Abstract
The recently described bacterium “Candidatus Methylomirabilis oxyfera” couples the oxidation of the important greenhouse gas methane to the reduction of nitrite. The ecological significance of “Ca. Methylomirabilis oxyfera” is still underexplored, as our ability to identify the presence of this bacterium is thus far limited to DNA-based techniques. Here, we investigated the lipid composition of “Ca. Methylomirabilis oxyfera” to identify new, gene-independent biomarkers for the environmental detection of this bacterium. Multiple “Ca. Methylomirabilis oxyfera” enrichment cultures were investigated. In all cultures, the lipid profile was dominated up to 46% by the fatty acid (FA) 10-methylhexadecanoic acid (10MeC16:0). Furthermore, a unique FA was identified that has not been reported elsewhere: the monounsaturated 10-methylhexadecenoic acid with a double bond at the Δ7 position (10MeC16:1Δ7), which comprised up to 10% of the total FA profile. We propose that the typical branched fatty acids 10MeC16:0 and 10MeC16:1Δ7 are key and characteristic components of the lipid profile of “Ca. Methylomirabilis oxyfera.” The successful detection of these fatty acids in a peatland from which one of the enrichment cultures originated supports the potential of these unique lipids as biomarkers for the process of nitrite-dependent methane oxidation in the environment.
INTRODUCTION
Methane (CH4) represents globally the second most important greenhouse gas. Methane contributes approximately 20% to the total greenhouse gas budget, with a global warming potential which is about 25 times as strong as that of CO2 (29). Understanding the sources and sinks of CH4 is paramount to enabling the development of adequate management strategies that aim to mitigate greenhouse gas emissions and global warming. Methane is one of the least reactive organic molecules, and it was long assumed that methane could only be oxidized aerobically, i.e., with the use of oxygen. However, in recent decades ample evidence has been collected for the occurrence of methane oxidation in anoxic environments. In marine systems, distributions of CH4 and sulfate in the water column and in sediments first led to the awareness of significant CH4 consumption in anoxic zones (38, 44). This resulted in the identification of anaerobic oxidation of methane (AOM) coupled to sulfate reduction, and since its first documentation, consortia of archaea and sulfate-reducing bacteria responsible for this process have been studied at many locations (3, 25, 27, 39, 40).
Besides oxygen and sulfate, nitrate or nitrite could theoretically also serve as a suitable, energetically favorable electron acceptor for the oxidation of CH4. The actual occurrence of AOM coupled to the reduction of nitrate and/or nitrite, however, remained elusive for a long time and was only discovered in 2006 (42). At that time, the methane-oxidizing enrichment culture still comprised both bacteria and archaea, which supported the hypothesis that, similar to what had been found for sulfate-dependent methane oxidation, a bacterial-archaeal consortium accounted for the nitrite-dependent methane-oxidizing activity. Only later did the archaea disappear from the culture (20), possibly influenced by higher nitrite loads (28). Subsequent metagenomic analysis resulted in the complete genome assembly of the bacterium responsible for the nitrite-dependent methane oxidation (19). Despite its anaerobic lifestyle, this microbe is thought to employ an intra-aerobic pathway for methane oxidation by producing its own oxygen from the dismutation of nitric oxide, for which reason it was named “Candidatus Methylomirabilis oxyfera” (19).
Human activities not only have led to increased atmospheric methane concentrations but also continue to have a major impact on the global nitrogen cycle. Industrial and agricultural intensification have led to a substantial increase in nitrogen loadings in present-day freshwater and coastal marine environments, leading to environmental problems like eutrophication. Nitrite-dependent methane oxidation thus could comprise an important link between the carbon and nitrogen cycle in various ecosystems, improving the total greenhouse gas balance while alleviating disturbed nitrogen budgets. However, the environmental contribution of “Ca. Methylomirabilis oxyfera” remains relatively unexplored. So far, our ability to rapidly identify “Ca. Methylomirabilis oxyfera” in the environment relies on molecular techniques (13, 21, 37). Specific primers targeting 16S rRNA and functional genes (i.e., methane mono-oxygenase, pmoA) have been developed for the environmental detection of “Ca. Methylomirabilis oxyfera,” but these may capture only a selection of organisms potentially contributing to nitrite-dependent methane oxidation (13, 21, 37).
In addition to genomic approaches, lipid analyses are regularly used to study microbial processes and communities, including the methane cycle. Especially in combination with stable carbon isotope signatures, lipid biomarkers have been successfully used to demonstrate methanotrophy (coupled to oxygen or sulfate reduction) in various ecosystems (2, 8, 9, 18, 22, 25, 30, 40, 43, 46, 47, 50, 52, 54).
The earliest enrichment culture that was shown to be capable of nitrite-dependent methane oxidation was also screened for its lipid composition (42). However, at that time only a single enrichment culture was available, in which the microbial community still comprised substantial levels of archaea (10 to 15%), which was also confirmed by the presence of archaeol. At present, multiple cultures have been enriched from different environments, the genome of the responsible bacterium has been assembled, archaea members are virtually absent, and no other species comprise a significant part of the community besides “Ca. Methylomirabilis oxyfera” (35). Therefore, here we have investigated the potential for new biomarkers for “Ca. Methylomirabilis oxyfera” based on its lipid composition. From multiple enrichment cultures of “Ca. Methylomirabilis oxyfera” originating from different ecosystems, we determined the typical lipid profile and identified characteristic compounds that may serve as biomarkers for the detection of “Ca. Methylomirabilis oxyfera”-like bacteria in the environment. In addition, we investigated environmental samples from a peatland from which one of the “Ca. Methylomirabilis oxyfera” enrichment cultures had originally been obtained to test the validity of the potential new lipid biomarkers.
MATERIALS AND METHODS
Sample description.
Biomass was obtained from multiple enrichment cultures of “Ca. Methylomirabilis oxyfera” and a related, unnamed “Ca. Methylomirabilis” species at Radboud University Nijmegen that originated from different environments. The first enrichment culture capable of nitrite-dependent methane oxidation was enriched from the freshwater canal Twentekanaal in the Netherlands (TWK) (42). Since then, several additional cultures have been enriched from different Dutch ecosystems which are examined here: three enrichment cultures originating from sediment from a freshwater ditch in Ooijpolder (Ooij1, Ooij2, and Ooij3) (19, 21); one culture enriched from sludge from a wastewater treatment plant (WWTP) (36); and one, the most recent, culture enriched from the peatland Brunssummerheide (BRH), with a dominant strain different from “Ca. Methylomirabilis oxyfera” but within the genus “Ca. Methylomirabilis” (59). The enrichment cultures were grown and kept under constant temperature conditions, i.e., 25, 30, 20 to 23, and 25°C for TWK, Ooij (1, 2, and 3), WWTP, and BRH, respectively.
All cultures actively oxidized CH4 anaerobically with nitrite at the moment of sampling. The most recent enrichment culture, BRH, was sampled over the time of its development (along with increasing CH4 oxidizing activity) at 11, 14, and 17 months after inoculation. The other cultures had reached and maintained a more or less constant level of activity and enrichment already prior to the first sampling, and they were sampled in two or three replicates over time coinciding with the sampling moments of BRH. Molecular analyses with specific primers targeting 16S rRNA and pmoA genes of NC10 phylum bacteria (21, 37) had confirmed the presence of “Ca. Methylomirabilis” in all enrichment cultures studied here. The degree of enrichment of the cultures was checked by fluorescence in situ hybridization (FISH) analyses with specific probes (42).
In addition to the enrichment culture biomass, samples were taken from the peatland where the inoculum of the most recently enriched culture originated from BRH. A soil profile was sampled from 51- to 102-cm depth, divided into nine sections of 5 to 9 cm (i.e., 51 to 60 cm, 5-cm increments from 60 to 95 cm, and 95 to 102 cm). In this section of the soil profile, a countergradient of methane and nitrate and a peak of abundance of “Ca. Methylomirabilis oxyfera”-like bacteria (as measured by quantitative PCR [qPCR]) were observed (59).
Lipid analyses.
Typically, 50 to 100 ml of the enrichment cultures was harvested and centrifuged, after which the obtained pellets were freeze-dried. For the environmental samples, approximately 5 g of field moist soil per section of the obtained soil core was taken and freeze-dried. The freeze-dried samples were stored dry and frozen (−20°C) until further use. Subsamples of the freeze-dried material was saponified with 1N potassium hydroxide (KOH) in methanol (MeOH; 96%) to analyze free and ester-bound lipids. The obtained extracts were methylated with boron trifluoride (BF3) in MeOH and subsequently separated by column chromatography over activated alumina (Al2O3) into apolar and polar fractions with dichloromethane (DCM) and DCM-MeOH (1:1, vol/vol) as the eluent, respectively. The polar fractions were subsequently silylated using bis(trimethylsilyl)trifluoroacetamide (BSTFA) in pyridine at 60°C for 20 min. An aliquot of the apolar fractions was separated into a saturated and an unsaturated fraction by column chromatography over Ag+-impregnated silica with DCM and ethyl acetate (EtOAc) as the eluent, respectively. Subsequently, an aliquot of the unsaturated fraction (in EtOAc) was hydrogenated using platinum oxide (PtO2) with a droplet of acetic acid, flushed with hydrogen for 2 h, and stirred overnight. The remainder of the unsaturated apolar fractions was dissolved in hexane and derivatized with dimethyl-disulfide (DMDS) (activated with iodine in diethyl ether at 40°C overnight) to determine the position of the double bond(s) (5, 53).
The total apolar and polar fractions were analyzed by gas chromatography (GC) and subsequently by gas chromatography-mass spectrometry (GC-MS). The fractions after separation of the saturated and unsaturated fraction and after hydrogenation and DMDS adduction were also analyzed by GC and GC-MS. Relative abundance of the fatty acid methyl esters was derived from the (integrated) GC profile of the apolar fraction.
RESULTS
Enrichment cultures.
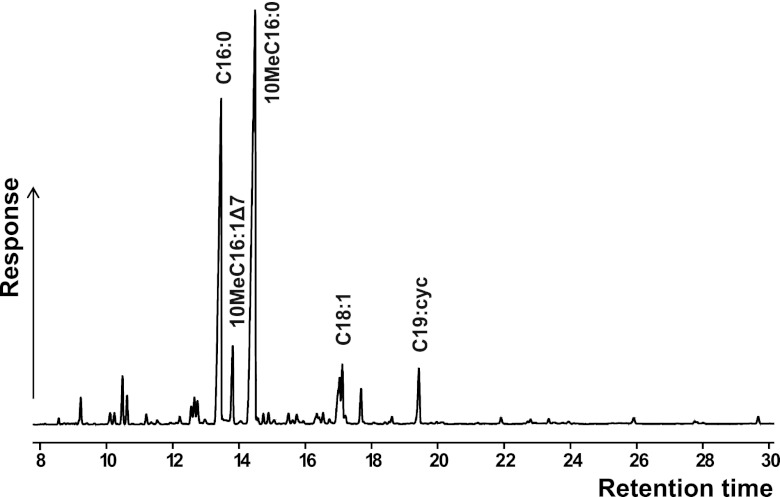
Figure 1 shows a representative gas chromatogram of the apolar fraction of “Ca. Methylomirabilis oxyfera” culture Ooij1. The apolar fraction comprised the majority of the total lipids, as analyses of the polar fraction did not reveal any other major lipids (data not shown). The lipids were identified by GC-MS analyses and comparison to established spectra of known compounds. The lipid profile was dominated by relatively short-chain fatty acids (FA) (C14 to C19). The most abundant FA was 10-methyl-hexadecanoic acid (10MeC16:0), followed by hexadecanoic acid (C16:0). In addition, GC-MS analyses revealed the presence of a previously unknown component, which eluted at a retention time between those of C16:0 and 10MeC16:0.
Fig 1.
Gas chromatogram of the apolar lipid fraction of the enrichment culture of the nitrite-dependent methane oxidizer “Ca. Methylomirabilis oxyfera” Ooij1. Major fatty acids are indicated; cyc denotes a cyclopropyl moiety.
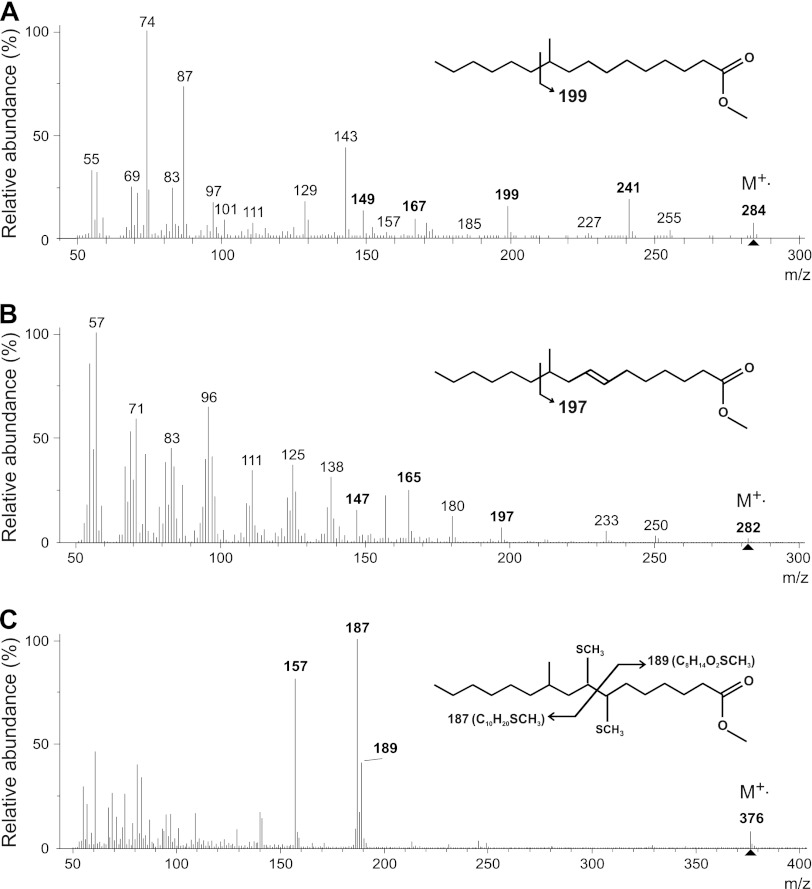
Figure 2 presents the mass spectra of the most abundant FA of the lipid profile, identified as a 10MeC16:0 (48) and the unknown compound observed in “Ca. Methylomirabilis oxyfera” enrichment lipid extracts. The spectrum of the unknown compound (Fig. 2B) showed a molecular ion at m/z 282, suggesting a monounsaturated C17 fatty acid. This compound was indeed recovered in the unsaturated fraction, and after hydrogenation it was converted to 10MeC16:0 FA, as confirmed by GC-MS. The mass spectrum of the unknown component is consistent with monounsaturated 10MeC16 FA: diagnostic fragment ions were observed from the preferential cleavage between the 10th and 11th C atom of the acyl chain at m/z 197, 165, and 147, all shifted by two Daltons relative to the equivalent fragment ions in the mass spectrum of its saturated counterpart, 10MeC16:0 FA (showing diagnostic fragment ions at m/z 199, 167, and 149, respectively). This identified the position of the double bond between the carboxyl carbon (C1) and the methyl group (at C10). GC-MS analyses after dimethyl-disulfide (DMDS) adduction resulted in diagnostic fragments of m/z 187, 189, and 157, representing ωC10, ΔC7, and ΔC7-32 fragments, respectively (Fig. 2C), thus locating the double bond at the Δ7 position. Consequently, this unknown FA was identified as 10-methyl-hexadec-7-enoic acid, or 10MeC16:1Δ7.
Fig 2.
Mass spectra of the fatty acids (FAs) 10MeC16:0 (A) and 10MeC16:1Δ7 (B) (analyzed as methyl ester derivatives), two key components of the lipid profile of enrichment cultures of Methylomirabilis oxyfera. The 10MeC16:1Δ7 FA thus far has been reported only in these enrichment cultures. (C) The position of the double bond in the 10MeC16:1Δ7 FA was deduced from the mass spectrum of the FA methyl ester after DMDS adduction.
The lipid profile of “Ca. Methylomirabilis oxyfera” enrichment cultures Ooij2, Ooij3, and WWTP was similar overall to the distribution described for Ooij1. Based on FISH analyses of the total microbial community, these enrichment cultures were found to be dominated by “Ca. Methylomirabilis oxyfera” by approximately 70 to 80% without any other single species making up a significant amount of the bacterial community (35). The relative abundances of the major FAs in the different enrichment cultures are given in Table 1. The most abundant FAs, 10MeC16:0 and C16:0, comprised approximately 35 to 46% and 18 to 26% of the total fatty acids, respectively. The unique FA 10MeC16:1Δ7 made up 4 to 10% of the total FA profile. Repeated sampling over time of a single enrichment culture showed only minor changes in the FA profile.
Table 1.
Approximate degree of enrichment in “Ca. Methylomirabilis oxyfera” and relative abundance of fatty acids in the lipid profile of the “Ca. Methylomirabilis oxyfera” enrichment culturesf
| Enrichment culture | % enrichment in “Ca. Methylomirabilis oxyfera”a | Relative abundance (%) of: |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | 10MeC14:0 | isoC15 | aiC15 | isoC16 | C16:1tot | C16:0 | 10MeC16:1Δ7 | 10MeC16:0 | C18:1tot | C18:0 | C19:cyc | C20:0 | C22:0 | C24:0 | C26:0 | C28:0 | Sum | Branchede | ||
| Ooij1b | 70–80 | 0.8 | 0.4 | 1.8 | 1.0 | 1.1 | 2.5 | 26.2 | 4.2 | 45.9 | 6.2 | 1.9 | 5.2 | 97.2 | 56.0 | |||||
| Ooij2b | 70–80 | 0.6 | 0.8 | 2.3 | 1.0 | 3.9 | 2.0 | 19.0 | 6.6 | 45.6 | 6.7 | 2.3 | 5.8 | 96.6 | 62.3 | |||||
| Ooij3b | 70–80 | 0.5 | 0.7 | 1.3 | 2.4 | 2.6 | 3.2 | 17.8 | 7.3 | 34.9 | 4.8 | 3.0 | 18.0 | 96.6 | 50.9 | |||||
| WWTPb | 70–80 | 1.5 | 0.9 | 2.3 | 1.5 | 4.3 | 5.8 | 21.1 | 10.1 | 37.8 | 8.6 | 1.4 | 2.3 | 97.6 | 58.3 | |||||
| TWKc | 0.8 | 2.3 | 6.8 | 11.4 | 5.5 | 28.9 | 14.3 | 2.8 | 12.7 | 85.5 | 42.9 | |||||||||
| BRH 11 md | 15–25 | 1.0 | 0.8 | 0.5 | 13.2 | 8.9 | 3.4 | 9.4 | 38.6 | 3.1 | 4.1 | 3.2 | 2.2 | 2.6 | 2.6 | 1.9 | 95.6 | 15.8 | ||
| BRH 14 md | 45–55 | 1.0 | 1.0 | 1.3 | 1.1 | 0.5 | 16.6 | 14.9 | 9.4 | 21.9 | 19.4 | 2.2 | 2.2 | 1.4 | 0.9 | 1.1 | 1.1 | 0.7 | 96.5 | 36.4 |
| BRH 17 md | 65–75 | 0.8 | 0.7 | 0.7 | 0.7 | 14.0 | 19.0 | 10.5 | 27.4 | 14.5 | 3.1 | 2.2 | 0.9 | 0.5 | 95.5 | 42.7 | ||||
Approximate enrichment of the culture based on FISH analyses.
Averaged data of replicate lipid analyses: n = 3 for Ooij1 and Ooij3 and n = 2 for Ooij2 and WWTP.
Data are from Raghoebarsing et al. (42).
11 m, 14 m, and 17 m denote 11, 14, and 17 months of enrichment of the BRH culture, respectively.
Branched FA as a percentage of the sum of FA (in the preceding column).
Compounds comprising <0.5% of the total FA are omitted.
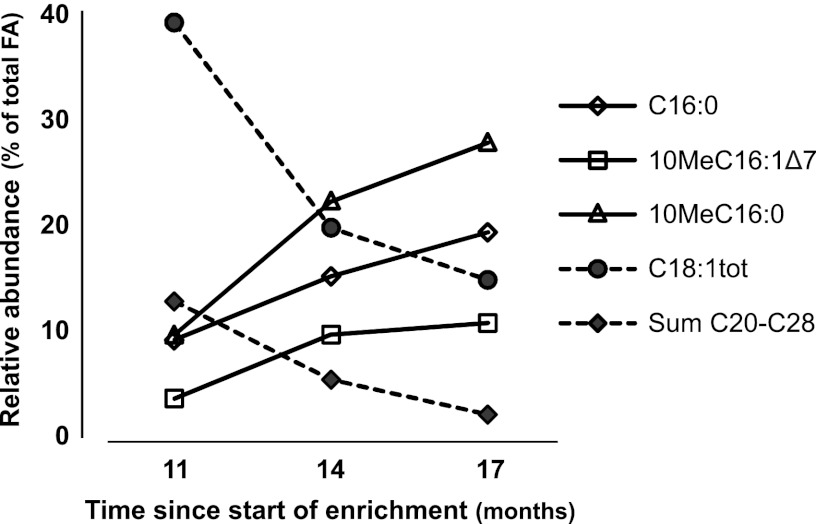
In contrast to these stable enrichments, distinct changes in the profile over time were observed for the BRH culture that was sampled over the time course of enrichment (Table 1 and Fig. 3). Sampling over time coincided with increased activity and degree of enrichment of the culture. FISH analyses estimated the abundance of “Ca. Methylomirabilis” in the culture to be approximately 15 to 25, 45 to 55, and 65 to 75% after 11, 14, and 17 months of enrichment, respectively (Table 1). At the earliest sampling moment after 11 months of enrichment, the lipid profile from the BRH reactor contained considerable amounts of monounsaturated octadecenoic acids (C18:1; 38.6%) and longer saturated even-carbon-number FAs (C20:0 to C28:0; totaling 12.5%). These FAs likely are derived from plant tissue (e.g., plant waxes or sphingolipids) in the original sample from the environment (16). At later sampling times, these FAs had become much less abundant (totaling 14.5 and 1.9%, respectively, after 17 months), and the relative contribution of the typical FA 10MeC16:0 and 10MeC16:1Δ7 had increased from 9.4 and 3.4% after 11 months of enrichment to 27.4 and 10.5% after 17 months, respectively (Table 1 and Fig. 3).
Fig 3.
Changes over time in the relative contribution of key FAs of Methylomirabilis enrichment culture BRH. During the depicted period since the start of enrichment, the degree of enrichment and activity of methane oxidation coupled to nitrite reduction in the culture increased.
Environmental samples.
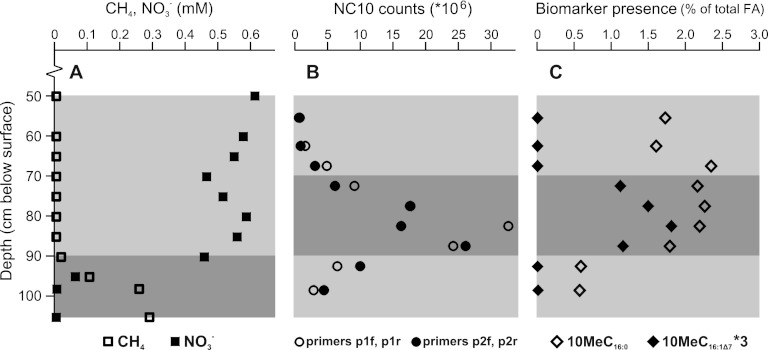
Samples from a 51- to 102-cm soil core from the Brunssummerheide peatland were examined for the presence of the FA 10MeC16:0 and 10MeC16:1Δ7. Lipid analyses revealed the presence of the FA 10MeC16:0 throughout the 51- to 102-cm soil profile (Fig. 4). The FA 10MeC16:1Δ7 was identified in the sections from 70 to 90 cm. At their highest concentrations, the 10MeC16:0 and 10MeC16:1Δ7 FA comprised approximately 2.2 and 0.5% of the total fatty acids, respectively (Fig. 4). Relative to the C16:0 FA, the 10MeC16:0 and 10MeC16:1Δ7 FA were present at approximately 10 and 3% of the amount of C16:0. Together with the characteristic FA 10MeC16:0 and 10MeC16:1Δ7, all other major FA observed in the lipid profile of “Ca. Methylomirabilis oxyfera” enrichment cultures were also present throughout the 50- to 100-cm soil profile.
Fig 4.
Profile of the proposed biomarker lipids together with cell numbers of Methylomirabilis and methane and nitrate concentrations in a peatland (Brunssummerheide, the Netherlands). (A) Methane (CH4) and nitrate (NO3−) concentrations throughout the profile (June 2010). (B) Cell numbers per g wet soil obtained by qPCR analysis with two independent primer sets targeting the 16S rRNA gene of “Ca. Methylomirabilis”-like bacteria (data are from Zhu et al. [59]). (C) Relative abundance of 10MeC16:0 and 10MeC16:1Δ7 as percentages of the total fatty acids recovered from the apolar fraction after lipid extraction (note that values for 10MeC16:1Δ7 are multiplied by three) (data are from this study). The darker gray bars indicate the methane-nitrate countergradient (A), and just above that is the peak in NC10 abundance (B) and the peak in abundance of the proposed biomarker lipids (C).
DISCUSSION
The two C17 fatty acids 10MeC16:0 and 10MeC16:1Δ7 characterize the lipid profile of the “Ca. Methylomirabilis oxyfera” enrichment cultures. To the best of our knowledge, the latter so far has only been observed in “Ca. Methylomirabilis oxyfera” enrichment cultures. One other unsaturated 10MeC16:1 has been reported before (10MeC16:1Δ8 [2]), but the nitrite-dependent “Ca. Methylomirabilis” bacteria provide the only account of 10MeC16:1Δ7. Interestingly, lipid analyses of the first available nitrite-dependent methane-oxidizing enrichment culture (TWK) already showed these remarkable lipids (42). However, at that time the microbial community in the culture still comprised substantial levels of archaea (10 to 15%), which was confirmed by the presence of the typical archaeol. At present, multiple cultures have been enriched from different ecosystems, and after prolonged enrichment these cultures have become 70 to 80% enriched in “Ca. Methylomirabilis oxyfera,” while archaea are virtually absent (confirmed by FISH analyses and the absence of archaeol) and no other species comprises a significant part of the community besides “Ca. Methylomirabilis oxyfera” (35).
Our new data reveal the presence of the characteristic fatty acids 10MeC16:0 and 10MeC16:1Δ7 in all investigated “Ca. Methylomirabilis” enrichment cultures. Moreover, we could successfully detect both FA in environmental samples from a field site from which one of the enrichment cultures originated (Fig. 4). In the stable enrichment cultures, these two fatty acids together make up more than 40% of the total lipids (Table 1). The change over time in the fatty acid composition of the BRH enrichment culture shows that both 10MeC16:0 and 10MeC16:1Δ7 became more abundant while the abundance of C18:1 and the longer-chain fatty acids decreased (Table 1 and Fig. 3). This reflects the increased enrichment of the culture and demonstrates that both of these FA are key constituents of the lipid profile of “Ca. Methylomirabilis oxyfera.” This is further supported by the observation that, over the time of development, the ratio of the abundance of 10MeC16:1Δ7 FA to 10MeC16:0 FA remains more or less the same. Intrinsically, analyses of enrichment culture biomass rather than pure cultures will not provide the pure lipid profile of the organism of interest but will always include lipids of the members of the side community. We believe, however, that the abundance of the fatty acids 10MeC16:0 and 10MeC16:1Δ7 in all enrichment cultures from different origins and their increase over the time of development clearly show that the anaerobic nitrite-reducing methanotroph “Ca. Methylomirabilis” is the most likely source organism of these fatty acids.
As temperature can affect the degree of saturation of fatty acids (10), the question arises of whether the unprecedented 10MeC16:1Δ7 will be a significant fatty acid of “Ca. Methylomirabilis” under all environmental conditions. The setup of our study did not allow us to thoroughly test this for “Ca. Methylomirabilis oxyfera.” However, the ratio of saturated to unsaturated FA is generally thought to increase with temperature. As our enrichment temperatures are relatively high compared to those in the environment, the relative abundance of the novel unsaturated 10MeC16:1Δ7 FA is more likely to be higher rather than lower in the environment than in our enrichment cultures.
Its high abundance denotes that the 10MeC16:0 fatty acid is a characteristic component of the “Ca. Methylomirabilis oxyfera” lipids, but it is not exclusive to “Ca. Methylomirabilis oxyfera.” 10MeC16:0 FA has been proposed to be characteristic of sulfate-reducing bacteria (SRB) of the genera Desulfobacter and Desulfobacula, where it is found to make up various amounts of approximately 5 to 25% of the total lipids (4, 15, 32, 33, 45, 51). Besides these sulfate reducers, 10MeC16:0 FA is also occasionally, but generally in much smaller amounts, found in other bacteria; the literature reports its presence in several actinobacteria (6, 12, 23, 31, 41, 56, 58), in anammox bacteria and other planctomycetes (48, 49), in an iron-reducing Geobacter species (34, 57), in a Marinobacter species (55), and in the marine denitrifier Pseudomonas nautica (14). Several studies report the presence of 10MeC16:0 FA in relation to the occurrence of anaerobic methane oxidation (AOM) coupled to sulfate reduction (1, 2, 7, 17, 26, 39). One study on the methane-oxidizing community in landfill cover soils reports remarkably large amounts of 10MeC16:0 FA of up to 16% of the total extracted phospholipid fatty acids. However, this distinct fatty acid was not discussed to be characteristic for the (methane-oxidizing) bacterial population, despite the fact that this 10MeC16:0 FA became significantly enriched in 13C after incubation with 13C-CH4 (11). Based on the high abundance of 10MeC16:0 FA in “Ca. Methylomirabilis oxyfera” identified in our study, we speculate that the presence and 13C labeling of this FA observed in the aforementioned study is indicative of the involvement of “Ca. Methylomirabilis”-like microorganisms in those ecosystems.
The association of the signature lipid 10MeC16:0 with both sulfate- and nitrite-dependent AOM may obscure the diagnostic value of this FA as a biomarker for either bacterial process. In studies where the presence of this FA is used to substantiate the involvement of sulfate reducers in methane oxidation, a potential role of “Ca. Methylomirabilis oxyfera”-like bacteria thus far has gone unnoticed. This would not pose a major concern as long as the two processes are thought to occur in separated environments. AOM coupled to sulfate reduction has mainly been studied in marine ecosystems, while all “Ca. Methylomirabilis oxyfera” enrichment cultures enriched thus far have been from freshwater areas. Only one study has reported the potential presence of “Ca. Methylomirabilis oxyfera”-like bacteria in marine sediments based on genomic analyses, but this was only based on 16S rRNA reads and no similarity with functional genes (for methane mono-oxygenase) of “Ca. Methylomirabilis oxyfera” could be found in the metagenome (24). The contribution of “Ca. Methylomirabilis oxyfera” to methane oxidation in a nitrite/nitrate-limited and sulfate-rich environment may be insignificant and vice versa. However, some ecosystems may be suitable for the cooccurrence of both types of AOM, for example, estuaries that are influenced by sulfate-rich seawater and that simultaneously experience high nitrogen inputs from river water.
Fortunately, the unprecedented finding of the fatty acid 10MeC16:1Δ7 in “Ca. Methylomirabilis” offers a unique biomarker for these methane-oxidizing bacteria. However, the likely low abundance of “Ca. Methylomirabilis oxyfera” in a complex microbial community may complicate the detection of this single FA 10MeC16:1Δ7 in environmental samples. Nevertheless, we were able to successfully detect both of the FA 10MeC16:0 and 10MeC16:1Δ7 in environmental samples. The observed presence of the characteristic lipids in the field samples corresponds with qPCR analyses on the same soil core: abundance of NC10 phylum bacteria (to which “Ca. Methylomirabilis” belongs) based on qPCR data and the presence of the 10MeC16:0 and 10MeC16:1Δ7 FA both peak between 80 and 90 cm depth in the soil profile. Correspondingly, in this zone countergradients of methane and nitrate are observed, indicating the activity of anaerobic methanotrophs (Fig. 4) (59). The 10MeC16:0 and 10MeC16:1Δ7 FA made up approximately 2.2 and 0.5% of the total FA in the environmental samples, which, compared to their quantities in enrichment cultures (35 to 46% and 4 to 10%, respectively), could represent an approximate maximum abundance of “Ca. Methylomirabilis” of about 4 to 5% of the community. This is in good accordance with the share of NC10 phylum bacteria in the total bacterial community of 3 to 8% at a depth of 80 to 90 cm as assessed by qPCR (59). We propose that the combined search for the unique FA 10MeC16:1Δ7 and the foremost abundant 10MeC16:0 FA provide a strong tool to study the presence and significance of “Ca. Methylomirabilis oxyfera”-like bacteria in the environment.
Conclusion.
The characteristic fatty acids 10MeC16:0 and 10MeC16:1Δ7 may offer a useful tool in studying “Ca. Methylomirabilis oxyfera” and the process of nitrite-dependent methane oxidation in the environment. The successful detection of these fatty acids in samples from the ecosystem from which one of the “Ca. Methylomirabilis oxyfera” enrichment cultures originated validates the potential of these lipids as biomarkers. These new lipid biomarkers can complement genomic techniques to study the diverse players in the methane cycle.
ACKNOWLEDGMENTS
We thank Francisca Luesken and Mingliang Wu for access to the enrichment culture biomass and Gijs van Dijk, Christian Fritz, and Alfons Smolders for collaboration on the Brunssummerheide samples.
This research was funded by a personal Rubicon grant to D.M.K. from the Netherlands Science Foundation (NOW; project 825.10.018) and the Darwin Centre for Biogeosciences in support of D.M.K. and K.F.E. (projects 142.16.3071 and 142.16.3072, publication number DW-2012-1006). B.Z. is financially supported by a CAS-KNAW grant and M.S.M.J. by the ERC (grant 232937).
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Alain K, et al. 2006. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ. Microbiol. 8:574–590 [DOI] [PubMed] [Google Scholar]
- 2. Blumenberg M, Seifert R, Reitner J, Pape T, Michaelis W. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. U. S. A. 101:11111–11116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boetius A, et al. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626 [DOI] [PubMed] [Google Scholar]
- 4. Bühring SI, Elvert M, Witte U. 2005. The microbial community structure of different permeable sandy sediments characterized by the investigation of bacterial fatty acids and fluorescence in situ hybridization. Environ. Microbiol. 7:281–293 [DOI] [PubMed] [Google Scholar]
- 5. Buser HR, Arn H, Guerin P, Rauscher S. 1983. Determination of double-bond position in mono-unsaturated acetates by mass-spectrometry of dimethyl disulfide adducts. Anal. Chem. 55:818–822 [Google Scholar]
- 6. Campbell IM, Naworal J. 1969. Composition of saturated and mono-unsaturated fatty acids of Mycobacterium phlei. J. Lipid Res. 10:593–598 [PubMed] [Google Scholar]
- 7. Chevalier N, et al. 2011. Authigenic carbonates at cold seeps in the Marmara Sea (Turkey): a lipid biomarker and stable carbon and oxygen isotope investigation. Mar. Geol. 288:112–121 [Google Scholar]
- 8. Collister JW, Summons RE, Lichtfouse E, Hayes JM. 1992. An isotopic biochemical study of the green river oil-shale. Org. Geochem. 19:265–276 [DOI] [PubMed] [Google Scholar]
- 9. Coolen MJL, et al. 2004. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change. Org. Geochem. 35:1151–1167 [Google Scholar]
- 10. Cronan JE. 1975. Thermal regulation of membrane lipid-composition of Escherichia coli–evidence for direct control of fatty-acid synthesis. J. Biol. Chem. 250:7074–7077 [PubMed] [Google Scholar]
- 11. Crossman ZM, Abraham F, Evershed RP. 2004. Stable isotope pulse-chasing and compound specific stable carbon isotope analysis of phospholipid fatty acids to assess methane oxidizing bacterial populations in landfill cover soils. Environ. Sci. Technol. 38:1359–1367 [DOI] [PubMed] [Google Scholar]
- 12. Dastager SG, Lee J-C, Ju Y-J, Park D-J, Kim C-J. 2008. Nocardioides koreensis sp nov., Nocardioides bigeumensis sp nov and Nocardioides agariphilus sp nov., isolated from soil from Bigeum Island, Korea. Int. J. Syst. Evol. Microbiol. 58:2292–2296 [DOI] [PubMed] [Google Scholar]
- 13. Deutzmann JS, Schink B. 2011. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl. Environ. Microbiol. 77:4429–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doumenq P, et al. 1999. Changes in fatty acids of Pseudomonas nautica, a marine denitrifying bacterium, in response to n-eicosane as carbon source and various culture conditions. FEMS Microbiol. Ecol. 28:151–161 [Google Scholar]
- 15. Dowling NJE, Widdel F, White DC. 1986. Phospholipid ester-linked fatty-acid biomarkers of acetate-oxidizing sulfate-reducers and other sulfide-forming bacteria. J. Gen. Microbiol. 132:1815–1825 [Google Scholar]
- 16. Eglinton G, Hamilton RJ. 1967. Leaf epicuticular waxes. Science 156:1322–1335 [DOI] [PubMed] [Google Scholar]
- 17. Elvert M, Boetius A, Knittel K, Jorgensen BB. 2003. Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane. Geomicrobiol. J. 20:403–419 [Google Scholar]
- 18. Elvert M, Suess E, Whiticar MJ. 1999. Anaerobic methane oxidation associated with marine gas hydrates: superlight C-isotopes from saturated and unsaturated C-20 and C-25 irregular isoprenoids. Naturwissenschaften 86:295–300 [Google Scholar]
- 19. Ettwig KF, et al. 2010. Nitrate-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 20. Ettwig KF, et al. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164–3173 [DOI] [PubMed] [Google Scholar]
- 21. Ettwig KF, Van Alen T, Van de Pas-Schoonen KT, Jetten MSM, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeman KH, Hayes JM, Trendel JM, Albrecht P. 1990. Evidence from carbon isotope measurements for diverse origins of sedimentary hydrocarbons. Nature 343:254–256 [DOI] [PubMed] [Google Scholar]
- 23. Gonzalez I, Ayuso-Sacido A, Anderson A, Genilloud O. 2005. Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54:401–415 [DOI] [PubMed] [Google Scholar]
- 24. Havelsrud OE, Haverkamp THA, Kristensen T, Jakobsen KS, Rike AG. 2011. A metagenomic study of methanotrophic microorganisms in Coal Oil Point seep sediments. BMC Microbiol. 11:221 doi:10.1186/1471-2180-11-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805 [DOI] [PubMed] [Google Scholar]
- 26. Hinrichs KU, Summons RE, Orphan V, Sylva SP, Hayes JM. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685–1701 [Google Scholar]
- 27. Hoehler TM, Alperin MJ, Albert DB, Martens CS. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Glob. Biogeochem. Cycle 8:451–463 [Google Scholar]
- 28. Hu S, Zeng RJ, Keller J, Lant PA, Yuan Z. 2011. Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ. Microbiol. Rep. 3:315–319 [DOI] [PubMed] [Google Scholar]
- 29. IPCC 2007. Climate change 2007: the physical science basis. Contribution of Working Group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 30. Jahnke LL, Summons RE, Dowling LM, Zahiralis KD. 1995. Identification of methanotrophic lipid biomarkers in cold-seep mussel gills: chemical and isotopic analysis. Appl. Environ. Microbiol. 61:576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kampfer P, Kroppenstedt RM. 2004. Pseudonocardia benzenivorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:749–751 [DOI] [PubMed] [Google Scholar]
- 32. Kuever J, Könneke M, Galushko A, Drzyzga O. 2001. Reclassification of Desulfobacterium phenolicum as Desulfobacula phenolica comb. nov. and description of strain Sax(T) as Desulfotignum balticum gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 51:171–177 [DOI] [PubMed] [Google Scholar]
- 33. Londry KL, Jahnke LL, Marais DJD. 2004. Stable carbon isotope ratios of lipid biomarkers of sulfate-reducing bacteria. Appl. Environ. Microbiol. 70:745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lovley DR, et al. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 35. Luesken FA, et al. 2011. A metagenomics-based in situ detection and phylogenetic composition of an enrichment culture that performs nitrite-driven anaerobic methane oxidation, p 43–57 Applied aspects of nitrite-dependent methane oxidation. Ph.D. thesis Radboud University Nijmegen, Nijmegen, the Netherlands [Google Scholar]
- 36. Luesken FA, et al. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. 92:845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luesken FA, et al. 2011. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murray JW, Grundmanis V, Smethie WM. 1978. Interstitial water chemistry in sediments of Saanich Inlet. Geochim. Cosmochim. Acta 42:1011–1026 [Google Scholar]
- 39. Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487 [DOI] [PubMed] [Google Scholar]
- 40. Pancost RD, Sinninghe Damsté JS, De Lint S, Van der Maarel MJEC, Gottschal JC. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl. Environ. Microbiol. 66:1126–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park SW, Park ST, Lee JE, Kim YM. 2008. Pseudonocardia carboxydivorans sp. nov., a carbon monoxide-oxidizing actinomycete, and an emended description of the genus Pseudonocardia. Int. J. Syst. Evol. Microbiol. 58:2475–2478 [DOI] [PubMed] [Google Scholar]
- 42. Raghoebarsing AA, et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 43. Raghoebarsing AA, et al. 2005. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436:1153–1156 [DOI] [PubMed] [Google Scholar]
- 44. Reeburgh WS. 1980. Anaerobic methane oxidation-rate depth distributions in Skan Bay sediments. Earth Planet. Sci. Lett. 47:345–352 [Google Scholar]
- 45. Rütters H, Sass H, Cypionka H, Rullkötter J. 2002. Phospholipid analysis as a tool to study complex microbial communities in marine sediments. J. Microbiol. Methods 48:149–160 [DOI] [PubMed] [Google Scholar]
- 46. Schubert CJ, et al. 2006. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ. Microbiol. 8:1844–1856 [DOI] [PubMed] [Google Scholar]
- 47. Schubert CJ, et al. 2011. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol. Ecol. 76:26–38 [DOI] [PubMed] [Google Scholar]
- 48. Sinninghe Damsté JS, Rijpstra WIC, Geenevasen JAJ, Strous M, Jetten MSM. 2005. Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox). FEBS J. 272:4270–4283 [DOI] [PubMed] [Google Scholar]
- 49. Sittig M, Schlesner H. 1993. Chemotaxonomic investigation of various prosthecate and or budding bacteria. Syst. Appl. Microbiol. 16:92–103 [Google Scholar]
- 50. Spooner N, et al. 1994. Stable carbon isotopic correlation of individual biolipids in aquatic organisms and a lake bottom sediment. Org. Geochem. 21:823–827 [Google Scholar]
- 51. Taylor J, Parkes RJ. 1983. The cellular fatty-acids of the sulfate-reducing bacteria, Desulfobacter sp., Desulfobulbus sp. and Desulfovibrio desulfuricans. J. Gen. Microbiol. 129:3303–3309 [Google Scholar]
- 52. Thiel V, et al. 1999. Highly isotopically depleted isoprenoids: Molecular markers for ancient methane venting. Geochim. Cosmochim. Acta 63:3959–3966 [Google Scholar]
- 53. Vincenti M, Guglielmetti G, Cassani G, Tonini C. 1987. Determination of double-bond position in di-unsaturated compounds by mass-spectrometry of dimethyl disulfide derivatives. Anal. Chem. 59:694–699 [Google Scholar]
- 54. Wakeham SG, Lewis CM, Hopmans EC, Schouten S, Sinninghe Damsté JS. 2003. Archaea mediate anaerobic oxidation of methane in deep euxinic waters of the Black Sea. Geochim. Cosmochim. Acta 67:1359–1374 [Google Scholar]
- 55. Yoon J-H, Lee M-H, Kang S-J, Oh T-K. 2007. Marinobacter salicampi sp. nov., isolated from a marine solar saltern in Korea. Int. J. Syst. Evol. Microbiol. 57:2102–2105 [DOI] [PubMed] [Google Scholar]
- 56. Yoon J-H, et al. 2000. Rhodococcus koreensis sp. nov., a 2,4-dinitrophenol-degrading bacterium. Int. J. Syst. Evol. Microbiol. 50:1193–1201 [DOI] [PubMed] [Google Scholar]
- 57. Zhang CLL, et al. 2003. Carbon isotope signatures of fatty acids in Geobacter metallireducens and Shewanella algae. Chem. Geol. 195:17–28 [Google Scholar]
- 58. Zhao G-Z, et al. 2011. Pseudonocardia serianimatus sp. nov., a novel actinomycete isolated from the surface-sterilized leaves of Artemisia annua L. Antonie Van Leeuwenhoek 100:521–528 [DOI] [PubMed] [Google Scholar]
- 59. Zhu B, et al. 2012. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl. Environ. Microbiol. 78:8657–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]