Abstract
In clinical microbiology, bacterial identification is labor-intensive and time-consuming. A solution for this problem is the use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). In this study, we evaluated a modified protein extraction method of identification performed on target plates (on-plate extraction method) with MALDI-TOF (Bruker Microflex LT with Biotyper version 3.0) and compared it to 2 previously described methods: the direct colony method and a standard protein extraction method (standard extraction method). We evaluated the species of 273 clinical strains and 14 reference strains of staphylococci. All isolates were characterized using the superoxide dismutase A sequence as a reference. For the species identification, the on-plate, standard extraction, and direct colony methods identified 257 isolates (89.5%), 232 isolates (80.8%), and 173 isolates (60.2%), respectively, with statistically significant differences among the three methods (P < 0.05). In conclusion, the on-plate extraction method is at least as good as standard extraction in identification rate and has the advantage of a shorter processing time.
INTRODUCTION
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been used to identify various microorganisms in clinical laboratories, including Gram-positive bacteria, Gram-negative bacteria, yeasts, and even filamentous fungi (1, 8, 10, 12, 14, 16–19, 21, 22, 24). It is a rapid and inexpensive alternative to molecular identification that offers equivalent accuracy (6). Additionally, complete bacterial identification by MALDI-TOF MS is more cost-effective than by conventional methods (9, 15, 22). Two preparatory methods have been reported for MALDI-TOF MS-based identification (1, 4, 5): the direct colony method and the standard protein extraction method (standard extraction method). The direct colony method is performed by picking up colonies from a culture plate and placing them, as a thin layer of sample, onto the target plate and then subjecting the matrix to MALDI-TOF MS analysis (1). This method is simple and can be performed rapidly, but it is inferior in accuracy to the standard extraction method. This deficiency is particularly evident for Gram-positive organisms because of insufficient cell wall disruption (1). The standard extraction method, in contrast, employs formic acid and acetonitrile to disrupt the cells before they are placed onto the target plate for MALDI-TOF MS analysis. The standard extraction method involves 2 centrifugation steps and requires approximately 6-fold-more processing time than does the direct colony method.
We evaluated an on-plate extraction method that could be performed on the target plates without any centrifugation steps. Our study compared the on-plate extraction method with both the direct colony and the standard extraction methods for MALDI-TOF MS-based identification of staphylococcal clinical isolates.
MATERIALS AND METHODS
Bacterial isolates.
This study used 273 staphylococci of nonduplicate clinical isolates and 14 type and reference strains (Staphylococcus aureus NCTC 8325, S. epidermidis ATCC 14490T, Staphylococcus capitis subsp. capitis CCUG 7326T, Staphylococcus capitis subsp. ureolyticus ATCC 49326, Staphylococcus haemolyticus ATCC 29970T, Staphylococcus lugdunensis ATCC 43809T, Staphylococcus saprophyticus JCM 2427T, Staphylococcus cohnii ATCC 29974T, Staphylococcus hominis ATCC 27844T, Staphylococcus pettenkoferi CCUG 51279T, Staphylococcus schleiferi subsp. schleiferi ATCC 43808T, Staphylococcus schleiferi subsp. coagulans JCM 7470, Staphylococcus warneri ATCC 27836T, and Staphylococcus caprae ATCC 33538T). All strains were stored at −80°C until use, precultured for 12 to 24 h, and cultured aerobically overnight on 5% sheep blood agar at 37°C.
Species identification of staphylococci.
Isolated species were identified by partial sequencing of superoxide dismutase A (sodA) by using primers d1 and d2, as previously described (20, 23). All isolates were considered correctly identified when the sodA sequence yielded ≥98% sequence similarity with the closest species sequence in the GenBank database. The sodA sequence profiles and the origin of the clinical isolates are presented in the supplemental material.
MALDI-TOF MS.
For the direct colony method, bacteria were applied, using sterile toothpicks, as thin films on 96-spot, polished, stainless steel target plates (Bruker Daltonik GmbH, Leipzig, Germany). The bacteria were then left to dry at room temperature for 1 min. Subsequently, 1.5 μl of the matrix solution, comprising a saturated α-cyano-4-hydrocinnamic acid (Bruker Daltonik) in 50% acetonitrile (Wako Pure Chemical Industries, Osaka, Japan) and 2.5% trifluoroacetic acid (Wako), was applied to the samples and cocrystallized with them at room temperature for 10 min.
For the on-plate extraction method, each strain was applied and dried on the target plate as in the direct colony method. Following this, 0.5 μl of 70% formic acid (Wako) was mixed with the sample on the plate by pipetting, followed by 0.5 μl of acetonitrile, and the resultant mixture was dried at room temperature for approximately 10 min. Finally, 1.5 μl of the matrix solution was applied onto the spot as in the direct colony method.
For the standard extraction method, a small sample of each colony was suspended in 300 μl of distilled water and adjusted at McFarland 2 standard, and 900 μl of absolute ethanol was added. The suspension was vortexed vigorously and centrifuged at 20,000 × g for 2 min. The supernatant was then discarded, and the pellet was dried at 55°C for at least 30 min. Fifty microliters of 70% formic acid was then added and thoroughly mixed by pipetting. Next, 50 μl of acetonitrile was added, and the sample was centrifuged again at 20,000 × g for 2 min. Subsequently, 1 μl of supernatant was placed onto the target plate and left to dry for approximately 10 min at room temperature. Finally, 1.5 μl of matrix solution was applied onto the spot, as in the direct colony procedure.
The samples prepared by each method were applied to a MicroFlex LT mass spectrometer (Bruker Daltonik), and the results were analyzed by MALDI Biotyper 3.0 software (Bruker Daltonik). Each measurement was performed only once for each culture. Escherichia coli DH5α was used as a quality control as recommended by the manufacturer on each experiment.
Data analysis.
The manufacturer's recommended log score identification criteria were used as follows: a score of 2.000 to 3.000 indicated species-level identification, a score of 1.700 to 1.999 indicated genus-level identification, and a score of <1.700 indicated an unreliable identification. Duplicate experiments were performed. For genus- and species-level identification, a less stringent identification criterion was used for analysis. If 1 strain attained genus-level identification in the first experiment and species-level identification in the second experiment, genus-level identification was used for analysis. If 2 experiments resulted in different results, such as genus-level identification and unreliable identification, the unreliable-identification result was used. Additionally, the rates for different criteria (genus-level, species-level, or unreliable identification) were calculated by the number of different criteria in first and second experiments divided by the total number of isolates. Different results between MALDI-TOF MS and molecular identification were categorized as erroneous identifications. Additionally, when the Biotyper ascertained only genus-level identification (log score, 1.70 to 2.00), probable species identification was estimated in accordance with the log score order. Different species results between the highest log score candidate and molecular identification were categorized as discordant results. In the case of such results, the protein signature profile was analyzed. A dendrogram was constructed using the correlation distance measure and the average-linkage algorithm settings of the Biotyper 3.0 software. All processes related to MALDI-TOF MS identification were performed by trained personnel only.
Statistical analysis.
Comparisons of genus- and species-level identifications among the 3 preparatory methods were performed using chi-square tests. A P value of <0.05 was considered statistically significant.
RESULTS
Identification rates for the three different methods.
The identification rates obtained using the 3 different methods are shown in Table 1. The on-plate method identified 283 (98.6%) isolates at the genus level and 257 (89.5%) isolates at the species level. By comparison, the standard extraction method identified 283 (98.6%) isolates at the genus level and 232 (80.8%) isolates at the species level, while the direct colony method identified 256 (89.2%) isolates at the genus level and 173 (60.2%) isolates at the species level. There were no statistically significant differences in genus-level identifications between the on-plate method and the standard extraction method (P = 0.1545), but a significantly higher identification rate was achieved by the on-plate method at the species level than by the standard extraction method (P < 0.0001). On the other hand, the direct colony method yielded lower identification rates at the species level than did both the standard extraction (P < 0.0001) and on-plate extraction methods (P = 0.0450). The rates for different identification criteria in the first and second experiments were 30 (10.4%), 23 (8%), and 79 (27.5%) for standard extraction, on-plate extraction, and the direct method, respectively (data not shown). In species identification, there was a large difference between on-plate extraction and standard extraction for S. caprae (75% and 4.2%, respectively) and S. saprophyticus (78.9% and 5.2%, respectively). Other strains have smaller identification rate differences or only a small number of isolates. The species identification rates for coagulase-negative staphylococci (CoNS) were 169 (75.8%), 194 (87.0%), and 117 (52.5%) for standard extraction, on-plate extraction, and the direct colony method, respectively.
TABLE 1.
Comparison of three methods for identification of Staphylococcus to the genus and species levels
| Organism | No. of isolates | No. of isolates (% correct) identified bya: |
No. of spectra in database | |||||
|---|---|---|---|---|---|---|---|---|
| Standard extraction |
On-plate extraction |
Direct colony |
||||||
| Genus level | Species level | Genus level | Species level | Genus level | Species level | |||
| Staphylococcus aureus | 64 | 64 (100) | 63 (98.4) | 64 (100) | 63 (98.4) | 64 (100) | 56 (87.5) | 12 |
| Staphylococcus epidermidis | 93 | 93 (100) | 87 (93.5) | 93 (100) | 89 (95.7) | 93 (100) | 64 (68.8) | 9 |
| Staphylococcus capitis | 20 | 20 (100) | 20 (100) | 20 (100) | 18 (90) | 20 (100) | 16 (80) | 6 |
| Staphylococcus caprae | 24 | 22 (91.6) | 1 (4.2) | 24 (100) | 18 (75) | 21 (87.5) | 6 (25) | 2 |
| Staphylococcus haemolyticus | 19 | 19 (100) | 18 (94.7) | 18 (94.7) | 17 (89.4) | 16 (88.9) | 7 (36.8) | 8 |
| Staphylococcus lugdunensis | 21 | 21 (100) | 21 (100) | 20 (95.2) | 19 (90.4) | 21 (100) | 17 (80.9) | 6 |
| Staphylococcus saprophyticus | 19 | 17 (89.4) | 1 (5.2) | 19 (100) | 15 (78.9) | 5 (26.3) | 0 (0) | 8 |
| Staphylococcus cohnii | 3 | 3 (100) | 0 (0) | 1 (33.3) | 0 (0) | 0 (0) | 0 (0) | 5 |
| Staphylococcus hominis | 7 | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 5 (71.4) | 6 |
| Staphylococcus pettenkoferi | 5 | 5 (100) | 2 (40) | 5 (100) | 5 (100) | 5 (100) | 1 (20) | 5 |
| Staphylococcus schleiferi | 6 | 6 (100) | 6 (100) | 6 (100) | 4 (66.7) | 2 (33.3) | 1 (16.6) | 6 |
| Staphylococcus warneri | 6 | 6 (100) | 6 (100) | 6 (100) | 2 (33.3) | 2 (33.3) | 0 (0) | 4 |
| Total | 287 | 283 (98.6) | 232 (80.8)* | 283 (98.6) | 257 (89.5)** | 256 (89.2) | 173 (60.2) | |
*, P < 0.05 against direct colony for species-level identification;
, P < 0.05 against both direct colony and standard extraction for species-level identification.
Time required for each method.
The mean times required for 48 duplicate samples (24 strains) in each procedure were 30 min for the direct colony method, 60 min for on-plate extraction, and 180 min for the standard extraction method.
Erroneous or discordant identification.
No erroneous identifications were made, but 5 discordant results were obtained (Table 2).
TABLE 2.
Strains with discordant identification by each method
| Organism | Strain | Expt | ID and score of MALDI-TOF MSa |
|||||
|---|---|---|---|---|---|---|---|---|
| Standard extraction |
On-plate extraction |
Direct colony |
||||||
| ID | Score | ID | Score | ID | Score | |||
| Staphylococcus caprae | 54709 | 1 | Staphylococcus caprae | 2.051 | Staphylococcus caprae | 2.188 | Staphylococcus caprae | 1.797* |
| 2 | Staphylococcus pasteuri | 1.909** | Staphylococcus caprae | 2.015 | Staphylococcus caprae | 1.802* | ||
| PG1037 | 1 | Staphylococcus caprae | 2.105 | Staphylococcus caprae | 2.162 | Staphylococcus caprae | 1.876* | |
| 2 | Staphylococcus epidermidis | 1.741** | Staphylococcus caprae | 2.137 | Staphylococcus caprae | 1.751* | ||
| PG2043 | 1 | Staphylococcus caprae | 2.014 | Staphylococcus caprae | 2.16 | Staphylococcus caprae | 1.888* | |
| 2 | Staphylococcus pasteuri | 1.882** | Staphylococcus caprae | 2.123 | Staphylococcus caprae | 2.094 | ||
| PG2075 | 1 | Staphylococcus caprae | 1.749* | Staphylococcus caprae | 2.22 | Staphylococcus caprae | 1.909* | |
| 2 | Staphylococcus pasteuri | 1.703** | Staphylococcus caprae | 2.076 | Staphylococcus caprae | 1.999* | ||
| Staphylococcus warneri | 54826 | 1 | Staphylococcus warneri | 2.04 | Staphylococcus pasteuri | 1.841** | Staphylococcus warneri | 1.812* |
| 2 | Staphylococcus warneri | 2.084 | Staphylococcus warneri | 1.889 | Unreliable identification | 1.64 | ||
*, genus-level identification with correct species identification (ID);
, genus-level ID with discordant species ID.
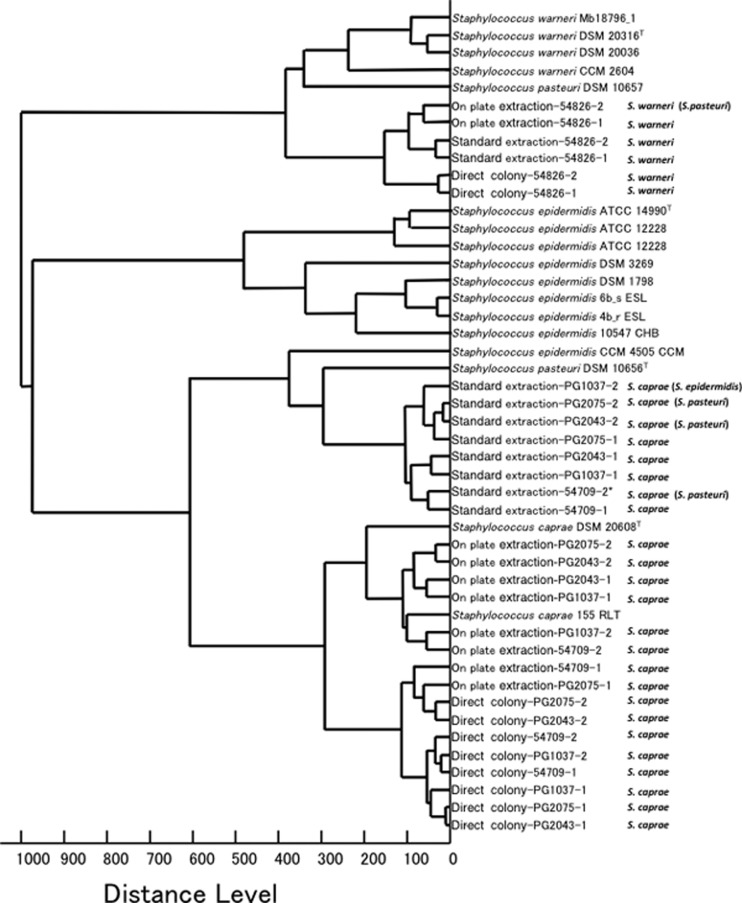
Dendrogram.
A dendrogram was created with the strains that showed discordant results and related reference strains that were installed in Biotyper 3.0 (Fig. 1). The protein signature profiles of S. caprae strains created by the standard extraction method comprised a cluster with S. epidermidis CCM4505 and S. pasteuri DSM 10656T. In contrast, the profiles obtained by the direct colony and on-plate extraction methods were closely related to the profile of S. caprae DSM 20608T (Fig. 1). The profiles of S. warneri 54826 as determined by the 3 methods were clustered into the same group as the profile of S. warneri DSM 20316T.
Fig 1.
Dendrogram derived from the MALDI-TOF MS-specific protein signatures for strains with discordant identification results, including the profiles of S. warneri, S. pasteuri, S. epidermidis, and S. caprae from the MALDI Biotyper 3.0 database. Species names in this study defined by sodA sequence are shown after sample names. Additional species in parentheses are the discordant species identified by MALDI-TOF-based identification. Distance values were relative and normalized to a maximal value of 1,000.
DISCUSSION
Since MALDI-TOF MS was first applied for the identification of microorganisms as early as 1975 (2), this system has been developed for clinical microbiology laboratories as a high-throughput apparatus (3, 4, 24). The direct identification of positive blood cultures by MALDI-TOF MS was recently reported with a species identification rate of 91% (13). Improvements in the Biotyper 3.0 software and its database may change the clinical microbiology laboratory workflow (5, 6).
For MALDI-TOF MS-based bacterial identification, 2 preparatory methods have been introduced by the manufacturer. One of these is the direct colony method, which is suited for routine workflows in the modern clinical microbiology laboratory, requiring a shorter period (less than 30 min for 48 samples) and an easier procedure. However, the method presents the problem of a low identification rate, especially for Gram-positive bacteria (1, 5). Bizzini et al. reported species identification rates by the direct colony method for Streptococcus agalactiae as 58%, Streptococcus pneumoniae as 73%, S. aureus as 79%, and S. epidermidis as 58%, using MALDI Biotyper 2.0 software (4). In this study, we used MALDI Biotyper 3.0 and achieved only slightly higher rates, i.e., 87.5% and 68.8%, respectively, for S. aureus and S. epidermidis by the direct colony method. Our higher identification rates may be due to the improvement of the Biotyper version.
Another method, the standard extraction method, has been used in the construction of a database for Biotyper and has been recommended by the manufacturer as a reference method for identifications (1, 4, 7). In this study, the identification rate of the standard extraction method was 80.8% at the species level, a better result than previously reported (1). The variability of identification rates among different studies may be explained by differences in growth conditions, sample preparation, number of reference strains, version of Biotyper software, and study design (3). In our protocol, the standard extraction method consisted of approximately 13 steps, including completion of 2 rounds of centrifugation for 2 min each, requiring approximately 180 min for 48 samples. Because this is difficult to automate, the routine use of the standard extraction method may not be suitable for primary preparation (1, 4). Standard extraction may be more suitable as a reference method for use when the on-plate method or direct colony method fails to identify the species.
The on-plate extraction method offers the advantage of a simple and easy-to-use procedure. It requires only 4 steps and 60 min for a complete identification of 48 samples. Bacterial identification rates achieved with the on-plate extraction method were at least equal to those achieved using the standard extraction method. We found that S. aureus had a higher identification rate than CoNS. Further study is needed to identify the cause of this difference.
The identification rate of S. caprae and S. saprophyticus by the on-plate method was shown to be higher than that by the standard extraction method. The other species showed smaller identification differences between the on-plate method and standard extraction methods. This indicates that for some species of staphylococci, the on-plate method may have a better identification rate than the standard extraction method. Our protocol of complete drying up of samples for standard extraction at 55°C may provide an explanation. This method may cause the degradation of proteins, shifting the protein profile used for identification. If we had chosen a different protocol, such as a dry-up time of 10 min at room temperature, the total time required for protein extraction would be shortened, potentially producing a better identification rate for standard extraction.
Furthermore, the on-plate method may be improved as Haigh et al. reported (16). They introduced a method that did not use acetonitrile, demonstrating a 10.9% improvement in the genus-level identification rates of various clinical strains (16). A shorter method will be a great help in the installation of MALDI-TOF-based bacterial identification.
There were 5 discordant results in the data set. S. caprae was identified as S. pasteuri and S. epidermidis in experiment 2 using the standard extraction method (Table 2). S. warneri was identified as S. pasteuri in experiment 1 using the on-plate extraction method. According to results from the MALDI Biotyper 3.0 database, the protein profile dendrograms for standard extractions of 4 strains of S. caprae showed clustering with S. epidermidis CCM4045 and S. pasteuri DSM10656T and separation from the cluster with S. caprae DSM 20608T (Fig. 1). The protein profile of S. warneri DSM 20316T was closely related to that of S. pasteuri DSM10657 but was separate from that of S. pasteuri DSM10656T. This discrepancy of phylogeny between the MALDI-TOF MS profile and the sodA sequence may be correlated with the discordant results obtained in our experiment. Similarly, the close genetic and protein relatedness between S. pasteuri and S. warneri might have caused the discordant results with S. warneri 54826.
The MALDI Biotyper 3.0 included only 2 strains of S. caprae as references, compared with 4 strains of S. warneri and 9 of S. epidermidis (Table 1). This small number of reference strains might have limited the accuracy in identifying S. caprae. Indeed, Seng et al. previously described the correlation between the accuracy of identification by MALDI-TOF MS and the number of reference strains (22). Similar observations have been reported by several other researchers (1, 18). Further, Lista et al. showed that additional reference strains should more accurately represent the genetic diversity of the strain (19). Therefore, proper representatives from various genetic backgrounds for each species should improve the species-level identification rates. Future studies using an improved and extended database should overcome this issue.
In conclusion, we demonstrated that the on-plate extraction method offers species identification rates at least equivalent to the results of the standard extraction method, with advantages in convenience and faster processing time, and a significantly better species identification rate than the direct colony method in Staphylococcus analysis.
Supplementary Material
ACKNOWLEDGMENT
This study was supported in part by a grant-in-aid (S0991013) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (MEXT), for the Foundation of Strategic Research Projects in Private Universities.
Footnotes
Published ahead of print 19 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anhalt JP, Fenselau C. 1975. Identification of bacteria using mass spectrometry. Anal. Chem. 47:219–225 [Google Scholar]
- 3. Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424 doi:10.1371/journal.pone.0016424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 6. Bizzini A, et al. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carbonnelle E, et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 8. Cherkaoui A, et al. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubois D, et al. 2010. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dupont C, et al. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 12. Eigner U, et al. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 13. Ferroni A, et al. 2010. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedrichs C, Rodloff AC, Chhatwal GS, Schellenberger W, Eschrich K. 2007. Rapid identification of viridans streptococci by mass spectrometric discrimination. J. Clin. Microbiol. 45:2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaillot O, et al. 2011. Cost-effectiveness of switch to matrix-assisted laser desorption ionization time of flight mass spectrometry for routine bacterial identification. J. Clin. Microbiol. 49:4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haigh J, Degun A, Eydmann M, Millar M, Wilks M. 2011. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry system in the clinical microbiology laboratory. J. Clin. Microbiol. 49:3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Konrad R, et al. 2010. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 15(43):pi=19699 [DOI] [PubMed] [Google Scholar]
- 18. Lartigue MF, et al. 2009. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 47:2284–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lista F, et al. 2011. Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol. 11:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poyart C, Quesne G, Boumaila C, Trieu-Cuot P. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saffert RT, et al. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seng P, et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 23. Sivadon V, et al. 2004. Use of sodA sequencing for the identification of clinical isolates of coagulase-negative staphylococci. Clin. Microbiol. Infect. 10:939–942 [DOI] [PubMed] [Google Scholar]
- 24. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.