Abstract
Staphylococcus aureus clonal complex 398 (CC398) isolates colonize livestock and can spread to human contacts. Genetic analysis of isolates epidemiologically associated with human-to-human, but not livestock, transmission in multiple countries and continents identified a common clade that was negative for tet(M) and positive for bacteriophage ϕ3. Another group of human-to-human-transmitted isolates belonged to the common livestock-associated clade but had acquired a unique ϕ7 bacteriophage.
TEXT
Staphylococcus aureus is a commensal organism and opportunistic pathogen of humans and animals. Methicillin-resistant S. aureus (MRSA) is particularly difficult to control, and major clones are prevalent in hospital and community reservoirs (1, 2). Livestock-associated MRSA (LA-MRSA) is an emerging problem, particularly with pig farming in Europe, North America, and Asia (5, 7, 15, 23, 28). Infection in livestock is rare, but transmission to humans and subsequent infection is of public health concern (4).
The most prevalent lineage of S. aureus in livestock is clonal complex 398 (CC398) (9). Several studies have identified variants of CC398, particularly variants associated with different hosts and geography (11, 14, 17). Most of the variation is in the mobile genetic element (MGE) content, including resistance and virulence genes carried on bacteriophage, S. aureus pathogenicity islands (SaPIs), plasmids, and transposons. These studies have predominantly included CC398 isolates from livestock and from humans with livestock contact.
Uhlemann et al. (19, 20) recently identified a unique set of CC398 isolates from patients in New York and the Dominican Republic with no documented livestock contact, which is strong evidence of human-human transmission, The isolates formed a genetically distinct clade and varied from LC-CC398 in carriage of MGEs and functional surface proteins. The aim of this study was to identify additional isolates of CC398 associated with human-to-human transmission from multiple countries and continents and to identify universal genetic markers of human-to-human transmission by using whole-genome analysis.
In total, 110 CC398 isolates were compared (Table 1). Isolates from humans without pig contact (n = 31; 12 MRSA and 19 methicillin-sensitive S. aureus [MSSA] isolates) came from five studies from The Netherlands (21), Belgium (24), New York/Dominican Republic/Martinique (20), Belgium (unpublished data), and Denmark (unpublished data). Control isolates from pigs (n = 50; 45 MRSA and 5 MSSA) and from humans with pig contact (n = 29; 26 MRSA and 3 MSSA) were matched from similar times and geographical locations (11), including both infection and colonizing isolates and MRSA and MSSA isolates.
Table 1.
CC398 isolates analyzed by microarray
| Host | Country | No. of CC398 colonization isolatesa | No. of CC398 infection isolatesa |
|---|---|---|---|
| Human with pig contact (H) | Belgium | 2 (2) | 4 (4) |
| Denmark | 9 (7) | 3 (3) | |
| The Netherlands | 4 (3) | 7 (7) | |
| Pig (P) | Belgium | 6 (6) | 0 |
| Denmark | 22 (19) | 3 (1) | |
| The Netherlands | 13 (13) | 0 | |
| Italy | 3 (3) | 0 | |
| Canada | 3 (3) | 0 | |
| Human without pig contact (HWP) | Belgium | 1 (0) | 7 (0) |
| Denmark | 8 (8) | 2 (2) | |
| The Netherlands | 2 (0) | 3 (0) | |
| USA/Dominican Republic/Martinique | 4 (0) | 4 (0) |
Values in parentheses are the number of MRSA isolates.
Pig contact was defined as direct or known contact with pigs at the time of sampling; however, 2 individuals from Denmark reported peripheral pig contact (Fig. 1). The details for these individuals include the following: one individual has two stepchildren who have contact with their biological father who works with pigs, and the other individual worked with pigs until 1 month before sampling.
Fig 1.
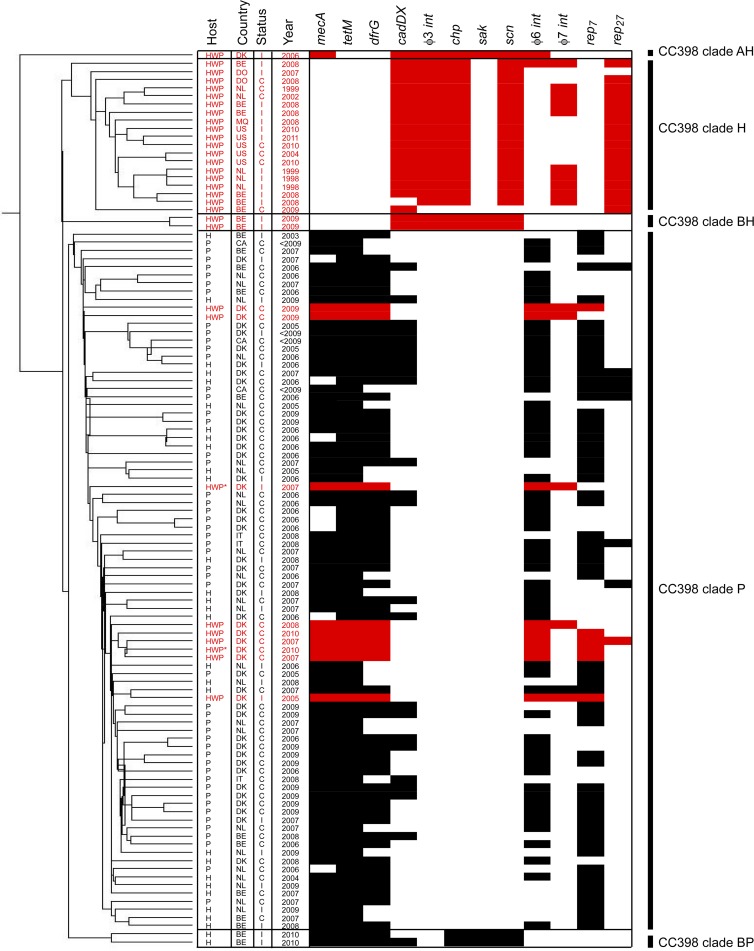
Microarray clustering analysis of CC398 isolates based on 11,715 60-mer oligonucleotides representing MGE genes. The clustering on the left shows the evolutionary relatedness of the isolates. For each isolate, the host origin (human with pig contact = H, pig = P, and human without pig contact = HWP), country origin (Belgium = BE, Canada = CA, Denmark = DK, Dominican Republic = DO, Italy = IT, Martinique = MQ, The Netherlands = NL, and United States = US), status (colonization = C and infection = I), and year of isolation is shown. Isolates from humans without pig contact are highlighted in red, while isolates from pigs and humans with pig contact are shown in black. Multiple MGEs varied between isolates, and representative genes of those MGEs with the strongest association with a clade are shown as present (black/red boxes) or absent (white boxes) Each MGE gene was significantly associated (P < 0.05, χ2 test) with either the major pig-associated CC398 clade (clade P) or the major human-specific CC398 clade (clade H). Two humans without pig contact may have had previous exposure to reservoirs of pig bacteria; they are denoted with an asterisk (HWP*).
Comparative genomics analysis using a 62-strain S. aureus microarray (SAM-62) was performed as previously described (11). SAM-62 contains 29,739 60-mer oligonucleotide probes, representing 6,520 genes, and an additional 579 gene variants, from the first 62 sequenced S. aureus genomes and from 153 sequenced plasmid genomes. The array design is available in BμG@Sbase (accession number A-BUGS-38 [http://bugs.sgul.ac.uk/A-BUGS-38]) and ArrayExpress (accession number A-BUGS-38). All analyses were performed using GeneSpring GX v11.0 (Agilent Technologies).
Independent clades of S. aureus CC398.
Five distinct clades of CC398 were generated by clustering based on 11,715 60-mer oligonucleotide probes that represent genes found in all MGEs (Fig. 1). A highly consistent tree was also generated by clustering based on 10,386 60-mer oligonucleotide probes representing genes found in all MGEs except for SCCmec elements (data not shown).
All CC398 isolates originating from pigs (n = 50; both MRSA and MSSA), as well as the majority of isolates from humans in contact with pigs (n = 27 of 29), were closely related and formed the pig clade (clade P). This clade represents the well-described pig-associated population of CC398, consistent with reports of transient human colonization and occasional opportunistic infections in humans (11, 22).
CC398 isolates associated with human-to-human transmission were present in four different clades. Belgian, Dutch, North American, and Caribbean MSSA CC398 isolates (n = 19) from humans without pig contact were closely related and formed a distinct clade (clade H). This clade is separate from the pig-associated clade (clade P). Clade H therefore appears to represent human-specific populations of CC398 found in both Europe and the Americas. Two Belgian isolates from humans without pig contact clustered separately to form clade BH, and a Chinese clade of Panton-Valentine leukocidin (PVL)-positive MRSA CC398 that has been previously described formed clade AH (17).
Our findings for pig and human CC398 clades are consistent with a concurrent whole-genome sequencing study of 89 S. aureus CC398 isolates, which suggests a human origin of the original CC398 lineage (14), followed by divergent evolution of the clades.
It is notable that both major CC398 clades have crossed the Atlantic and continued to evolve independently but maintained their host specificity.
Genetic markers of the human-specific CC398 clade H.
S. aureus CC398 is easily detected by using the restriction-modification (RM) test (16) and PCR detection (26). The majority of isolates in clade H, as well as 2 isolates from clade P, were spa t571, indicating that spa type was not a clade marker. This is consistent with a previous study (14). Therefore, to identify genetic markers of the distinct CC398 clades, we compared the distribution of MGE genes between the major human-specific clade (clade H; n = 19) and major pig-associated clade (clade P; n = 86).
The best genetic marker for assigning an isolate to clade H was the ϕ3 bacteriophage (Fig. 1). This element was found in 94.7% of clade H but 0% of clade P isolates (χ2 = 81.47; P < 0.001). ϕ3 in clade H carried the human-specific immune evasion cluster (IEC) genes chp and scn, but not sak (18, 25). ϕ3 was also found in human-specific isolates from clades BH and AH; however, these isolates carried ϕ3 with chp, scn, and sak. sak may additionally be a suitable marker for differentiating clade H from clades AH and BH.
Other genetic markers of human-specific CC398 were cadDX and rep27. cadDX was found in 95.73% of the human-specific population (clade H) but only 25.58% of the pig-associated population (clade P) (χ2 = 19.53; P < 0.001), while plasmid replication gene rep27 was present in 94.70% of the human-specific population (clade H) and in 9.30% of the pig-associated population (clade P) (χ2 = 45.87; P < 0.001). PVL genes were present in the Chinese isolate of clade AH, as well as all isolates in clade BH and clade BP.
Genetic markers of the pig-associated CC398 clade P.
The best genetic marker of the pig-associated clade was the tet(M) gene encoding tetracycline resistance, present in 100% of clade P and 0% of clade H (χ2 = 19.00; P < 0.001). Other resistance genes were common in clade P but largely absent among clade H, including mec(A) (χ2 = 17.23; P < 0.001) and dfr(G) (χ2 = 15.24; P < 0.001), encoding methicillin and trimethoprim resistance, respectively. Multiple SCCmec types were found in clade P. These findings agree with a report that SCCmec type does not correlate with clade (14).
Other genetic markers of clade P were the previously described plasmid replication gene rep7 (χ2 = 14.14; P < 0.001) and ϕ6 (χ2 = 12.24; P < 0.01) (6, 10, 12). The ϕ6 bacteriophage is present in the sequenced genome of a human CC398 isolate (S0385, clade P), and in the genomes of cow (RF122; CC151) and sheep (ED133 and O11 and O46; CC133 and CC1173, respectively) isolates (12).
Human-to-human transmission of the pig-associated clade P.
Danish MRSA CC398 isolates from humans without pig contact (n = 9) clustered into clade P, suggesting that MRSA CC398 from clade P may have adapted to human-to-human transmission in Denmark (Fig. 1). These isolates carried the genes typical of clade P isolates [such as tet(M), dfr(G), rep7, and ϕ6) but surprisingly did not carry the ϕ3 bacteriophage or any of the human-specific IEC genes.
ϕ7 was carried in 5/9 (55.5%) of the Danish isolates from humans without pig contact and was only present in one other pig-associated isolate (Fig. 1). Importantly, ϕ7 was also present in 10/11 Belgian and Dutch isolates from the human-specific clade H. It is therefore possible that ϕ7 of CC398 in Europe may have transferred horizontally to clade P and contributed to human adaptation. It may also be a suitable marker of human-to-human transmission capability in clade P isolates. Only one fully annotated ϕ7 bacteriophage sequence of human origin is in the public domain (Newman strain, lineage CC8), but no human specificity factor has been characterized. Sequencing of the ϕ7 bacteriophage is under way, and preliminary studies indicate that this bacteriophage is highly unstable during laboratory culture.
Conclusions.
This study showed S. aureus CC398 isolates associated with human-to-human transmission belong to an independently evolving clade H that is distinct from the well-described pig-associated clade P. This suggests clades are host specific, but there is also a clear difference in resistance gene carriage between clades H and P; mec(A), dfr(G), and tet(M) are common in the pig-associated clade but rare among the human-specific clades. It cannot be ruled out that methicillin and tetracycline resistance may be responsible for selection of clade P in pig populations, reflecting the higher use of these antibiotics in pigs compared to human populations (13).
Each S. aureus CC398 clade possessed a unique MGE profile. Therefore, the presence of specific MGEs is a potentially powerful tool for rapidly determining the epidemiological origin of CC398 isolates. This is supported by recent studies that demonstrated MGEs can appear at different frequencies in different host populations (11) and that MGE profiling can identify close epidemiological relationships (8). The best genetic marker of human-human transmission of CC398 was ϕ3, while the best genetic marker of the livestock-associated CC398 was tet(M).
The prospect that clade P isolates can evolve to successfully transfer from human to human is alarming. Currently, clade P MRSA CC398 from patients with livestock contact appears to have reduced transmissibility in hospitals compared to typical hospital-acquired MRSA (27). An expanding host range of LA-MRSA would elevate its significance for human health, potentially increasing the incidence of human infection and encouraging continued evolution to increased virulence and resistance. In countries with intense pig farming, MRSA CC398 from clade P successfully adapted for human-to-human transmission would necessitate the implementation of costly controls to reduce the MRSA incidence in livestock production (3).
Microarray data accession number.
Fully annotated microarray data are available in BμG@Sbase (accession number E-BUGS-124 [http://bugs.sgul.ac.uk/E-BUGS-124]) and also ArrayExpress (accession number E-BUGS-124).
ACKNOWLEDGMENTS
We thank Jason Hinds, Kate Gould, Denise Waldron, and Adam Witney from BμG@S (the Bacterial Microarray Group at St. George's, University of London) for supply of the microarrays and advice and The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. We thank Arshnee Moodley and Luca Guardabassi (Faculty of Life Sciences, University of Copenhagen) for providing CC398 S. aureus isolates. We are thankful to Andreas Voss (Department of Medical Microbiology and Infection Control, Radboud University Nijmegen Medical Centre and Canisius-Wilhelmina Hospital) and Els Broens (Quantitative Veterinary Epidemiology Group, Wageningen Institute of Animal Sciences) for providing CC398 S. aureus isolates.
This work was supported by the PILGRIM FP7 grant from the EU.
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Chambers HF. 2005. Community-associated MRSA: resistance and virulence converge. N. Engl. J. Med. 352:1485–1487 [DOI] [PubMed] [Google Scholar]
- 2. Enright MC, et al. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. European Food Safety Authority Panel on Biological Hazards 2009. Assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA J. 993:1–73 [Google Scholar]
- 4. Fitzgerald JR. 2012. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20:192–198 [DOI] [PubMed] [Google Scholar]
- 5. Huijsdens XW, et al. 2006. Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jensen LB, et al. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80:25–43 [DOI] [PubMed] [Google Scholar]
- 7. Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 30:298–303 [DOI] [PubMed] [Google Scholar]
- 8. McCarthy AJ, Breathnach AS, Lindsay JA. 2012. Detection of mobile-genetic-element variation between colonizing and infecting hospital-associated methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 50:1073–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCarthy AJ, Lindsay JA, Loeffler A. 2012. Are all meticillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Vet. Dermatol. 23:267–275 [DOI] [PubMed] [Google Scholar]
- 10. McCarthy AJ, Lindsay JA. 2012. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 12:104 doi:10.1186/1471-2180-12-19-04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCarthy AJ, et al. 2011. The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biol. Evol. 3:1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarthy AJ, Witney AA, Lindsay JA. 2012. Staphylococcus aureus temperate bacteriophage: carriage and horizontal gene transfer is lineage associated. Front. Cell. Infect. Microbiol. 2:6 doi:10.3389/fcimb.2012.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phillips I, et al. 2004. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 53:28–52 [DOI] [PubMed] [Google Scholar]
- 14. Price LB, et al. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith TC, et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258 doi:10.1371/journal.pone.0004258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stegger M, et al. 2011. Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J. Clin. Microbiol. 49:732–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stegger M, Lindsay JA, Sørum M, Gould KA, Skov R. 2010. Genetic diversity in CC398 methicillin-resistant Staphylococcus aureus isolates of different geographical origin. Clin. Microbiol. Infect. 16:1017–1019 [DOI] [PubMed] [Google Scholar]
- 18. Sung JM, Lloyd DH, Lindsay JA. 2008. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154:1949–1959 [DOI] [PubMed] [Google Scholar]
- 19. Uhlemann AC, et al. 2011. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. Eur. J. Clin. Microbiol. Infect. Dis. 31:505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uhlemann AC, et al. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3:e00027–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Belkum A, et al. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Cleef BA, et al. 2011. Persistence of livestock-associated methicillin-resistant Staphylococcus aureus in field workers after short-term occupational exposure to pigs and veal calves. J. Clin. Microbiol. 49:1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Cleef BA, et al. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg. Infect. Dis. 17:502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vandendriessche S, Kadlec K, Schwarz S, Denis O. 2011. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincosamide-streptogramin B resistance gene erm(T) in Belgian hospitals. J. Antimicrob. Chemother. 66:2455–2459 [DOI] [PubMed] [Google Scholar]
- 25. van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Wamel WJ, et al. 2010. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur. J. Clin. Microbiol. Infect. Dis. 29:119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wassenberg MW, Bootsma MC, Troelstra A, Kluytmans JA, Bonten MJ. 2011. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin. Microbiol. Infect. 17:316–319 [DOI] [PubMed] [Google Scholar]
- 28. Wulf M, Voss A. 2008. MRSA in livestock animals: an epidemic waiting to happen? Clin. Microbiol. Infect. 14:519–521 [DOI] [PubMed] [Google Scholar]