production of recombinant fungal endo-beta-1,4-xylanase in the methylotrophic yeast Pichia pastoris. Protein Expr. Purif. 19:179–187 [DOI] [PubMed] [Google Scholar]
- 2. Berrin JG, Ajandouz EH, Georis J, Arnaut F, Juge N. 2007. Substrate and product hydrolysis specificity in family 11 glycoside hydrolases: an analysis of Penicillium funiculosum and Penicillium griseofulvum xylanases. Appl. Microbiol. Biotechnol. 74:1001–1010 [DOI] [PubMed] [Google Scholar]
- 3. Bonnina E, et al. 2001. Aspergillus niger I-1472 and Pycnoporus cinnabarinus MUCL39533, selected for the biotransformation of ferulic acid to vanillin, are also able to produce cell wall polysaccharide-enzymes and feruloyl esterases. Enzyme Microb. Technol. 28:70–80 [DOI] [PubMed] [Google Scholar]
- 4. Burton RA, Fincher GB. 2009. (1,3;1,4)-Beta-d-glucans in cell walls of the Poaceae, lower plants, and fungi: a tale of two linkages. Mol. Plant 2:873–882 [DOI] [PubMed] [Google Scholar]
- 5. Cantarel BL, et al. 2009. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, et al. 2012. High-level expression of a novel Penicillium endo-1,3(4)-β-d-glucanase with high specific activity in Pichia pastoris. J. Ind. Microbiol. Biotechnol. 39:869–876 [DOI] [PubMed] [Google Scholar]
- 7. Coutinho PM, et al. 2009. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet. Biol. 46:S161–S169 [DOI] [PubMed] [Google Scholar]
- 8. Couturier M, et al. 2011. Podospora anserina hemicellulases potentiate the Trichoderma reesei secretome for saccharification of lignocellulosic biomass. Appl. Environ. Microbiol. 77:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Couturier M, et al. 2012. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genomics 13:57 doi:10.1186/1471-2164-13-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Espagne E, et al. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fontaine T, et al. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 8:27594–27607 [DOI] [PubMed] [Google Scholar]
- 12. Hrmova M, Fincher GB. 1998. Barley beta-d-glucan exohydrolases. Substrate specificity and kinetics properties. Carbohydr. Res. 305:209–221 [Google Scholar]
- 13. Hrmova M, Fincher GB. 2007. Dissecting the catalytic mechanism of a plant beta-d-glucan glucohydrolase through structural biology using inhibitors and substrate analogues. Carbohydr. Res. 342:1613–1623 [DOI] [PubMed] [Google Scholar]
- 14. Ishida T, et al. 2009. Crystal structure of glycoside hydrolase family 55 beta-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 284:10100–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kollár R, et al. 1997. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→)3-glucan, and chitin. J. Biol. Chem. 272:17762–17775 [DOI] [PubMed] [Google Scholar]
- 16. Matsui I, et al. 1991. Subsite structure of Saccharomycopsis alpha-amylase secreted from Saccharomyces cerevisiae. J. Biochem. 109:566–569 [DOI] [PubMed] [Google Scholar]
- 17. Nakatani Y, Lamont IL, Cutfield JF. 2010. Discovery and characterization of a distinctive exo-1,3/1,4-β-glucanase from the marine bacterium Pseudoalteromonas sp. strain BB1. Appl. Environ. Microbiol. 76:6760–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Navarro D, et al. 2010. Automated assay for screening the enzymatic release of reducing sugars from micronized biomass. Microb. Cell Fact. 9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]
- 20. Nobe R, et al. 2003. Purification and characterization of laminaran hydrolases from Trichoderma viride. Biosci. Biotechnol. Biochem. 67:1349–1357 [DOI] [PubMed] [Google Scholar]
- 21. O'Connell E, Piggott C, Tuohy M. 2011. Purification of exo-1,3-beta-glucanase, a new extracellular glucanolytic enzyme from Talaromyces emersonii. Appl. Microbiol. Biotechnol. 89:685–696 [DOI] [PubMed] [Google Scholar]
- 22. Pitson SM, Seviour RJ, McDougall BM, Stone BA, Sadek M. 1996. Purification and characterization of an extracellular (1→6)-beta-glucanase from the filamentous fungus Acremonium persicinum. Biochem. J. 316:841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Planas A. 2000. Bacterial 1,3-1,4-beta-glucanases: structure, function and protein engineering. Biochim. Biophys. Acta 1543:361–382 [DOI] [PubMed] [Google Scholar]
- 24. Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 9:945–967 [Google Scholar]
- 25. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto Y, Nakade K, Konno N. 2011. Endo-β-1,3-glucanase GLU1, from the fruiting body of Lentinula edodes, belongs to a new glycoside hydrolase family. Appl. Environ. Microbiol. 77:8350–8354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahashi M, Konishi T, Takeda T. 2011. Biochemical characterization of Magnaporthe oryzae β-glucosidases for efficient β-glucan hydrolysis. Appl. Microbiol. Biotechnol. 91:1073–1082 [DOI] [PubMed] [Google Scholar]
- 28. Takeda T, et al. 2010. Characterization of endo-1,3-1,4-β-glucanases in GH family 12 from Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 88:1113–1123 [DOI] [PubMed] [Google Scholar]
- 29. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]