Abstract
Iron fertilization experiments in high-nutrient, low-chlorophyll areas are known to induce phytoplankton blooms. However, little is known about the response of the microbial community upon iron fertilization. As part of the LOHAFEX experiment in the southern Atlantic Ocean, Bacteria and Archaea were monitored within and outside an induced bloom, dominated by Phaeocystis-like nanoplankton, during the 38 days of the experiment. The microbial production increased 1.6-fold (thymidine uptake) and 2.1-fold (leucine uptake), while total cell numbers increased only slightly over the course of the experiment. 454 tag pyrosequencing of partial 16S rRNA genes and catalyzed reporter deposition fluorescence in situ hybridization (CARD FISH) showed that the composition and abundance of the bacterial and archaeal community in the iron-fertilized water body were remarkably constant without development of typical bloom-related succession patterns. Members of groups usually found in phytoplankton blooms, such as Roseobacter and Gammaproteobacteria, showed no response or only a minor response to the bloom. However, sequence numbers and total cell numbers of the SAR11 and SAR86 clades increased slightly but significantly toward the end of the experiment. It seems that although microbial productivity was enhanced within the fertilized area, a succession-like response of the microbial community upon the algal bloom was averted by highly effective grazing. Only small-celled members like the SAR11 and SAR86 clades could possibly escape the grazing pressure, explaining a net increase of those clades in numbers.
INTRODUCTION
Vast areas of the ocean feature low concentrations of chlorophyll a despite their high concentrations of nutrients. These so-called “high-nutrient, low-chlorophyll” (HNLC) areas cover 25 to 30% of the world's oceans, mainly in the northern and equatorial Pacific and the Southern Ocean. Within these regions, bioavailable iron was found to be the limiting factor for algal growth (36). Since the early 1990s, several iron fertilization experiments have been conducted in HNLC areas, showing that phytoplankton blooms can be induced (37). Most of these blooms consisted of diatoms (5), but in silicate-depleted waters (e.g., during SOFeX-North) the blooms consisted mostly of Phaeocystis-like nanoplankton encompassing prymnesiophytes, pelagophytes, and dinoflagellates (5). The enhanced primary production within such blooms triggers the biological carbon pump and may lead to a transport of biomass into the deep ocean (10, 28, 56). Heterotrophic nanoplankton is potentially counteracting the net deposition of fixed carbon by mineralization of algal biomass, resulting in an immediate release of carbon dioxide back to the atmosphere. However, not much is known about the direct and indirect effects of iron fertilization on the Archaea and Bacteria. For those, iron can also be a limiting nutrient, and thus its addition might have a direct effect on their growth. For example, heterotrophic bacteria accounted for 20 to 45% of the biological iron uptake in the subarctic Pacific Ocean (60). Alternatively, microorganisms might be indirectly affected by the increased primary production of the algal blooms induced by iron fertilization (6, 21). Besides such a bottom-up control by nutrient and substrate availability, the microbial community will also be top-down controlled by viral lysis and protozoan grazing (13, 45, 46). Viruses are highly specific and change the microbial community by infecting discrete abundant members (59), while heterotrophic predators rather feed on microorganisms within a particular size range (45). Both bottom-up and top-down control mechanisms need to be considered in interpreting microbial successions during iron fertilizations.
In an earlier study, (62), the microbial succession had been observed during a phytoplankton bloom induced by natural iron fertilization on the Kerguelen Plateau, where a natural inflow of iron-rich deep waters led to a major bloom. Using 16S rRNA clone libraries and single-strand conformation polymorphism (SSCP), a community structure consisting mainly of the clades Roseobacter NAC11-7 (Alphaproteobacteria), SAR92 (Gammaproteobacteria), and Agg58 (Bacteroidetes) was found in the naturally fertilized area. This is supported by the finding of major parts of the bacterial production assigned to high-nucleic-acid-containing bacteria (HNA) (42). Outside the fertilized area, the SAR11 clade (Alphaproteobacteria) and Polaribacter (Bacteroidetes) dominated the microbial community (62). In contrast, no major changes in the microbial community composition of the fertilized patch were found by terminal restriction fragment length polymorphism (T-RFLP) analyses during the iron fertilization experiment EisenEx (where “Eisen” is German for “iron” and “Ex” is the abbreviation for “experiment”), although a few days after each of the three subsequent iron additions to the water, the microbial production was 2- to 3-fold increased (3).
Since all artificial iron fertilization experiments had so far focused on the onset of the phytoplankton bloom, the iron fertilization experiment LOHAFEX (where “loha” is Indian for “iron” and “FEX” represents “fertilization experiment”) aimed at monitoring the phytoplankton bloom in an iron-fertilized eddy until its senescence and decay, thereby closing a gap in our understanding of carbon transport. This study also allowed for more continuous monitoring of changes in the microbial diversity and community composition. The hypothesis tested in this study was that succession patterns of bacterial and archaeal populations would occur that are caused either directly or indirectly by the iron fertilization. Monitoring of the microbial community composition during the iron fertilization experiment LOHAFEX was done using 16S rRNA gene tag pyrosequencing and catalyzed reporter deposition fluorescence in situ hybridization (CARD FISH) techniques and semiautomated microscopy.
MATERIALS AND METHODS
Iron fertilization.
The iron fertilization experiment was done during RV Polarstern cruise ANT XXV/3 (12 January to 6 March 2009). A cold core eddy in the area between 47°12′S to 49°12′S and 14°00′W to 16°24″W (Fig. 1) along the polar front was chosen based on satellite imagery and in situ measurements of physical and chemical parameters (Patrick Martin, submitted for publication) for the iron fertilization experiment. In two fertilization steps, an acidified iron(II) sulfate solution was released into the water at days 0 (27 January 2009) and 18 (14 February 2009) of the experiment. We used drifter buoys, determined the photosynthetic efficiency index (Fv/Fm) of the surface water with a fast-repetition-rate fluorometer (FRRF), and measured online the concentration of the inert tracer gas SF6 (sulfur hexafluoride), which was added with the iron solution to the water to monitor the movement of the fertilized patch. At a later stage of the experiment, when enough biomass had accumulated, chlorophyll a concentrations were measured to determine the location of the fertilized patch and to define “IN” (fertilized) and “OUT” (nonfertilized) stations.
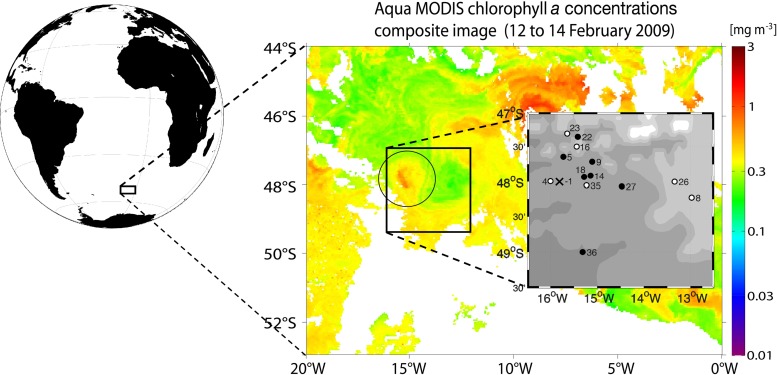
Fig 1.
Map of the fertilized area. The chlorophyll a picture shows the LOHAFEX bloom (encircled). Stations and experiment days of both the IN (black) and OUT stations (white) are shown in the small map. X marks day −1, the beginning of the experiment. The globe and the inset map were generated with the M_Map package for Matlab (version 7.12.0.635; MathWorks, Natick, MA). The chlorophyll a data were downloaded from the NASA website http://oceancolor.gsfc.nasa.gov/.
Sampling.
Samples from different depths of the wind-mixed layer (WML; from surface to depth of ∼80 m in steps of 10 m) were taken inside the fertilized patch on days −1, 5, 9, 14, 18, 22, 27, and 36 (IN stations), while OUT station samples were taken arbitrarily outside the fertilized area but inside the eddy on days −1, 4, 8, 16, 23, 26 and 35 (Fig. 1). On days −1, 9, and 16 (OUT), 18 and 36 additional samples were taken at depths of 100 m, 300 m, and 500 m (except for day 18). Water was collected using 12-liter Niskin bottles on a rosette. Depending on the cell abundance, duplicates of 20 to 100 ml of water were fixed with 1% formaldehyde final concentration and filtered on 0.22-μm-pore-size polycarbonate filters (Millipore, Eschborn, Germany) for CARD FISH analyses. These samples were stored at −20°C. In addition, samples were taken for DNA analysis and 454 tag pyrosequencing on days −1, 9, 18, and 36 by filtering 90, 85, 67, and 85 liters, respectively, of 5-μm-pore-prefiltered water on 0.22-μm-pore-size cellulose acetate filters (Sartorius, Göttingen, Germany), which were subsequently stored at −80°C.
Bacterial production.
Bacterial production rates were estimated from the measurements of [methyl-3H]thymidine (specific activity, 18,000 mCi mM−1; Amersham Corp., Amersham, England) incorporation rates (TdR) following the method described by Fuhrman and Azam (17). Samples were taken from depths of 5, 10, 20, and 50 m in three replicates of 20 ml from each depth. An aliquot of 100 μl of a 60 nM working solution of [3H]thymidine was added to each tube and incubated in the dark at 4 to 5°C for 3 h. The thymidine uptake was stopped by the addition of 300 μl formaldehyde. Cells were collected on 0.22-μm-pore cellulose acetate filters (Millipore India, Ltd., Bangalore, India) and rinsed three times with cold trichloroacetic acid (TCA) (10% [wt/vol]) and once with ethanol (96% [vol/vol]; final rinse). The filters were stored individually in 8-ml scintillation vials under moisture-free conditions, and 5 ml scintillation cocktail Ultima Golds RX (PerkinElmer, Waltham, MA) was added a day prior to the radioassay in a liquid scintillation counter (PerkinElmer, Waltham, MA) onboard RV Polarstern. The amount (pM liter−1 h−1) of incorporated tritiated thymidine (TdR) was calculated using the formula given by Fuhrman and Azam (17).
The quantification of [4,5-3H]leucine incorporation was done by following the protocol of Kirchman (23). [4,5-3H]leucine (specific activity, 50,000 mCi mM−1; Amersham Corp., Amersham, England) was added to 2-ml aliquots of water sampled at depths of 5, 10, 20, and 50 m to a final concentration of 0.5 nM and incubated in the dark at 4 to 5°C for 1.5 h. At the end of the incubation, TCA was added (5% final concentration). The tubes were then centrifuged at 16,000 rpm for 10 min, and then the supernatant was discarded. Before the scintillation cocktail was added for the radioassay, all tubes were stored at 4°C for 48 h to reduce the moisture content. The amount (pM liter−1 h−1) of incorporated tritiated leucine was calculated following the protocol of Kirchman (24).
454 tag pyrosequencing.
For 454 tag pyrosequencing, we extracted DNA from the <5-μm fraction of IN station samples on days −1, 9, 18, and 36 according to the method described by Zhou et al. (64) combined with an initial FastPrep step (MPBiomedicals, Illkirch Cedex, France) to destroy the filter. After DNA extraction, a PCR was done using the primer pair Bakt341/Bakt805_W (22) (Table 1). The amplicon was purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany). 454 tag pyrosequencing of 16S rRNA amplicons was done by LGC genomics GmbH (Berlin, Germany) using Roche/454 GS FLX Titanium technology. The retrieved sequence reads had an average read length of 422 ± 52 bp and were classified after a quality check using a BLAST-based search against SILVA database release 108 (50, 57). Chao1, Simpson, and Shannon indexes were calculated on the genus level using the R standard library, including the vegan suite (R Development Core Team, 2011).
Table 1.
List of oligonucleotide probes and primers used
| Probe or primer | Target organism(s) | Sequence (5′→3′) | E. coli positiona | FA (%)b | Reference |
|---|---|---|---|---|---|
| EUB 338c | Most Bacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 35 | 2 |
| EUB IIc | Planctomycetales | GCAGCCACCCGTAGGTGT | 338–355 | 35 | 8 |
| EUB IIIc | Verrucomicrobiales | GCTGCCACCCGTAGGTGT | 338–355 | 35 | 8 |
| NON338 | Control | ACTCCTACGGGAGGCAGC | 338–355 | 35 | 61 |
| CREN554 | Thaumarchaeota | TTAGGCCCAATAATCMTCCT | 554–573 | 0 | 39 |
| ALF968 | Alphaproteobacteria except Rickettsiales | GGTAAGGTTCTGCGCGTT | 968–985 | 35 | 41 |
| SAR11-441 | SAR11 clade | TACAGTCATTTTCTTCCCCGAC | 441–463 | 25 | 52 |
| ROS537 | Roseobacter, SAR83 | CAACGCTAACCCCCTCC | 537–553 | 35 | 12 |
| RCA1000 | Roseobacter clade DC5-80-3 (formerly RCA) | ATCTCTGGTAGTAGCACA | 1000–1017 | 20 | 57 |
| RCA1000-comp | Competitor to RCA1000 | ATCTCTGGTAGTGGCACA | 1000–1017 | 20 | 57 |
| RCA1000-h3 | Helper to RCA1000 | CGTCCCCGAAGGGAACGTACC | 1018–1038 | 20 | 57 |
| RCA1000-h5 | Helper to RCA1000 | GGATGTCAAGGGTTGGTAAGG | 979–999 | 20 | 57 |
| ROCT1004 | Roseobacter clade OCT cluster | CTCCATCTCTGGAGCGAC | 1004–1021 | 20 | This study |
| ROCT1004-h3 | Helper to ROCT1004 | CTCGTCCAGCCKARCTGAAAG | 1022–1042 | 20 | This study |
| ROCT1004-h5 | Helper to ROCT1004 | GACGAGTATGTCAAGGGTTGG | 984–1004 | 20 | This study |
| GAM42ad | Gammaproteobacteria | GCCTTCCCACATCGTTT | 1027–1043 | 35 | 35 |
| SAR86-1245 | SAR86 clade | TTAGCGTCCGTCTGTAT | 1245–1262 | 35 | 67 |
| SAR86-1245-h3 | Helper to SAR86 1245 | GGATTRGCACCACCTCGCGGC | 1263–1283 | 35 | 67 |
| SAR86-1245-h5 | Helper to SAR86 1245 | CCATTGTAGCACGTGTGTAGC | 1222–1242 | 35 | 67 |
| CF319a | Most Flavobacteria, some Bacteroidetes, some Sphingobacteria | TGGTCCGTGTCTCAGTAC | 319–336 | 35 | 34 |
| CF6-1267 | Marine Flavobacteria, DE2 clade (including NS5) | GAAGATTCGCTCCTCCTC | 1267–1284 | 35 | 25 |
| FORM181A | Formosa-related clade A | GATGCCACTCTAAGAGAC | 181–199 | 25 | 57 |
| FORM181B | Competitor to FORM181A | GATGCCACTCTTAGAGAC | 181–199 | 35 | 57 |
| POL740 | Polaribacter | CCCTCAGCGTCAGTACATACG T | 740–761 | 35 | 33 |
| BAKT_341_F | Primer for Bacteria | CCTACGGGNGGCWGCAG | 341–358 | 22 | |
| BAKT_805_R | Primer for Bacteria | GACTACHVGGGTATCTAATCC | 805–785 | 22 |
Probe target position on Escherichia coli 16S rRNA according to reference 16.
Formamide concentration in the CARD FISH hybridization buffer.
Used in the mix of EUB I to III.
Used with the unlabeled competitor BETA42a.
CARD FISH.
To investigate the changes in the microbial community, CARD FISH was done on samples from the wind-mixed layer at depths between 30 m and 60 m, depending on station, as well as from depths of 100, 300, and 500 m using 11 oligonucleotide probes in a nested approach (Table 1). Since the WML is a homogenous water body, samples from different depths were used as duplicates. CARD FISH was done according to the method described by A. Pernthaler as modified by S. Thiele (44, 58). In brief, after the embedding of filters with agarose (0.1%) and lysozyme treatment for cell permeabilization, the hybridization was done using a probe (50 ng μl−1)/hybridization buffer mix ratio of 1:100 for 2 h. Signal amplification was done using a 5- (and 6-)-carboxyfluorescein-labeled tyramide (1 mg ml−1)/amplification buffer mix ratio of 1:100 for 45 min. Both hybridization and amplification of the filters were done on glass slides in humidity chambers. All filters were stained with DAPI (4′,6-diamidino-2-phenylindole) and counted manually (minimum of 1,000 DAPI signals) or using an automatic counting machine based on an epifluorescence microscope (Zeiss Axioplan II; Carl Zeiss AG, Jena, Germany) (47). The semiautomatic mode was used to count at least 2,000 DAPI signals with a size of between 8 and 200 pixels with a signal/noise (S/N) ratio of 12. A linear regression between three data sets of all stations did not show significant differences between semiautomated and manual counts (R2 = 0.92, P < 0.001, n = 110). Thus, semiautomatic counting was used for all probes besides EUB I to III, SAR11-441, POL740, and FORM181A. The total cell counts (TCC) were calculated as an average from all DAPI counts per station and depth.
Probe design.
A new probe, ROCT1004, targeting the Roseobacter clade OCT was designed using the ARB tool ProbeDesign (29) (Table 1). Two helper probes (Table 1) increase the target site accessibility (16). ROCT1004 currently covers 74% of the OCT clade when analyzed against SILVA release 108 (50). This comparison also indicated 108 outgroup hits mainly within the Antarctobacter clade of the Rhodobacteraceae. Since members of this clade are found in hypersaline lakes (26), false positives are unlikely to be detected within our study. The probe was used in an equimolar mixture with helpers at a formamide concentration of 20%.
Statistics.
Different analysis of variance (ANOVA) tests were used to evaluate the changes in cell abundance inside and outside the fertilized patch over time. If the normality tests failed, a nonparametric ANOVA on ranks or ANOVA on measurements was performed using the multiple comparison method recommended for every single case by SigmaStat 3.5 (Statcon, Witzenhausen, Germany). The effects of time and location (IN versus OUT stations) and their interactions were examined using linear (time or location) and quadratic regressions (time and location). We used linear models as suggested by permutation tests of R2 (30). Generally no interactions between time and patch location were detected. Statistical calculations were implemented with the R standard library, including the vegan suite (R Development Core Team, 2011).
RESULTS
Induction of a phytoplankton bloom.
The addition of acidified iron(II) sulfate solution to a clockwise-rotating cold core eddy at days 0 and 18 raised the iron concentration in the upper 30 to 40 m of the water column to 0.3 to 1.5 nM (L. M. Laglera, personal communication). This is only slightly higher than the concentrations of between 0.1 and 1.0 nM reported for iron-depleted waters in the Southern Ocean (9). In response to the fertilization, a nearly circular phytoplankton bloom of a size of ∼300 km2 was induced (Fig. 1). The chlorophyll a concentration increased from 0.5 μg liter−1 before fertilization to a maximum of 1.25 μg liter−1 at day 23 (M. Gauns, personal communication). The temperature in the eddy was increasing slightly from 6.5°C to 7.5°C during the course of the experiment. The fertilized patch circled twice with the eddy in the first 2 weeks of the experiment, before it moved south. The waters in the eddy were limited in silicic acid (<2 μM). Consequently, the bloom mainly consisted of Prymnesiophytes and not of diatoms.
Microbial cell numbers and production.
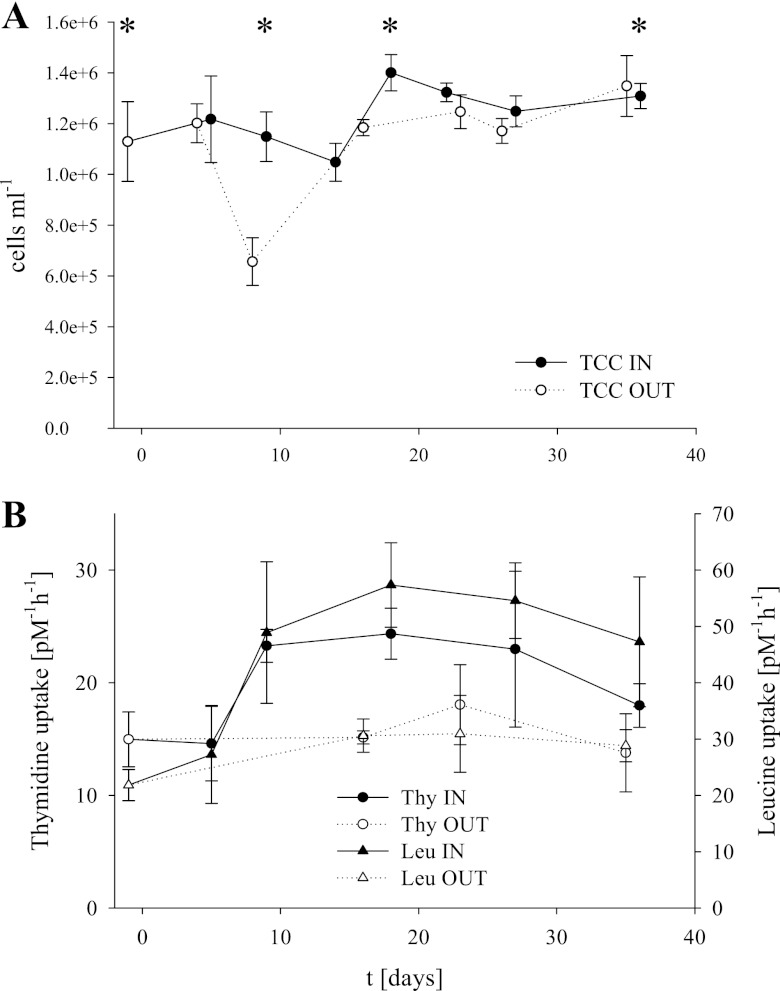
Total cell numbers (Fig. 2; see Table S1 in the supplemental material) inside the fertilized patch increased slightly but significantly from 1.1 × 106 on day −1 to 1.4 × 106 cells ml−1 on day 18 (R2 = 0.053, P = 3.73 ×10−4) and were on that date significantly higher than the 1.1 × 106 cells ml−1 in the corresponding OUT stations on day 16 (R2 = 0.141, P = 0.017). The low values on day 8 (OUT) might result from errors in eddy determination, and thus samples might be from a different water body and should be treated carefully.
Fig 2.
Total cell counts calculated from DAPI counts after CARD FISH (A) with error bars showing standard deviations of all counts. Microbial production was measured as thymidine (Thy) and leucine (Leu) uptake rates (B), with error bars showing the standard deviations of all measurements. Asterisks mark sampling points for 454 tag pyrosequencing.
The microbial production measured by leucine and thymidine uptake inside the fertilized patch increased from day 5 onwards, while no significant increase was measured outside the fertilized patch (Fig. 2). Thymidine uptake rates increased significantly (P = 0.002) 1.6-fold from about 15 pM liter−1 h−1 to 24 pM liter−1 h−1 on day 18, compared to 15 pM liter−1 h−1 on day 16 (OUT), and then decreased again beginning on day 27. Leucine uptake rates doubled from 27 pM liter−1 h−1 on day 5 to 57 pM liter−1 h−1 on day 18 (P = <0.001), compared to 31 pM liter−1 h−1 outside the fertilized patch on day 16.
Bacterial diversity and community composition.
Community richness based on Chao1 values decreased from day −1 (126.0 ± 12.4) to day 9 (111.7 ± 3.4) and increased subsequently toward day 18 (134.9 ± 17.0) and day 36 (157.7 ± 18.7). However, these minute changes in the diversity of the community were not reflected by the Shannon and Simpson indices. The Shannon index ranged from 3.26 (day 18) to 3.39 (day 36), while the Simpson index ranged from 0.936 (day 18) to 0.944 (day 9).
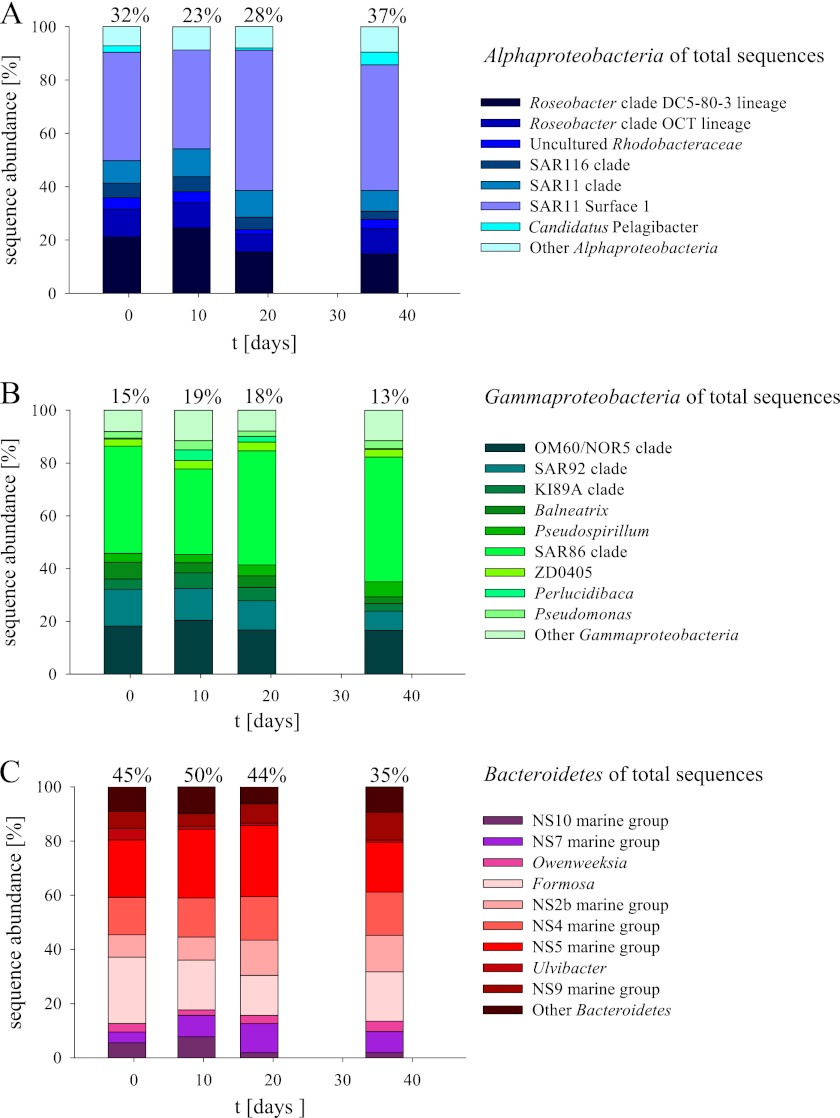
Within the Alphaproteobacteria, the Roseobacter clade, including the clades DC5-80-3 and OCT and the SAR11 clade, in particular the surface 1 clade, were dominant encompassing >80% of the sequences (Fig. 3). The fraction of SAR11 clade sequences increased from 49% to 67% and 62% of alphaproteobacterial sequences from day 9 to days 18 and 36, respectively, while the Roseobacter clade sequences decreased from 35% to 23% and 25% on the same days. The Gammaproteobacteria were dominated by the SAR86 clade and Alteromonadales (including the OM60/NOR5 and SAR92 clades) (Fig. 3). An initial decrease of SAR86 clade sequences was followed by increases from 32% to 43% and 47% on days 9, 18, and 36, respectively. This relative increase was mirrored by a decrease of Alteromonadales sequences from 35% to 29% and 25% on the same days. The diversity within the Bacteroidetes community was rather constant. It was dominated by Flavobacteriales, including the slightly increasing Formosa clade and the slightly decreasing DE2/NS5 clade (Fig. 3). Polaribacter sequences made up less than 3% of the Bacteroidetes sequences.
Fig 3.
Results from 454 tag pyrosequencing of the WML from stations on days −1 (control), 9 (IN), 18 (IN), and 36 (IN). Alphaproteobacteria (A), Gammaproteobacteria (B), and Bacteroidetes (C) are separately shown, while the percentage of each group of the total sequences is stated above the columns. For simplicity, only clades with more than 3% of group sequences on at least one of the days are shown. All clades below that threshold are summed up as “Other.” t, time.
Abundances of key bacterial clades.
Since the 454 tag pyrosequencing did not show significant changes in microbial community during the course of the experiment, and given the uncertainties in reflecting the abundances of microbial clades (65), specific oligonucleotide probes (Table 1) were selected to quantify representative phylogenetic groups by CARD FISH in the iron-fertilized patch and the surrounding waters. We used the probe mix EUB I to III and CREN554 to determine the abundance of Bacteria and Crenarchaea. In the WML, Bacteria made up 8.7 × 105 to 1.3 × 106 cells ml−1 in the fertilized patch and were significantly higher around day 18 than in the comparable OUT station at day 16 (R2 = 0.081, P = 0.033). Crenarchaeal numbers did not exceed 4.8 × 104 cells ml−1 and did not differ significantly between the IN and OUT stations. Both probes together targeted ∼90% of the DAPI counts, with the exception of day 26 at the OUT station and day 36 at the IN station, where only 78% and 71% were covered.
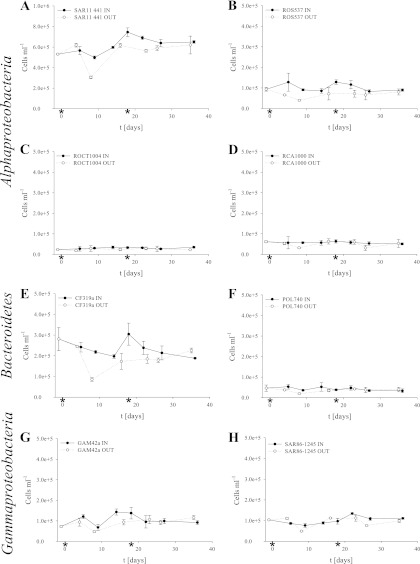
The SAR11 clade, as detected by probe SAR11-441 (Fig. 4A), was the most abundant group, accounting frequently for more than 50% of total cell counts. It increased significantly within the fertilized patch from 5.5 × 105 cells ml−1 to 7.7 × 105 cells ml−1 at day 18 (R2 = 0.15, P = 0.005), staying on this level until the end of the experiment. Cell numbers of SAR11 were generally higher inside the fertilized patch compared to those at the corresponding OUT stations (R2 = 0.09, P = 0.016). Outside the patch, a slight increase was found at the beginning of the experiment, but cell numbers reached a lower maximum of 6.4 × 105 cells ml−1 on day 5 compared to those at the IN stations.
Fig 4.
Absolute cell numbers calculated from total cell numbers (Fig. 2) and abundances retrieved in triplicate from CARD FISH analyses of 3 independent filters. For simplification, error bars show the standard errors of the CARD FISH triplicates. Absolute cell numbers of IN (black, straight lines) and OUT (white, dotted lines) stations are shown for SAR11 (A), Roseobacter (B), the Roseobacter subgroup clades OCT (C) and DC5-80-3 (D), Bacteroidetes (E) and the subgroup Polaribacter (F), and Gammaproteobacteria (G) with the subgroup SAR86 (H). We used a different scale for the SAR11 clade numbers. The days of iron fertilization events are marked with asterisks. t, time.
Cell numbers of the Roseobacter clade did not change throughout the course of the experiment in both IN stations (around 1.0 × 105 cells ml−1) and OUT stations (around 8.5 × 104 cells ml−1; probe ROS537) (Fig. 4B). The IN counts were significantly higher than the OUT counts (R2 = 0.31, P = 7.7 × 10−5 cells ml−1). Within the Roseobacter clade, the cell number of OCT clade was also significantly higher at the IN stations compared to those outside the patch (probe ROCT1004 [Fig. 4C]) (R2 = 0.1, P = 0.022), while the Roseobacter DC5-80-3 clade showed no significant changes (probe RCA1000 [Fig. 4D]) (R2 < 0.01, P = 0.321).
The total cell numbers of Bacteroidetes showed a significant difference between the IN and OUT stations between days 8 and 27 (probe CF319a [Fig. 4E]) (R2 = 0.13, P = 8.5 × 10−3). Inside the fertilized patch, the highest cell numbers were found on day 18 (3.1 × 105 cells ml−1). This maximum was followed by a decrease in cell numbers toward the end of the experiment. In the OUT sample from day 4, cell numbers were as high as those at the IN station on day 5 (2.5 × 105 cells ml−1) but decreased to 2.0 × 105 cells ml−1 and below. From day 16 on, bacteroidetal numbers remained stable at all of the OUT stations (1.8 × 105 cells ml−1). These abundance patterns were not followed by Polaribacter (probe POL740 [Fig. 4F]), which showed cell numbers around 4 × 104 cells ml−1 both inside and out of the fertilized patch over the entire course of the experiment. Formosa (probe FORM181A) was found in low abundances of around 2.4 × 104 cells ml−1. The DE2/NS5 clade (probe CF6-1267) was detected at numbers between 1.1 × 104 cells ml−1 and 3.1 × 104 cells ml−1.
Similar to the abundance of Alphaproteobacteria and Bacteroidetes, the abundances of Gammaproteobacteria as assessed by probe GAM42a were between 7.0 × 104 cells ml−1 and 1.5 × 105 cells ml−1 (Fig. 4G). The dominant group within the Gammaproteobacteria class was the SAR86 clade (probe SAR86-1245 [Fig. 4H]), accounting for 60 to 100% of the gammaproteobacterial cell counts. Significant changes were found neither during the progression of the iron fertilization nor between the IN and OUT samples.
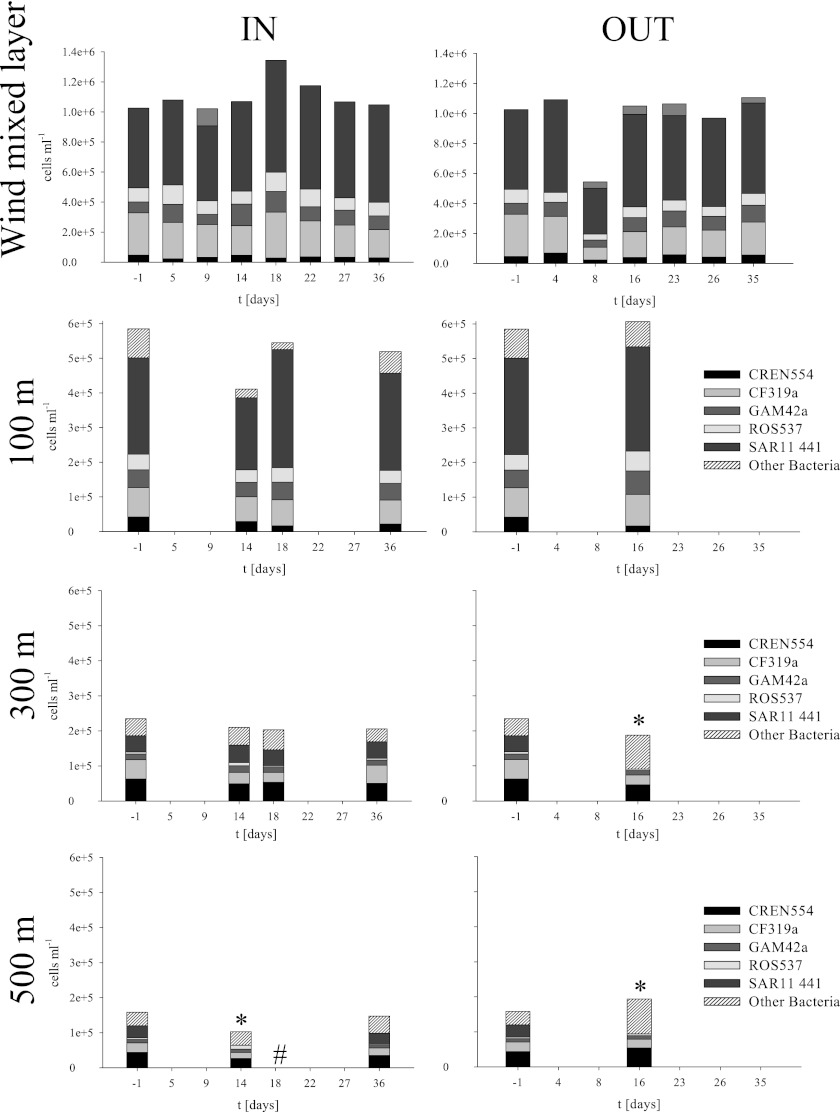
Depth profiles.
Bacterial abundances beneath the WML were quantified at 5 stations at depths of 100, 300, and 500 m (Fig. 5). A decrease in bacterial cell numbers on all taxonomic levels of an average of 73% was found from the 100-m depth to the 300-m depth, followed by a minor decrease of an average of 25% toward 500 m. The major decrease in total cell numbers was mainly due to a decrease in the numerically dominant SAR11 clade from an average of 6.6 × 105 cells ml−1 in the WML to an average of 3.3 × 104 cells ml−1 at 500 m. Bacteroidetes decreased less on days −1 and 36 and subsequently showed a significantly (P = 0.004) higher absolute abundance at a depth of 300 m. While bacterial cell numbers decreased, Crenarchaea increased an average of 2.1-fold between depths of 100 m (2.5 × 104 cells ml−1) and 300 m (5.2 × 104 cells ml−1) on all days and were stable toward a depth of 500 m.
Fig 5.
Depth profiles of the total cell numbers of the first 500 m of the water column. Days −1, IN station days 9, 18, and 36, and OUT station day 16 were investigated additionally to the WML at depths of 100, 300, and 500 m. Missing values (*) for SAR11 at days 14 and 16 (OUT) are due to limited sample quantities. In addition, no sampling was done at 500 m on day 18 (#). Error bars are omitted for simplicity. t, time.
DISCUSSION
In contrast to most other iron fertilization experiments (3, 6, 43), the total cell numbers of bacterioplankton and of the major clades were rather constant throughout the LOHAFEX experiment, although a positive correlation of chlorophyll a with bacterial production implied active growth of the bacterial community (Thy, P = 0.033; Leu, P = 0.003). This is unlike the recently described dynamic succession of mostly bacteroidetal and gammabacterial genera after a spring phytoplankton bloom in the North Sea (57). Instead we observed a rather subtle change in numbers for the individual clades. Presumably, these changes are comparable to coastal late spring/summer blooms, rather than spring blooms. The reaction of grazers to spring blooms is rather slow, compared to the reaction to summer blooms, as suggested by Wiltshire and Manly (63).
For SAR11 and Roseobacter, and to a lesser extent for Bacteroidetes, a stimulatory effect on the cell numbers during the iron fertilization could be detected. SAR11 is usually dominant in oligotrophic waters outside algal blooms (40, 62), but it is also highly abundant and productive in coastal and open ocean waters (31, 32). This clade increased significantly inside the fertilized patch from day 18 of the experiment on and showed high numbers until the end of the bloom. SAR11 uses low-molecular-weight substrates (18, 52), which might have been more abundant in the later phase of the bloom, providing an advantage in resource competition for this group. In addition, members of clade SAR11 might escape protist grazing due to their small cell size.
Roseobacter strains as bacterial generalists are known to accompany and benefit from phytoplankton blooms (4, 66) and attach often to particles and dinoflagellates (53). Inside the LOHAFEX bloom, significantly higher cell numbers were found compared to those at the OUT stations. However, the Roseobacter abundance was lower in the LOHAFEX bloom than those of other phytoplankton blooms before (66). This might be due to relatively low chlorophyll a concentrations indicating a rather weak phytoplankton bloom. Microscopic observations already onboard showed that the Roseobacter cells, including cells from the OCT and DC5-80-3 clades, were always >1 μm in diameter. This is within the size range of potential grazers, and we could occasionally find them inside larger flagellate cells (see Fig. S1 in the supplemental material). Therefore, we assume that the grazer community exerts a strict top-down control on Roseobacter, keeping their cell numbers fairly constant.
Most marine Gammaproteobacteria, besides members of the SAR86 clade, are r-strategists that react quickly to sudden nutrient availability (51). They are also rather susceptible to enhanced mortality by grazing or viral lysis (45). Since the bacterial production was increased during the experiment, but no increase of Gammaproteobacteria was found, a mechanism to control the gammaproteobacterial numbers similar to that of the Roseobacter clades can be assumed. Only SAR86 cells, which are about as small as SAR11 cells, might have escaped this grazing. SAR86 is known to be abundant in oligotrophic waters (38, 44). In comparison to SAR11, SAR86 is specialized on lipid degradation (11). Thus, SAR86 might have profited from grazing pressure on potential competitors for this substrate, such as other Gammaproteobacteria or Bacteroidetes. In addition, sloppy feeding from those grazers might have led to a release of fatty acids, providing more resources to SAR86.
Bacteroidetes have been reported to be highly abundant in Phaeocystis blooms in the Southern Ocean (1, 55). Marine members of the Bacteroidetes utilize higher-molecular-weight carbon sources, such as polysaccharides (20) and proteins (7, 48, 49), and are often present in high numbers during and after algal blooms (27, 49). During LOHAFEX, the Bacteroidetes were present in fairly high numbers (∼20% of all cells) compared to those found in late summer situations with similar water temperatures and chlorophyll a concentrations in arctic regions (19, 54). They decreased with the onset of the phytoplankton bloom after the first fertilization campaign, but also at the OUT stations, suggesting depletion of substrate originating from a pre-LOHAFEX phytoplankton bloom and grazing by protozoa. The onset of the decay of the LOHAFEX-induced Prymnesiophytes might have increased the substrate availability and enabled Bacteroidetes to slightly increase in numbers.
All bacterial groups examined decreased with depth at any time point during the course of the experiment. Similar to the WML, the deeper layers showed no clear change in community composition within the respective layer. There was no accumulation of cells in deeper layers, as one would expect from a sinking and decaying algae bloom (14, 56). Only for the Bacteroidetes could a slightly higher cell abundance be monitored at a depth of 300 m on days −1 and 36 compared to days 14 and 18 (both IN), as well as day 16 (OUT). These higher numbers might result from Bacteroidetes cells attached to marine snow particles from the hypothesized bloom prior to LOHAFEX (day −1) and the LOHAFEX bloom (day 36). Such an accumulation of marine snow particles, measured as particulate organic carbon (POC), has been found in previous iron fertilization experiments. During the EIFEX iron fertilization experiment, POC accumulated in a deeper water layer inside the patch at the late phase of the bloom as well as outside the patch as a result of slower-sinking particles from natural blooms that occurred prior to the experiment (56).
The analysis of iron fertilization experiments relies strongly on the exact localization of the IN and OUT stations, since only the former belong to a homogeneous water mass with a common history. Therefore, only data from these stations can be compared to each other. In turn, the OUT stations might be placed not only outside the fertilized patch, but also outside the eddy, belonging to entirely different water masses originating, e.g., from the Southern Atlantic Gyre. Along those lines, the large deviation in cell numbers on day 8 of LOHAFEX compared to other OUT stations might be due to an uncertainty in the determination of the eddy, and therefore this station should be treated cautiously. This is also true for day 26, even though total cell counts and CARD FISH counts are similar to those of the preceding and following days.
In general, our findings are similar to findings from the EisenEx experiment, where no changes in the microbial community were found by terminal restriction fragment length polymorphism (T-RFLP) analyses (3). During EisenEx, SOIREE, and IronEx II, hints of a negative correlation between bacterial abundance and the abundance of heterotrophic nanoflagellates (HNFs) were found, giving rise to the assumption of a top-down-controlled system by bacteriovorous grazers (3, 6, 21). Most likely in LOHAFEX, an effective grazer population had been established during a preceding austral summer bloom which responded directly to the increasing microbial cell numbers after the iron fertilization. Onboard flow cytometry and microscopic counting showed a nanoplankton abundance of ∼10,000 cells ml−1 (S. Thiele, I. Schulz, and B. M. Fuchs, unpublished data), compared to an average of ∼1,000 cells ml−1 in open ocean waters of the Southern Gyre (68). Assuming that most nanoplankton cells are mixotrophs (15), this would result in a predator/prey ratio of ∼1:100 during LOHAFEX, which is 10-fold higher than the commonly observed ratio of ∼ 1:1,000 (13). This well-established community of nanoflagellates might have exerted a high grazing pressure onto the microbial community, keeping cell numbers more or less constant despite increased bacterial production. The control of the microbial community by grazers is congruent with findings from SOFeX North, where a bloom of nanoflagellates was established in silicate-depleted waters (5). Furthermore, the typical grazing range of HNFs is between cell lengths of 1 and 3 μm (45). During LOHAFEX, the cell sizes of Roseobacter (length, ∼1.6 μm; n = 10), Gammaproteobacteria (length, ∼1.5 μm; n = 10) and Bacteroidetes (length, ∼1.0 μm; n = 10) were well within this edible size range. The tiny SAR11 cells (length, ∼0.4 μm; n = 10) and the similar-size SAR86 cells would escape the HNF grazing, explaining why they showed slight increases. These preliminary observations have to be tested in future grazing experiments during similar summer blooms with strong grazer communities.
The overall aim of LOHAFEX was to investigate the sequestration of a phytoplankton bloom induced by iron fertilization. As a consequence of the phytoplankton-bacterioplankton coupling, we hypothesized that algal exudates and decaying algal biomass would result in a dynamic bacterial succession of distinct genera (57). Based on our results indicating a general lack of a substrate-driven succession or direct effects of iron fertilization on the bacterioplankton composition, we speculate that—similar to the tight top-down control of the LOHAFEX phytoplankton bloom—any changes in the bacterioplankton abundance and composition were tightly controlled by flagellate grazing. In retrospect, the late summer situation, in which the LOHAFEX experiment was conducted, was characterized by a well-balanced microbial loop. Extra nutrient pulses as induced by the iron fertilization were effectively processed through the microbial food web.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Smetacek and S. W. A. Naqvi, the chief scientists of the project, the captain and crew of the RV Polarstern, and all collaborators within LOHAFEX. We thank J. Wulf for sampling during the cruise, A. Sakar, D. Baraniya, A. Galina, and M. Fernández Méndez for help and discussions, A. Ramette for help with statistical analyses, J. Peplies and C. Quast for help with the 454 tag pyrosequencing data analyses, and J. Köhler for help with the map.
This project was funded by the Max Planck Society.
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alderkamp AC, Sintes E, Herndl GJ. 2006. Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. Aquat. Microb. Ecol. 45:237–246 [Google Scholar]
- 2. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arrieta JM, Weinbauer MG, Lute C, Herndl GJ. 2004. Response of bacterioplankton to iron fertilization in the southern ocean. Limnol. Oceanogr. 49:799–808 [Google Scholar]
- 4. Buchan A, Gonzalez JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coale KH, et al. 2004. Southern Ocean iron enrichment experiment: carbon cycling in high- and low-Si waters. Science 304:408–414 [DOI] [PubMed] [Google Scholar]
- 6. Cochlan WP. 2001. The heterotrophic bacterial response during a mesoscale iron enrichment experiment (IronEx II) in the eastern equatorial Pacific Ocean. Limnol. Oceanogr. 46:428–435 [Google Scholar]
- 7. Cottrell MT, Kirchman D. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 9. deBaar HJW, deJong JTM. 2001. Distributions, sources and sinks of iron in seawater, p 123–253 In Turner D, Hunter KA. (ed), The biogeochemistry of iron in seawater. Wiley, New York, NY [Google Scholar]
- 10. Ducklow HW, Steinberg DK, Buessler KO. 2001. Upper ocean carbon export and the biological pump. Oceanography 14:50–58 [Google Scholar]
- 11. Dupont CL, et al. 2011. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6:1186–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eilers H, et al. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67:5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenchel T. 1986. The ecology of heterotrophic microflagellates. Adv. Microb. Ecol. 9:57–97 [Google Scholar]
- 14. Frette L, Winding A, Kroer N. 2010. Genetic and metabolic diversity of Arctic bacterioplankton during the post-spring phytoplankton bloom in Disko Bay, western Greenland. Aquat. Microb. Ecol. 60:29–41 [Google Scholar]
- 15. Frias-Lopez J, Thompson A, Waldbauer J, Chisholm SW. 2009. Use of stable isotope-labeled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ. Microbiol. 11:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuchs BM, Glöckner FO, Wulf J, Amann R. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhrman JA, Azam F. 1980. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl. Environ. Microbiol. 39:1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giovannoni SJ, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
- 19. Gomez-Pereira PR, et al. 2010. Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J. 4:472–487 [DOI] [PubMed] [Google Scholar]
- 20. Gómez-Pereira PR, et al. 2012. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ. Microbiol. 14:52–66 [DOI] [PubMed] [Google Scholar]
- 21. Hall JA, Safi K. 2001. The impact of in situ Fe fertilisation on the microbial food web in the Southern Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 48:2591–2613 [Google Scholar]
- 22. Herlemann DP, et al. 2011. Transitions in bacterial communities along the 2000-km salinity gradient of the Baltic Sea. ISME J. 5:1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirchman D, K'nees E, Hodson R. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirchman DL, Hoch MP. 1988. Bacterial production in the Delaware Bay estuary estimated from thymidine and leucine incorporation rates. Mar. Ecol. Prog. Ser. 45:169–178 [Google Scholar]
- 25. Kirchman DL, Yu L, Cottrell MT. 2003. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware Estuary. Appl. Environ. Microbiol. 69:6587–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labrenz M, et al. 1998. Antarctobacter heliothermus gen. nov., sp. nov., a budding bacterium from hypersaline and heliothermal Ekho Lake. Int. J. Syst. Evol. Microbiol. 48:1363–1372 [DOI] [PubMed] [Google Scholar]
- 27. Lamy D, et al. 2009. Temporal changes of major bacterial groups and bacterial heterotrophic activity during a Phaeocystis globosa bloom in the eastern English Channel. Aquat. Microb. Ecol. 58:95–107 [Google Scholar]
- 28. Lochte K, Bjørnsen PK, Giesenhagen H, Weber A. 1997. Bacterial standing stock and production and their relation to phytoplankton in the Southern Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 44:321–340 [Google Scholar]
- 29. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makarenkov V, Legendre P. 2002. Nonlinear redundancy analysis and canonical correspondence analysis based on polynomial regression. Ecology 83:1146–1161 [Google Scholar]
- 31. Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL. 2005. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71:2979–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL. 2007. Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aquat. Microb. Ecol. 47:45–55 [Google Scholar]
- 34. Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097–1106 [DOI] [PubMed] [Google Scholar]
- 35. Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 36. Martin JH. 1990. Glacial-interglacial CO2 change: the iron hypothesis. Paleoceanography 5:1–13 [Google Scholar]
- 37. Martin JH, et al. 1994. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature 371:123–129 [Google Scholar]
- 38. Mary I, et al. 2006. Seasonal dynamics of bacterioplankton community structure at a coastal station in the western English Channel. Aquat. Microb. Ecol. 42:119–126 [Google Scholar]
- 39. Massana R, Murray AE, Preston CM, DeLong EF. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morris RM, et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810 [DOI] [PubMed] [Google Scholar]
- 41. Neef A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Technische Universität München, Lehrstuhl für Mikrobiologie, Munich, Germany [Google Scholar]
- 42. Obernosterer I, et al. 2008. Rapid bacterial mineralization of organic carbon produced during a phytoplankton bloom induced by natural iron fertilization in the Southern Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 55:777–789 [Google Scholar]
- 43. Oliver JL, Barber RT, Smith WO, Ducklow HW. 2004. The heterotrophic bacterial response during the Southern Ocean Iron Experiment (SOFeX). Limnol. Oceangr. 49:2129–2140 [Google Scholar]
- 44. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537–546 [DOI] [PubMed] [Google Scholar]
- 46. Pernthaler J, Amann R. 2005. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol. Mol. Biol. Rev. 69:440–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pernthaler J, Pernthaler A, Amann R. 2003. Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pinhassi J, et al. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13–26 [Google Scholar]
- 49. Pinhassi J, et al. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Puddu A, et al. 2003. Bacterial uptake of DOM released from P-limited phytoplankton. FEMS Microbiol. Ecol. 46:257–268 [DOI] [PubMed] [Google Scholar]
- 52. Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 53. Rooney-Varga JN, et al. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49:163–175 [DOI] [PubMed] [Google Scholar]
- 54. Schattenhofer M, et al. 2011. Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst. Appl. Microbiol. 34:470–475 [DOI] [PubMed] [Google Scholar]
- 55. Simon M, Glöckner F, Amann R. 1999. Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat. Microb. Ecol. 18:275–284 [Google Scholar]
- 56. Smetacek V, et al. 2012. Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature 487:313–319 [DOI] [PubMed] [Google Scholar]
- 57. Teeling H, et al. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611 [DOI] [PubMed] [Google Scholar]
- 58. Thiele S, Fuchs BM, Amann RI. 2011. Identification of microorganisms using the ribosomal RNA approach and fluorescence in situ hybridization, p 171–189 In Wilderer P. (ed), Treatise on water science. Elsevier, Oxford, United Kingdom [Google Scholar]
- 59. Thingstad TF. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceangr. 45:1320–1328 [Google Scholar]
- 60. Tortell PD, Maldonado MT, Price NM. 1996. The role of heterotrophic bacteria in iron-limited ocean ecosystems. Nature 383:330–332 [Google Scholar]
- 61. Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143 [DOI] [PubMed] [Google Scholar]
- 62. West NJ, Obernosterer I, Zemb O, Lebaron P. 2008. Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ. Microbiol. 10:738–756 [DOI] [PubMed] [Google Scholar]
- 63. Wiltshire KH, Manly BFJ. 2004. The warming trend at Helgoland Roads, North Sea: phytoplankton response. Helgol. Mar. Res. 58:269–273 [Google Scholar]
- 64. Zhou J, Bruns M, Tiedje J. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou J, et al. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5:1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zubkov MV, et al. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304–311 [DOI] [PubMed] [Google Scholar]
- 67. Zubkov MV, Fuchs BM, Burkill PH, Amann R. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zubkov MV, Burkill PH, Topping JN. 2007. Flow cytometric enumeration of DNA-stained oceanic planktonic protists. J. Plankton Res. 29:79–86 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.