Abstract
We conducted active, laboratory-based surveillance for isolates from patients with invasive infections across China from August 2009 to July 2010. DNA sequencing methods were used to define species, and susceptibility to fluconazole and voriconazole was determined by the Clinical and Laboratory Standards Institute M44-A2 disk diffusion method but using up-to-date clinical breakpoints or epidemiological cutoff values. Candida spp. made up 90.5% of the 814 yeast strains isolated, followed by Cryptococcus neoformans (7.7%) and other non-Candida yeast strains (1.7%). Bloodstream isolates made up 42.9% of the strains, isolates from ascitic fluid made up 22.1%, but pus/tissue specimens yielded yeast strains in <5% of the cases. Among the Candida isolates, Candida albicans was the most common species from specimens other than blood (50.1%) but made up only 23% of the bloodstream isolates (P < 0.001). C. parapsilosis complex species were the most common Candida isolates from blood (33.2%). Uncommon bloodstream yeast strains included Trichosporon spp., C. pelliculosa, and the novel species C. quercitrusa, reported for the first time as a cause of candidemia. Most (>94%) of the isolates of C. albicans, C. tropicalis, and the C. parapsilosis complex were susceptible to fluconazole and voriconazole, as were all of the Trichosporon strains; however, 12.2% of the C. glabrata sensu stricto isolates were fluconazole resistant and 17.8% had non-wild-type susceptibility to voriconazole. Seven C. tropicalis strains were cross-resistant to fluconazole and voriconazole; six were from patients in the same institution. Resistance to fluconazole and voriconazole was seen in 31.9% and 13.3% of the uncommon Candida and non-Candida yeast strains, respectively. Causative species and azole susceptibility varied with the geographic region. This study provided clinically useful data on yeast strains and their antifungal susceptibilities in China.
INTRODUCTION
Invasive yeast infections are a major threat to the health of hospitalized patients, particularly the critically ill, causing substantial morbidity and hospital costs (3, 12, 23, 30). Although invasive candidiasis (IC), including candidemia, remains the most common yeast infection, previously uncommon pathogens such as Trichosporon and Geotrichum species, with reduced susceptibility to antifungal agents, have emerged (23). In addition, there have been important changes in the epidemiology of IC, with an overall shift toward Candida spp. other than Candida albicans, particularly Candida glabrata, with reduced susceptibility to the azole antifungals (1, 42).
Early and appropriate antifungal therapy is essential for optimum patient outcomes in invasive yeast infections (13, 26). However, initiation of culture-directed therapy is often delayed since standard diagnostic methods are slow (2 to 3 days) and instead, antifungal strategies such as preemptive or empirical approaches are often practiced. These are reliant on robust epidemiological data to inform the selection of the initial antifungal therapy. Knowledge of local epidemiological patterns is important since epidemiological data may not be generalizable between countries because of geographic variation in epidemiology (30, 32). Thus, accurate identification of yeast pathogens and determination of their antifungal susceptibility profiles, together with surveillance data, are essential in guiding clinical decisions (31, 32).
Fluconazole is an inexpensive, effective antifungal agent often used as first-line therapy of yeast infections (25, 28). Since it does not reliably cover an increasing number of Candida species and other yeast strains, the major consideration influencing the decision to use fluconazole or other antifungal—depending on species identification and susceptibility results—is the likelihood of a fluconazole-resistant species. Voriconazole, a broad-spectrum azole, may be used in place of fluconazole for initial therapy, yet many yeast strains are cross-resistant to all azoles (27, 34). Hence, data on susceptibility to antifungals are often required for definitive treatment decisions.
Antifungal resistance surveillance programs such as the ARTEMIS Global Antifungal Surveillance Program (1997 to 2007) have been instrumental in monitoring trends in causative species and susceptibility to antifungal agents among pathogenic yeast strains (31, 32). The ARTEMIS initiative further resulted in the development of standardized Clinical and Laboratory Standards Institute (CLSI) criteria for resistance or susceptibility to fluconazole and voriconazole using disk diffusion methodology as an alternative to broth microdilution (7, 8, 27, 28, 34, 35).
Although five Chinese hospitals participated in the ARTEMIS program, contemporary data on yeast susceptibility are lacking, with many clinicians relying on earlier data to guide treatment choices, with possible adverse patient outcomes (H. Wang, personal communication). Epidemiological data on invasive yeast infections in China have been restricted either to studies in single centers or to a limited number of yeast species (20, 41). To address these deficiencies, the multicenter nationwide China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study was initiated in July 2009 to prospectively monitor trends in the epidemiology of yeast infections and provide up-to-date data on susceptibility to antifungal drugs. Here we report the results of the first year (August 2009 to July 2010) of the study, focusing on the in vitro susceptibilities of yeast strains to fluconazole and voriconazole.
MATERIALS AND METHODS
Study design.
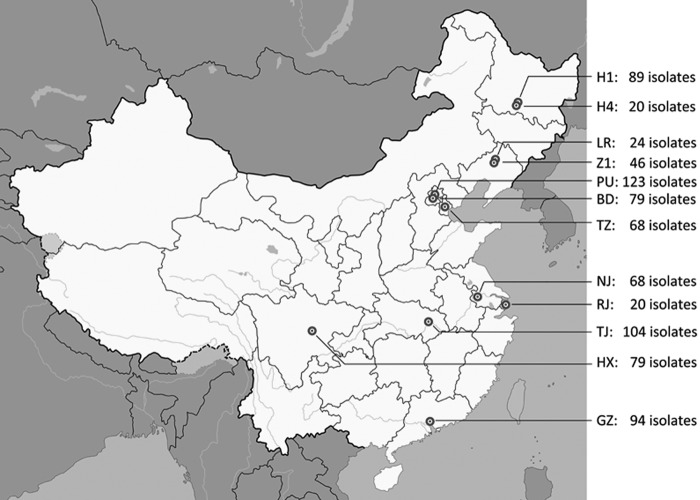
This study was a prospective, laboratory-based, multicenter study of invasive yeast infections with its inception in August 2009. A total of 12 “rank-A tertiary” hospitals (900 to 4,300 hospital beds with 1.09 to 3.15 million outpatient visits per year), serviced by a regional referral mycology laboratory, in nine major cities in China participated, i.e., two hospitals each from Beijing, Harbin, and Shenyang and one each from Chengdu, Guangzhou, Nanjing, Shanghai, Tianjin, and Wuhan (Fig. 1).
Fig 1.
Distribution of the 12 surveillance sites that participated in CHIF-NET 2010 in China. Hospital codes: BD, Peking University First Hospital; GZ, The First Affiliated Hospital of Sun Yat-Sen University; H1, The First Affiliated Hospital of Harbin Medical University; H4, the Fourth Affiliated Hospital of Harbin Medical University; HX, West China Hospital; LR, The People's Hospital of Liaoning Province; NJ, Nanjing General Hospital of PLA; PU, Peking Union Medical College Hospital; RJ, Ruijin Hospital, School of Medicine, Shanghai Jiaotong University; TJ, Tianjin Medical University General Hospital; Tongji Hospital; TZ, Tianjin Medical University General Hospital; Z1, The First Hospital of China Medical University.
For each episode of yeast isolation (see the criteria for study inclusion below), data were entered into a standard case report form. The information collected included the patient's age and gender, the patient's classification (inpatient or outpatient), and the ward location (e.g., emergency department [ED], surgical, medical, intensive care unit [ICU]) of the patient at the time of collection of the sample. The date of sample collection, the specimen type, the body site of isolation, and the initial species identification made by the referring laboratory were also recorded. A unique study identifier was assigned to each episode of yeast isolation.
Criteria for study inclusion.
All Candida, Cryptococcus, and other yeast strains recovered from blood; other sterile body fluids, including ascitic fluid and peritoneal dialysate fluid; pus; and tissue from patients with invasive yeast infections (11) were included in this study (Table 1). Yeast strains from bronchoalveolar lavage (BAL) fluid samples, central venous catheter (CVC) tips, and the gastrointestinal tracts (e.g., biliary tract fluid [aseptically collected]) of patients with invasive infections were tested; however, yeast strains from urine and the genital tract and others considered colonizers were excluded. Isolates of the same species and of the same susceptible or resistant biotype profile from the same site of a given patient that were recovered at a different time were considered duplicates and also excluded. All isolates were forwarded to a central laboratory (Department of Clinical Laboratory, Peking Union Medical College Hospital) for study.
TABLE 1.
Isolation of Candida, Cryptococcus, and other yeast species by specimen type, CHIF-NET study, 2010, China
| Specimen type(s) | No. (%) of isolates of: |
|||
|---|---|---|---|---|
| All spp. | Candida spp. | Cryptococcus spp. | Other yeast species | |
| Blood | 349 (42.9) | 322 (92.3) | 20 (5.7) | 7 (2.0) |
| Ascitic fluid | 180 (22.1) | 177 (98.3) | 1 (0.6) | 2 (1.1) |
| CVC tip | 62 (7.6) | 60 (96.8) | 1 (1.6) | 1 (1.6) |
| CSF | 62 (7.6) | 24 (38.7) | 37 (59.7) | 1 (1.6) |
| Pus (from abscesses)a | 39 (4.8) | 39 (100) | ||
| BAL fluid | 32 (3.9) | 31 (96.9) | 1 (3.1) | |
| Bile | 31 (3.8) | 31 (100) | ||
| Pleural fluid | 23 (2.8) | 22 (95.7) | 1 (4.3) | |
| Tissueb | 21 (2.6) | 16 (76.2) | 4 (19.0) | 1 (4.8) |
| Peritoneal dialysate fluid | 11 (1.4) | 11 (100) | ||
| Bone marrow | 2 (0.2) | 2 (0.3) | ||
| Eye | 2 (0.2) | 2 (0.3) | ||
| All | 814 (100) | 737 (100) | 63 (100) | 14 (100) |
Includes 29 isolates from intra-abdominal infections.
Includes 11 isolates from lung tissue, 6 from liver tissue, 1 from heart tissue, and 3 from other tissues.
Isolate identification.
Yeast strains were identified at each study center by routine mycological methods. By growth on Brilliance Candida agar (Oxoid Ltd., Hampshire, United Kingdom), Candida albicans, Candida tropicalis, Candida krusei, and C. parapsilosis complex were identified according to the manufacturer's instructions. Isolates that were not identifiable underwent analysis with the Vitek 2 Compact YST (bioMérieux, Marcy l'Etoile, France) and/or API 20C AUX (bioMérieux) system. These initial species identifications and isolates were forwarded to the central laboratory for species identity confirmation by DNA sequencing of the fungal internal transcribed spacer (ITS) region following ITS amplification using the primer pair ITS1/ITS4 as previously described (14). Sequencing of the D1/D2 domain of the 28S rRNA gene (amplified by primer pair F63/R635) (14) was performed for those isolates where ITS sequence analysis failed to produce a result (14% of the strains) (data not shown).
Agreement of the initial species identification and molecular identification results was evaluated as follows. A minor error was considered to have occurred when the (i) initial identification correctly identified an isolate to the genus level but was unable to identify a species (e.g., Candida catenulata identified as Candida spp.) or (ii) the initial identification correctly identified an isolate to the species complex level but not to the species level (e.g., Candida metapsilosis or Candida orthopsilosis identified as Candida parapsilosis complex and Candida nivariensis as C. glabrata complex) (21, 22, 24). A major error was defined as other disagreements between the initial identification and molecular identification. Definitive identification to the species level was that done by DNA sequencing.
Antifungal susceptibility testing.
Susceptibility to fluconazole and voriconazole was determined using the Clinical and Laboratory Standards Institute (CLSI) M44-A2 disk diffusion method (7). Briefly, agar plates containing BBL Mueller-Hinton II agar (BD, Sparks, MD) supplemented with 2% glucose and 0.5 μg of methylene blue per ml at a depth of 4.0 mm were used. The agar surface was inoculated by using a swab dipped in a cell suspension adjusted to a McFarland standard turbidity of 0.5 and standardized by the bioMérieux McFarland kit (bioMérieux). Fluconazole (25 μg) and voriconazole (1 μg) disks (BD) were placed onto the surfaces of the inoculated plates, and the plates were incubated in air at 37°C and read at 24 h. Slow-growing isolates, e.g., members of the genus Cryptococcus, were read after 48 h of incubation. Quality control was performed with each test run by using C. albicans ATCC 90029 and C. parapsilosis ATCC 22019.
The zone diameters of all isolates were recorded electronically. The species-specific interpretive criteria for fluconazole (susceptible [S], susceptible dose dependent [S-DD], and resistant [R]) and voriconazole (S, intermediate [I], and R) were applied to C. albicans, C. tropicalis, C. parapsilosis complex, and C. krusei isolates as described by Pfaller et al. (27, 28). C. glabrata complex isolates were categorized as S, S-DD, and R to fluconazole by species-specific clinical breakpoints (CBPs) (28), but with regard to susceptibility to voriconazole, they were categorized as wild type (WT) or non-WT (strains with acquired or mutational resistance mechanisms) by using species-specific epidemiologic cutoff values (ECVs) (27). For other yeast species, interpretation of fluconazole and voriconazole susceptibilities (S, S-DD, and R) was done in accordance with the CLSI M44-S3 guidelines (8).
Statistical analysis.
All comparisons were performed using SPSS software version 12.0 (SPSS Inc., Chicago, IL). Comparisons of continuous variables were performed by using the Mann-Whitney test, and comparisons of categorical variables were performed by using a chi-square test or Fisher's exact test, as appropriate. A P value of 0.05 was considered significant.
(He Wang presented results of the CHIF-NET10 study at the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom, 31 March to 3 April 2012.)
RESULTS
Isolates.
A total of 814 yeast isolates were studied (range, 20 to 123 isolates from each hospital; Fig. 1). Candida species were the most common yeast strains isolated (737/814; 90.5%), with Cryptococcus neoformans and non-Candida yeast strains comprising 7.7% (n = 63) and 1.7% (n = 14) of the isolates, respectively. Blood culture isolates made up 42.9% of the yeast strains, and ascitic fluid accounted for 22.1%, while pus from deep abscesses and tissue biopsy specimens yielded yeast strains uncommonly (<5%; Table 1).
Candida species isolates were broadly distributed and made up the majority of the blood culture isolates (92.3%; Table 1). Cryptococci and non-Candida yeast strains accounted for 7.7% of the yeast strains from blood cultures. The majority of the yeast strains from ascitic fluid (96.7%) and CVC tips (98.4%) were Candida spp., and Candida spp. were the only yeast isolates from pus. Conversely, 59.7% of the yeast strains from cerebrospinal fluid (CSF) samples were C. neoformans. There was a single isolate of Trichosporon asahii from a CSF sample (Table 1).
Patient location.
Of the isolates recovered, 92.4% were from hospital inpatients (ICUs, 34.3%; medical wards, 27.8%; surgical wards, 30.3%) and 7.6% were from the outpatient/ED setting (outpatient clinics, 2.6%; EDs, 5.6%) (Table 2). The predominant patient location varied with the pathogen group. Candida and non-Candida yeast strains were most often isolated from inpatients in ICUs and surgical wards (40% and 41.1% of the isolates, respectively) (Table 2). C. neoformans was more likely to be isolated from patients in medical wards (44/58 cases, 75.8%) than were Candida spp. (57/680 or 18.3%; P < 0.001) and were not recovered from ICU patients.
TABLE 2.
Distribution of Candida, Cryptococcus, and other yeast spp. by clinical service
| Organism | No. of isolates tested (% of total) | No. of isolates |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Inpatient |
Outpatient/emergency |
||||||||
| Total | ICU | Medicine | Surgery | Othera | Total | Emergency | Outpatient | ||
| Candida spp. | 737 (90.5) | 680 | 272 | 124 | 226 | 58 | 57 | 37 | 20 |
| Cryptococcus spp. | 63 (7.7) | 58 | 44 | 10 | 4 | 5 | 4 | 1 | |
| Other yeast spp. | 14 (1.7) | 14 | 7 | 1 | 4 | 2 | |||
| Total | 814 (100) | 752 | 279 | 169 | 240 | 64 | 62 | 41 | 21 |
Includes pediatrics, dermatology, obstetrics and gynecology, and ophthalmology services.
Yeast species.
A total of 27 yeast species were identified by sequencing of the ITS and/or D1/D2 regions of the rRNA gene during this study.
Initial species identification was concordant with the molecular identification of 688/814 (84.5%) isolates (Table 3; see Table S1 in the supplemental material). Major errors occurred in 99 identifications (12.2% of the cases, involving 21 species), and minor errors occurred in 27 (3.3%) isolate identifications (eight species). Initial identification of C. albicans, C. parapsilosis (sensu stricto), C. tropicalis, C. glabrata (sensu stricto), and C. neoformans showed good agreement (86.2 to 97.2%) with molecular identification; however, for the remaining yeast species (n = 113 isolates, 22 species), the overall agreement was significantly lower (mean, 35.4%); for example, all three Candida quercitrusa strains were identified as Candida lusitaniae (see Table S1 in the supplemental material). All isolates were identified to the species level by DNA sequencing.
TABLE 3.
Species distribution of 814 yeast strains isolates and comparison of molecular and initial identification results
| Organism | No. of isolates (% of all yeast isolates) | Molecular vs initial identification resultsa |
No. of blood/nonblood isolates | ||
|---|---|---|---|---|---|
| % agreement | % of identification errorsb |
||||
| Major | Minor | ||||
| Candida species | 737 (90.5) | 84.3 | 12.2 | 3.5 | 322/415 |
| C. albicans | 282 (34.6) | 97.2 | 2.8 | 74/208 | |
| C. parapsilosis species complex | |||||
| C. parapsilosis sensu stricto | 143 (17.6) | 91.6 | 7 | 1.4 | 90/53 |
| C. metapsilosis | 22 (2.7) | 36.4 | 63.6 | 11/11 | |
| C. orthopsilosis | 4 (0.5) | 100 | 4/0 | ||
| L. elongisporus | 3 (0.4) | 100 | 2/1 | ||
| C. tropicalis | 123 (15.1) | 86.2 | 11.4 | 2.4 | 48/75 |
| C. glabrata species complex | |||||
| C. glabrata sensu stricto | 90 (11.1) | 86.7 | 12.2 | 1.1 | 50/40 |
| C. nivariensis | 2 (0.2) | 50 | 50 | 1/1 | |
| C. krusei | 18 (2.2) | 83.3 | 16.7 | 5/13 | |
| C. guilliermondii | 12 (1.5) | 41.7 | 50 | 8.3 | 8/4 |
| C. pelliculosa | 12 (1.5) | 25 | 75 | 8/4 | |
| C. lipolytica | 10 (1.2) | 100 | 9/1 | ||
| C. lustitaniae | 6 (0.7) | 83.3 | 16.7 | 4/2 | |
| C. quercitrusa | 3 (0.4) | 100 | 3/0 | ||
| C. catenulata | 2 (0.2) | 100 | 2/0 | ||
| C. fabianii | 1 (0.1) | 100 | 1/0 | ||
| C. famata | 1 (0.1) | 100 | 1/0 | ||
| C. haemulonii | 1 (0.1) | 100 | 1/0 | ||
| C. kefyr | 1 (0.1) | 100 | 0/1 | ||
| C. norvegensis | 1 (0.1) | 100 | 0/1 | ||
| C. neoformans | 63 (7.7) | 93.7 | 6.3 | 20/43 | |
| Other yeast species | 14 (1.7) | 57.1 | 35.7 | 7.1 | 7/7 |
| Geotrichum capitum | 2 (0.2) | 100 | 1/1 | ||
| Kodamaea ohmeri | 2 (0.2) | 100 | 1/1 | ||
| Meyerozyma caribbica | 1 (0.1) | 100 | 1/0 | ||
| Rhodotorula mucilaginosa | 1 (0.1) | 100 | 1/0 | ||
| Trichosporon asahii | 7 (0.9) | 85.7 | 14.3 | 3/4 | |
| Trichosporon dermatis | 1 (0.1) | 100 | 0/1 | ||
| Total | 814 (100) | 84.5 | 12.2 | 3.3 | 349/465 |
Initial identification was performed at each surveillance site by phenotypic and biochemical methods; molecular identification was performed by sequencing of the ITS region and/or D1/D2 domain of the 28S rRNA gene.
A minor error was defined as (i) correct identification of an isolate to the genus level but inability to identify it to the species level (e.g., C. catenulata identified as Candida spp. by phenotypic methods) or (ii) initial correct identification of an isolate to the species complex level but not to the species level (e.g., C. metapsilosis or C. orthopsilosis identified as C. parapsilosis complex and C. nivariensis as C. glabrata complex). A major error was defined as other disagreements between the initial identification and molecular identification.
Overall, C. albicans was the most common Candida species (282/737 isolates, 38.3%), followed by C. parapsilosis complex (23.3%) and C. tropicalis (16.7%) (Table 3). All 63 strains of Cryptococcus were C. neoformans; there were no isolates of Cryptococcus gattii. Trichosporon species were uncommon (n = 8 isolates; 1% of all yeast strains). Of 172 isolates of the C. parapsilosis complex, 22 (12.8%) were C. metapsilosis, 4 (2.3%) were C. orthopsilosis, and 3 (1.8%) were Lodderomyces elongisporus. Only 2/92 C. glabrata complex isolates were C. nivariensis.
Comparison of blood isolates with those recovered from sites other than blood.
Among Candida spp., C. albicans was significantly more likely to be recovered from sites other than blood (50.1% of the Candida isolates) than from blood cultures (23%; P < 0.001). In contrast, the C. parapsilosis complex—C. parapsilosis sensu stricto (n = 90 isolates), C. metapsilosis (n = 11), C. orthopsilosis (n = 4), and L. elongisporus (n = 2)—the most common cause of candidemia, was disproportionately represented in blood cultures compared with nonblood specimens (107/322 isolates, 33.2%, versus 65/415 isolates, 15.6%; P < 0.001). The likelihood of isolating C. glabrata, including C. nivariensis (n = 2) and uncommon Candida spp. such as C. quercitrusa, from blood was also significantly higher (P = 0.01 and P < 0.001, respectively, Table 3). Three of eight Trichosporon isolates were blood culture isolates (all T. asahii), and the remaining five were from ascitic fluid (n = 2), BAL fluid, CSF, and a CVC tip (n = 1 each). Other rare yeast strains found in blood were Geotrichum capitum, Kodamaea ohmeri, Meyerozyma caribbica, and Rhodotorula mucilaginosa (n = 1 each).
In vitro susceptibilities.
The majority (>94%; Table 3) of the C. albicans, C. parapsilosis group, and C. tropicalis strains were susceptible to fluconazole. Among the C. glabrata complex strains, 12.2% (11/90) of the C. glabrata sensu stricto isolates were categorized as resistant to fluconazole and 87.8% were S-DD, as were both C. nivariensis isolates. All Trichosporon strains were susceptible. Fluconazole-resistant yeast isolates were Candida krusei and uncommon Candida species, including Candida pelliculosa and C. quercitrusa. The rate of resistance to fluconazole among uncommon Candida/non-Candida yeast strains was 31.9%.
Overall, most yeast strains were susceptible to voriconazole. Sixteen (17.4%) of 92 C. glabrata isolates were categorized as non-WT according to species-specific ECVs. In addition, voriconazole resistance was present in 7/123 C. tropicalis isolates (5.7%), 2 C. krusei isolates, C. catenulata, Candida lipolytica, C. pelliculosa, and R. mucilaginosa (Table 4). The rate of resistance to voriconazole among uncommon Candida/non-Candida yeast strains was 13.3%. Of 51 fluconazole-resistant non-C. glabrata isolates, nine strains of C. lipolytica, seven of C. tropicalis, two of C. catenulata and C. krusei, and one each of C. albicans, C. pelliculosa, and R. mucilaginosa (23 isolates in all) were cross-resistant to voriconazole. No isolate was voriconazole resistant but susceptible to fluconazole.
TABLE 4.
In vitro susceptibilities to fluconazole and voriconazole of 814 yeast strain isolates in the CHIF-NET study, 2010
| Organism |
In vitro susceptibilitya to: |
|||
|---|---|---|---|---|
| Fluconazole |
Voriconazole |
|||
| % S | % R | % S | % R | |
| C. albicans | 99.3 | 0.4 | 99.3 | 0.7 |
| C. parapsilosis complex | ||||
| C. parapsilosis sensu stricto | 98.6 | 1.4 | 100 | 0 |
| C. metapsilosis | 100 | 0 | 100 | 0 |
| C. orthopsilosis | 100 | 0 | 100 | 0 |
| L. elongisporus | 100 | 0 | 100 | 0 |
| C. tropicalis | 94.3 | 5.7 | 94.3 | 5.7 |
| C. glabrata complex | ||||
| C. glabrata sensu stricto | NAb | 12.2 | 82.2c | 17.8c |
| C. nivariensis | NA | 0 | 100c | 0c |
| C. krusei | 0 | 100 | 83.3 | 11.1 |
| C. guilliermondii | 100 | 0 | 100 | 0 |
| C. pelliculosa | 50.0 | 33.3 | 66.7 | 8.3 |
| C. lipolytica | 10.0 | 90.0 | 10.0 | 90.0 |
| C. lustitaniae | 83.3 | 0 | 100 | 0 |
| C. quercitrusa | 0 | 100 | 100 | 0 |
| C. catenulata | 0 | 100 | 0 | 100 |
| C. fabianii | 100 | 0 | 100 | 0 |
| C. famata | 100 | 0 | 100 | 0 |
| C. haemulonii | 100 | 0 | 100 | 0 |
| C. kefyr | 100 | 0 | 100 | 0 |
| C. norvegensis | 100 | 0 | 100 | 0 |
| C. neoformans | 90.5 | 6.3 | 100 | 0 |
| Other yeast species | ||||
| Geotrichum capitum | 100 | 0 | 100 | 0 |
| Kodamaea ohmeri | 100 | 0 | 100 | 0 |
| Meyerozyma caribbica | 100 | 0 | 100 | 0 |
| Rhodotorula mucilaginosa | 0 | 100 | 0 | 100 |
| Trichosporon asahii | 85.7 | 0 | 100 | 0 |
| Trichosporon dermatis | 100 | 0 | 100 | 0 |
Susceptibility data were interpreted (i) by species-specific interpretive criteria for C. albicans, C. tropicalis, C. parapsilosis complex, the C. glabrata complex, and C. krusei as described by Pfaller et al. (27, 28) and (ii) in accordance with CLSI document M44-S3 (8) for other yeast species.
The latest species-specific interpretive criteria for C. glabrata complex susceptibility to fluconazole were defined as ≤14 mm for R and ≥15 mm for S-DD (28); thus, category S is not applicable (NA) to the C. glabrata complex.
As CBPs for C. glabrata complex susceptibility to voriconazole are not available, the ECV (a zone diameter of ≥16 mm) was used to differentiate wild-type (listed in category S) from non-wild-type (listed in category R) strains of this complex, as described by Pfaller et al. (27).
Geographic variation in species distribution and in vitro susceptibilities.
C. albicans was the most common pathogen in all of the study centers, except site LR (northeastern China) and site HX (central China), where C. glabrata sensu stricto (8/24 yeast isolates, 33.3%) and C. parapsilosis sensu stricto (21/79 isolates, 26.6%) were predominant, respectively (Fig. 1 and 2). C. neoformans was recovered in <10% of the cases, except at sites HX and TJ (21.5% and 16.2% of the yeast isolations, respectively).
Fig 2.
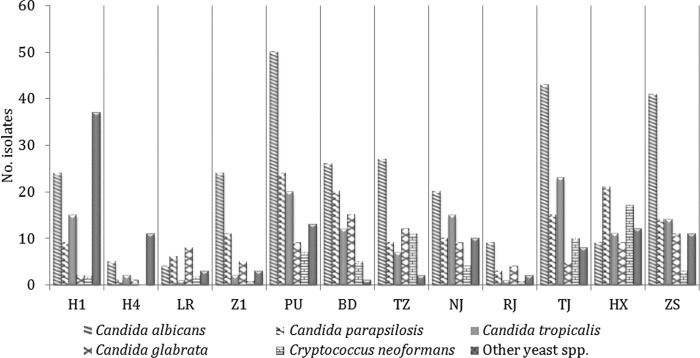
Geographic variations of C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, Cryptococcus neoformans, and other yeast species at 12 surveillance sites.
Of note, uncommon Candida or non-Candida yeast species were relatively common at sites H1 and H4, Harbin, northeastern China (Fig. 2). Further, a greater proportion of isolates from these two centers were fluconazole resistant (27% at H1 and 20% at H4), as well as being voriconazole resistant and in the non-WT category (19.1% at H1, 10% at H4). This compares with resistance rates of 2.5 to 15% for fluconazole and 0 to 16.7% for voriconazole at other centers (data not shown). Three fluconazole-resistant yeast species (C. catenulata, C. lipolytica, and C. quercitrusa, 14 isolates in all) were identified only at H1, and one species (C. pelliculosa, 4 isolates) was identified only at H4. Six of seven C. tropicalis isolates cross-resistant to fluconazole and voriconazole were identified at H1, including five isolates that were identified over 35 days (September to October in 2009) and one that was identified in July 2010.
DISCUSSION
The epidemiology of invasive yeast infections and the associated antifungal susceptibility patterns are poorly defined in China. Despite some data on susceptibility to antifungal agents reported through the ARTEMIS study (31, 32), these were not clearly separate from those of other Asia-Pacific regions, more recent data are lacking, and there is no ongoing surveillance. The results of the first year of this national surveillance program have provided clinically relevant data regarding causative pathogens and in vitro susceptibility to fluconazole and voriconazole; important regional differences were identified.
Accurate identification of yeast strains by using robust molecular methods (14, 15, 19) was essential to the aims of the study. Though the overall agreement between phenotypic and molecular identification was 84.5%, the rate of major errors was high (14.3 to 100%) for yeast strains, including C. krusei and rare Candida species (Table 3; see Table S1 in the supplemental material). This is not surprising, since yeast isolates may be “misidentified” on chromogenic media because of atypical color and because of database limitations of phenotypic identification systems, particularly for uncommon yeast strains. As with other studies, molecular but not phenotypic methods distinguished species within the C. parapsilosis complex (21, 22, 24) and the C. glabrata complex (4, 10). The results are also noteworthy for having identified three cases of bloodstream infection due to C. quercitrusa, highlighting the utility of molecular methods in identifying novel pathogens. Although molecular tools were used in the present study, their use in the routine identification of yeast strains is not feasible for most clinical microbiology laboratories in China because of high costs and a lack of standardized methods and quality control procedures. The contribution of active surveillance networks such as CHIF-NET is thus pivotal to providing these data.
IC is the most common invasive yeast infection, and the predominance of Candida isolates (90.5%) was expected. Yet although it was the most common Candida species overall, C. albicans made up only 38.3% of the Candida yeast strains (Table 3), a percentage substantially lower than those reported in other large surveys: 62.6% in the multicenter ARTEMIS study (1997 to 2007) (31, 32) and 46.1% in the SENTRY (2009) program (29). Instead, we noted a higher proportion of Candida species other than C. albicans, including species of the C. parapsilosis complex (23.3% of the Candida isolates, compared with 5.8% and 1.3% in the ARTEMIS and SENTRY surveys, respectively). While these large surveys analyzed species distribution according to the continent of origin, they did not consider potential differences within regions (30, 32). Additionally, in the present study, but not in the ARTEMIS study, sputum, urine, and genital tract samples were excluded from the analysis; C. albicans is the most common Candida species isolated from these clinical specimens (33).
A key finding of this study is the confirmation that trends in causative species of Candida bloodstream infections are not transferable between regions. While C. albicans has remained the most common etiological agent (37 to 70%) worldwide (2, 5, 9, 38, 39), we found the C. parapsilosis complex to be the most prevalent species in blood cultures (33.2% versus 23% for C. albicans). The high prevalence of C. parapsilosis candidemia in China is not readily explained. C. parapsilosis is associated with catheter-related fungemia (32, 40) and is known to colonize skin; evaluation of CVC care and infection control practices may be helpful in identifying risk factors for C. parapsilosis candidemia. C. glabrata candidemia (15.8% of the cases) was less prevalent than that reported in the United States (24 to 25%) (15, 17) but more so than in certain areas of Latin America and Europe (3 to 8% of the cases) (9, 37).
Few large surveys have described patterns of causative Candida species in nonbloodstream infections. The present study provides clinically relevant data, at least for Chinese physicians, in that 50.1% of the Candida isolates belonged to C. albicans, with C. tropicalis the second most common species (18.1%); C. parapsilosis complex and C. glabrata complex isolates were recovered in only 15.7% and 9.9% of the cases, respectively. It could be argued, although this should be supported by additional data, that in this context, selection of initial antifungal therapy need not necessarily encompass species with reduced susceptibility to azoles, e.g., C. glabrata.
Recently, a set of new interpretive criteria for disk diffusion testing with regard to fluconazole and voriconazole susceptibility has been recommended (27, 28). Using species-specific CBPs for C. albicans, C. parapsilosis complex, C. tropicalis, and C. krusei and ECVs for the voriconazole susceptibility of C. glabrata, overall, most of the yeast strains were susceptible to the two azoles, although susceptibility varied with the species. Azole resistance was uncommon in C. albicans, C. parapsilosis, and C. tropicalis (<10%), consistent with previous reports (29, 32). However, seven C. tropicalis strains were resistant to fluconazole, as well as voriconazole, and six of these seven were recovered from patients at a single location (site H1 in Harbin) within 35 days, all in the ICU. Clusters of infections due to C. tropicalis have been reported (6, 18). Other Candida species, e.g., C. krusei, have also been reported to cause clusters of infections (16). We are in the process of investigating the genetic relatedness of the C. tropicalis strains as the cause of a possible cluster with its infection control implications. Further, a larger proportion of resistant yeast species were from sites H1 and H4. This is due partly to the larger proportion of infections caused by uncommon Candida spp. at these sites (e.g., C. quercitrusa was isolated only at site H1, C. lipolytica was isolated from patients at site H1, and C. pelliculosa was isolated at site H4).
Interpretive criteria for azole susceptibility using disk diffusion methods have not yet been published for non-Candida yeast strains. We used the criteria in the CLSI M44-S3 document to assess susceptibility to enable result comparison (8, 31). Resistance to fluconazole was seen in only four C. neoformans isolates, and all were voriconazole susceptible. Historically, fluconazole resistance is uncommon in C. neoformans, although resistance rates may vary with the geographic region and be increasing (31, 36). In the ARTEMIS study, 7.3% of the strains were resistant to fluconazole from 1997 to 2000 and 11.7% were resistant to fluconazole from 2005 to 2007 (31).
In conclusion, this study has provided clinically useful data on the epidemiology of invasive yeast infections in China. Molecular methods are essential for the identification of uncommon yeast strains. C. parapsilosis was the most common pathogen in bloodstream infections due to Candida spp. but with geographical variation. Both fluconazole and voriconazole demonstrated good activity against C. albicans, C. parapsilosis, C. tropicalis, and C. neoformans but not against C. glabrata; cross-resistance to both azoles was noted in C. glabrata and uncommon yeast strains. Continued surveillance is warranted.
Supplementary Material
ACKNOWLEDGMENT
The CHIF-NET surveillance program is supported by an Investigator-Initiated Research grant from Pfizer, China.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 3 October 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Arendrup MC, et al. 2008. Semi-national surveillance of fungaemia in Denmark 2004-2006: increasing incidence of fungaemia and numbers of isolates with reduced azole susceptibility. Clin. Microbiol. Infect. 14:487–494 [DOI] [PubMed] [Google Scholar]
- 2. Arendrup MC, et al. 2011. Diagnostic issues, clinical characteristics, and outcomes for patients with fungemia. J. Clin. Microbiol. 49:3300–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold HM, et al. 2010. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy 30:361–368 [DOI] [PubMed] [Google Scholar]
- 4. Borman AM, et al. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 46:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen S, et al. 2006. Active surveillance for candidemia, Australia. Emerg. Infect. Dis. 12:1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chou HH, Lo HJ, Chen KW, Liao MH, Li SY. 2007. Multilocus sequence typing of Candida tropicalis shows clonal cluster enriched in isolates with resistance or trailing growth of fluconazole. Diagn. Microbiol. Infect. Dis. 58:427–433 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2009. Method for antifungal disk diffusion susceptibility testing of yeasts; approved guideline, 2nd ed, M44-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2009. Zone diameter interpretive standards, corresponding minimal inhibitory concentration (MIC) interpretive breakpoints, and quality control limits for antifungal disk diffusion susceptibility testing of yeasts; informational supplement, 3rd ed, M44-S3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Colombo AL, et al. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cuenca-Estrella M, et al. 2011. Prevalence of Candida bracarensis and Candida nivariensis in a Spanish collection of yeasts: comparison of results from a reference centre and from a population-based surveillance study of candidemia. Med. Mycol. 49:525–529 [DOI] [PubMed] [Google Scholar]
- 11. De Pauw B, et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Falagas ME, Apostolou KE, Pappas VD. 2006. Attributable mortality of candidemia: a systematic review of matched cohort and case-control studies. Eur. J. Clin. Microbiol. Infect. Dis. 25:419–425 [DOI] [PubMed] [Google Scholar]
- 13. Garey KW, et al. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin. Infect. Dis. 43:25–31 [DOI] [PubMed] [Google Scholar]
- 14. Guo LN, et al. 2011. Three-locus identification, genotyping, and antifungal susceptibilities of medically important Trichosporon species from China. J. Clin. Microbiol. 49:3805–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hajjeh RA, et al. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hautala T, et al. 2007. A cluster of Candida krusei infections in a haematological unit. BMC Infect. Dis. 7:97 doi:10.1186/1471-2334-7-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Horn DL, et al. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48:1695–1703 [DOI] [PubMed] [Google Scholar]
- 18. Jang SJ, et al. 2005. PFGE-based epidemiological study of an outbreak of Candida tropicalis candiduria: the importance of medical waste as a reservoir of nosocomial infection. Jpn. J. Infect. Dis. 58:263–267 [PubMed] [Google Scholar]
- 19. Leaw SN, et al. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C-R, Peng S-H, Li D, Li Y. 2002. A hospital acquired infection of deep fungus: a clinical investigation and drug resistance. Chin. J. Nosocomiol. 12:485–487 [Google Scholar]
- 21. Lockhart SR, Messer SA, Pfaller MA, Diekema DJ. 2008. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J. Clin. Microbiol. 46:2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lockhart SR, Messer SA, Pfaller MA, Diekema DJ. 2008. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of bloodstream infections. J. Clin. Microbiol. 46:374–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miceli MH, Diaz JA, Lee SA. 2011. Emerging opportunistic yeast infections. Lancet Infect. Dis. 11:142–151 [DOI] [PubMed] [Google Scholar]
- 24. Mirhendi H, et al. 2011. Differentiation of Candida glabrata, C. nivariensis and C. bracarensis based on fragment length polymorphism of ITS1 and ITS2 and restriction fragment length polymorphism of ITS and D1/D2 regions in rDNA. Eur. J. Clin. Microbiol. Infect. Dis. 30:1409–1416 [DOI] [PubMed] [Google Scholar]
- 25. Pappas PG, et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parkins MD, Sabuda DM, Elsayed S, Laupland KB. 2007. Adequacy of empirical antifungal therapy and effect on outcome among patients with invasive Candida species infections. J. Antimicrob. Chemother. 60:613–618 [DOI] [PubMed] [Google Scholar]
- 27. Pfaller MA, et al. 2011. Clinical breakpoints for voriconazole and Candida spp. revisited: review of microbiologic, molecular, pharmacodynamic, and clinical data as they pertain to the development of species-specific interpretive criteria. Diagn. Microbiol. Infect. Dis. 70:330–343 [DOI] [PubMed] [Google Scholar]
- 28. Pfaller MA, Andes D, Diekema DJ, Espinel-Ingroff A, Sheehan D. 2010. Wild-type MIC distributions, epidemiological cutoff values and species-specific clinical breakpoints for fluconazole and Candida: time for harmonization of CLSI and EUCAST broth microdilution methods. Drug Resist. Updat. 13:180–195 [DOI] [PubMed] [Google Scholar]
- 29. Pfaller MA, Castanheira M, Messer SA, Moet GJ, Jones RN. 2011. Echinocandin and triazole antifungal susceptibility profiles for Candida spp., Cryptococcus neoformans, and Aspergillus fumigatus: application of new CLSI clinical breakpoints and epidemiologic cutoff values to characterize resistance in the SENTRY Antimicrobial Surveillance Program (2009). Diagn. Microbiol. Infect. Dis. 69:45–50 [DOI] [PubMed] [Google Scholar]
- 30. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfaller MA, et al. 2009. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: 10.5-year analysis of susceptibilities of noncandidal yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 47:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfaller MA, et al. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfaller MA, et al. 2007. Results from the ARTEMIS DISK Global Antifungal Surveillance study, 1997 to 2005: an 8.5-year analysis of susceptibilities of Candida species and other yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 45:1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfaller MA, et al. 2006. Correlation of MIC with outcome for Candida species tested against voriconazole: analysis and proposal for interpretive breakpoints. J. Clin. Microbiol. 44:819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfaller MA, Diekema DJ, Sheehan DJ. 2006. Interpretive breakpoints for fluconazole and Candida revisited: a blueprint for the future of antifungal susceptibility testing. Clin. Microbiol. Rev. 19:435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfaller MA, et al. 2005. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004). J. Clin. Microbiol. 43:2163–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabino R, et al. 2010. Epidemiology of candidemia in oncology patients: a 6-year survey in a Portuguese central hospital. Med. Mycol. 48:346–354 [DOI] [PubMed] [Google Scholar]
- 38. Sandven P, et al. 2006. Candidemia in Norway (1991 to 2003): results from a nationwide study. J. Clin. Microbiol. 44:1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sofair AN, et al. 2006. Epidemiology of community-onset candidemia in Connecticut and Maryland. Clin. Infect. Dis. 43:32–39 [DOI] [PubMed] [Google Scholar]
- 40. Trofa D, Gacser A, Nosanchuk JD. 2008. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 21:606–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu X-L, Yang P-H, Sun Y-Q, Fan X. 2007. Deep fungi infection: flora distribution and drug resistance. Chin. J. Nosocomiol. 17:349–351 [Google Scholar]
- 42. Zilberberg MD, Shorr AF, Kollef MH. 2008. Secular trends in candidemia-related hospitalization in the United States, 2000-2005. Infect. Control Hosp. Epidemiol. 29:978–980 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.