Abstract
We detected a 3% prevalence rate for human parechovirus (HPeV) in 720 respiratory specimens collected from 637 children seen in our hospital in 2009. Fifteen of 20 were HPeV-3 and two were HPeV-1. Only nonspecific, modest respiratory symptoms were evident in patients with detectable HPeV in respiratory specimens. Seven patients had concurrent respiratory and central nervous system (CNS) HPeV-3 infection, suggesting a possible respiratory route of acquisition.
TEXT
The human parechoviruses (HPeVs) are increasingly detected worldwide as pathogens, particularly in infants and children (1–3, 5–7, 9, 13, 15–18, 20, 23). They are single-stranded, positive-sense RNA viruses previously categorized within the Enterovirus genus. However, genetic differences from enteroviruses led to reclassification in a separate genus in Picornaviridae (8). Sixteen HPeV types have been discovered, with varied tropisms ranging from the respiratory and gastrointestinal tracts to the central nervous system (CNS) (12). HPeV-1 and HPeV-2, formerly echoviruses 22 and 23, respectively, were initially the most commonly isolated in culture. Because culture has lower sensitivity than current molecular techniques, the true incidence of infection, distribution of genotypes, and clinical spectrum of HPeV disease remain unclear.
To better characterize HPeV infections in the United States, we performed a retrospective and descriptive study at the Children's Mercy Hospitals and Clinics (CMH) in Kansas City, MO and KS. We hypothesized that HPeV infects the respiratory tract of children in the United States but has not been frequently detected because it is not included in routine respiratory viral testing. We noticed a relatively high incidence of HPeV-3 CNS infections in 2009 (18a). Hence, we investigated the occurrence of HPeV respiratory infections during the same year. Individual respiratory specimens were eligible for HPeV testing if no other virus had been detected by Luminex xTAG multiplex respiratory viral panel PCR (RVP PCR) testing performed per routine clinical care. The RVP PCR panel included 12 viruses and subtypes: influenza virus (A, B, H1, H3, unsubtypeable), respiratory syncytial virus (A, B), parainfluenza virus (1, 2, 3), adenovirus, human metapneumovirus, and rhinovirus/enterovirus. The 2009 pandemic H1N1 influenza strain is reported by this RVP PCR as FluA unsubtypeable.
During calendar year 2009, a total of 2,359 respiratory specimens (either nasal aspirates or nasopharyngeal swabs) were submitted for RVP PCR testing. Frozen aliquots of total nucleic acid extracts from 720 RVP-negative specimens from 637 children were included in this study and tested by a two-step real-time HPeV reverse transcription (RT)-PCR assay as previously described (3, 18), with substitution of the AgPath-ID one-step RT-PCR enzyme for the second-step real-time PCR amplification. For all HPeV-positive specimens, the cDNA and an aliquot of nucleic acid extract were shipped to the Centers for Disease Control and Prevention (CDC) for confirmation by HPeV RT-PCR and for nucleotide sequencing of the VP1 region to determine HPeV type, as previously described (14). All patients had been evaluated by health care providers within the CMH system during their care in 2009. We reviewed records from that health care visit and their electronic medical record and collected clinical, laboratory, and epidemiologic characteristics of the patients to evaluate characteristics of patients with detectable HPeV in respiratory samples. The study was approved by the Institutional Review Board at CMH.
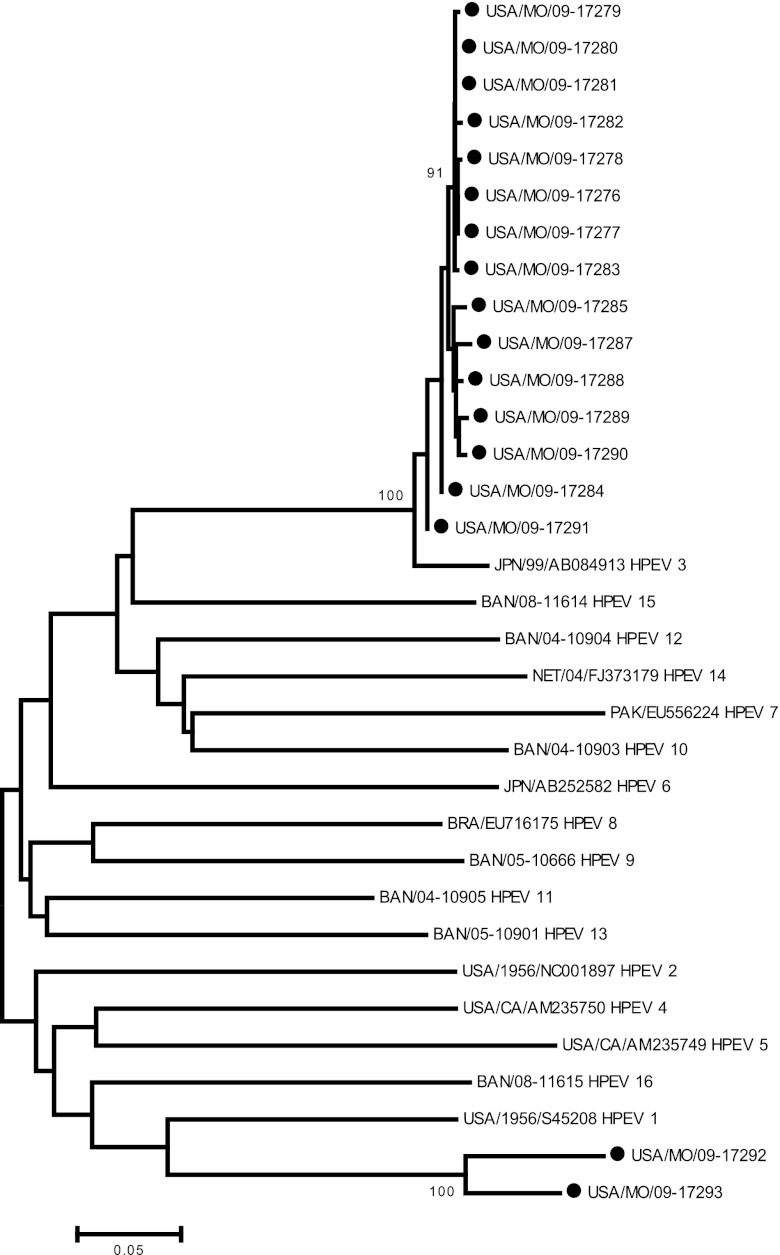
HPeV was detected from March to November with the highest prevalence in July and August. HPeV was detected in 20 of 720 specimens from 19/637 unique individuals, resulting in a 3% prevalence rate. Molecular typing identified 15/20 samples as HPeV-3 and 2 as HPeV-1. Three specimens could not be typed. The HPeV-3 VP1 sequences were closely related to one another phylogenetically (Fig. 1). All 19 HPeV-positive patients were less than 3 years of age (median, 34 days); 79% were less than 3 months old (Table 1). While 14 HPeV-positive patients presented with modest clinical symptoms, five patients exhibited signs that the clinician considered indicative of septic shock with poor perfusion, requiring volume resuscitation. Severe respiratory symptoms were universally absent, with mild symptoms being more common. Bacterial and/or fungal respiratory cultures were performed only on one subject with negative results. Chest radiograph abnormalities were noted on three of 20 patients; findings included one each with perihilar infiltrate, increased perihilar interstitial markings, and right middle lobe/lingular pneumonia versus atelectasis. Of the 14 patients who underwent diagnostic lumbar puncture, none demonstrated pleocytosis. Eleven of the 14 CSF specimens had also been tested by a two-step HPeV RT-PCR assay in the 2009 HPeV-CNS infection study (18a), and 7 CSF specimens were found to be positive for HPeV. Bacterial blood cultures from all 19 HPeV-positive subjects were negative. Bordetella pertussis was isolated from one HPeV-positive patient, and urine culture yielded methicillin-susceptible Staphylococcus aureus in another.
Fig 1.
Dendrogram of VP1 sequences from HPeV isolated from respiratory specimens in children from Kansas City and HPeV types 1 to 16. Evolutionary history was inferred using the neighbor-joining method. HPeV viral isolates are indicated by the country of origin: BAN, Bangladesh; BRA, Brazil; JPN, Japan; PAK, Pakistan; NET, Netherlands; USA, United States.
TABLE 1.
Clinical characteristics of patients with HPeV respiratory infectiona
| Characteristic | Value | Range |
|---|---|---|
| No. of patients with HPeV | 19 | |
| Age, mean ± 1 SD (in days) | 144 ± 256 | 10–817 |
| No. of males/females | 16/3 | |
| No. hospitalized (yes/no) | 17/2 | |
| Mean no. of hospital days | 5.3 ± 3.4 | 2–15 |
| No. with full-term gestation | 19 | |
| No. with fever, by history (yes/no) | 13/6 | |
| Mean admission temp (°C) | 37.6 ± 0.7 | 36.7–38.1 |
| Mean Tmax in hospital (°C) | 38.8 ± 1.0 | 37.0–40.3 |
| No. of days with fever | 3.2 ± 1.6 | 1–5 |
| No. with irritability | 15 | |
| No. with respiratory symptoms | 8 | |
| Congestion | 3/8 | |
| Cough | 2/8 | |
| Grunting | 3/8 | |
| Tachypnea | 1/8 | |
| No. with rash | 7 | |
| No. with emesis | 3 | |
| No. with chest X-ray abnormality | 3 | |
| No. with HPeV in CSF | 7/11 | |
| Mean CSF WBC (per μl) | 5.9 ± 5.4 | 1–20 |
| No. with CSF pleocytosis | 0 | |
| Mean peripheral WBC ×103 (per μl) | 9.4 ± 7.8 | 1.7–28.8 |
| Mean ANC ×103 (per μl) | 5.0 ± 6.0 | 0.75–25.7 |
| No. with antibiotic therapy | 15 |
CSF, cerebrospinal fluid; WBC, white blood cells; ANC, absolute neutrophil count; Tmax, maximum temperature.
This is the first systematic study in the United States to evaluate the prevalence, epidemiology, and clinical characteristics of patients in whom HPeV was detected in respiratory specimens. In a comparable study from 2007 in Edinburgh, Scotland, an HPeV prevalence rate in respiratory specimens of patients less than 5 years of age was 2.1% (27 of 1,299 study subjects) (6), compared to 3% prevalence in our study. In contrast to that study, we evaluated only respiratory specimens negative for the common respiratory viruses detected by RVP assay (12 virus types and subtypes) in an attempt to allow clinical and laboratory characteristics to more clearly be attributed to HPeV and not the result of the alternative organism in cases of multivirus infection. However, this approach precluded the determination of the true HPeV incidence in the clinical scenario of coinfection with any of the 12 viruses tested by our multiplex PCR.
The majority of HPeV respiratory infections (79%) occurred in children less than 3 months old, consistent with early acquisition of the virus. This is also consistent with seroprevalence studies from Finland and Japan and previous data regarding HPeV-CNS infections (7, 9, 10, 18, 19, 23). The peak seasonality was in late summer, similar to previous reports of HPeV in the CNS and the seasonality of enteroviruses (2, 17, 18, 21). Of the 20 HPeV-positive respiratory specimens identified in our study, 15 were typed as HPeV-3, in contrast to other reports in which types 1 and 4 to 6 were more common in respiratory specimens (1, 2, 4, 6, 13, 15, 16, 20).
Over the past 5 years, we had observed the highest prevalence of HPeV CNS infections (66/388 CSF specimens tested) in our region during summer 2009 (18a). In our current study, HPeV was also detected in 7/11 CSF specimens from children with HPeV respiratory infection. Sequencing analysis confirmed the HPeV-3 genotype in both CSF and respiratory specimens in each of the seven subjects. These seven patients were all less than 2 months of age at diagnosis and presented with fever and irritability but only modest, nonspecific respiratory symptoms. There appear to be different potential outcomes of HPeV respiratory tract infection. One is that HPeV in respiratory samples reflects the initial portal of entry for a subsequent CNS infection. Alternatively, infection may be confined to the respiratory tract. The duration of local shedding and frequency of asymptomatic respiratory shedding are currently undefined.
HPeV surveillance data are limited in the United States and depend on passive reporting through the National Enterovirus Surveillance System (NESS) (11, 22). From 2006 to 2008, HPeV-1 was among the 15 most common viruses reported to NESS (22). However, the passive and voluntary nature of the surveillance system, coupled with a limited network of laboratories, precludes estimation of the true HPeV prevalence in the U.S. population. Additionally, since HPeV cannot be detected by enterovirus-specific RT-PCR assays conventionally utilized in clinical laboratories, the CDC's Picornavirus Laboratory has performed the majority of HPeV detection and typing to date. Since most testing for EV and HPeV occurs during the summer months of traditional EV season, HPeV presence at other times of the year may be missed. To further categorize the prevalence as well as clinical characteristics of HPeV in other organ systems, an active surveillance system for HPeV infections would be ideal.
This study contributes to a growing understanding of HPeV prevalence and disease associations in U.S. children. In contrast to the published literature, HPeV-3 was the dominant type in respiratory specimens from our population of U.S. patients. Because there seems to be no specific respiratory presentation attributable to HPeV infection, e.g., bronchiolitis or pneumonia, we have insufficient evidence for a strong recommendation for routine HPeV testing on all respiratory specimens of infants and children. Additionally, the presence of HPeV in both respiratory and CNS samples blurs the distinction between the respiratory tract sample being a focus of viral shedding and it being a herald of true disease. Further, a relatively low HPeV prevalence in respiratory secretions compared to that of routinely tested viruses renders the addition of routine respiratory testing for HPeV unwarranted at this time. However, testing respiratory secretions for HPeV may be a reasonable adjunct to CSF testing in infants younger than 3 months of age with fever and irritability of unknown etiology.
ACKNOWLEDGMENTS
This study was funded by research residual funds from R.S. and funds from Section of Neonatology.
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Abed Y, Boivin G. 2006. Human parechovirus infections in Canada. Emerg. Infect. Dis. 12:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benschop KSM, et al. 2006. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin. Infect. Dis. 42:204–210 [DOI] [PubMed] [Google Scholar]
- 3. Benschop KS, Thomas X, Serpenti C, Molenkamp R, Wolthers K. 2008. High prevalence of human parechovirus (HPeV) genotypes in the Amsterdam region and identification of specific HPeV variants by direct genotyping of stool samples. J. Clin. Microbiol. 46:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chieochansin T, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y. 2011. Molecular epidemiology, genome characterization, and recombination event of human parechovirus. Virology 421:159–166 [DOI] [PubMed] [Google Scholar]
- 5. Drexler JF, et al. 2009. Novel human parechovirus from Brazil. Emerg. Infect. Dis. 15:310–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harvala H, et al. 2008. Epidemiology and clinical associations of human parchovirus respiratory infections. J. Clin. Microbiol. 46:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvala H, et al. 2009. Specific association of human parechovirus type 3 with sepsis and fever in young infants, as identified by direct typing of cerebrospinal fluid samples. J. Infect. Dis. 199:1753–1760 [DOI] [PubMed] [Google Scholar]
- 8. Hyppia T, et al. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. U. S. A. 89:8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K. 2004. Isolation and identification of a novel human parechovirus. J. Gen. Virol. 85:391–398 [DOI] [PubMed] [Google Scholar]
- 10. Joki-Korpela P, Hyypia T. 1998. Diagnosis and epidemiology of echovirus 22 infections. Clin. Infect. Dis. 26:29–36 [DOI] [PubMed] [Google Scholar]
- 11. Khetsuriani N, Lamonte A, Oberste MS, Pallansch M, Centers for Disease Control and Prevention 2006. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill. Summ. 55:1–20 [PubMed] [Google Scholar]
- 12. Knowles NJ, et al. 2011. Picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 13. Ljubin-Sternak S, et al. 2011. Clinical and molecular characterization of a parechovirus type 1 outbreak in neonates in Croatia. J. Med. Virol. 83:137–141 [DOI] [PubMed] [Google Scholar]
- 14. Nix WA, et al. 2008. Detection of all known parechoviruses by real-time PCR. J. Clin. Virol. 46:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pajkrt D, et al. 2009. Clinical characteristics of human parechoviruses 4–6 infections in young children. Ped. Infect. Dis. J. 28:1008–1010 [DOI] [PubMed] [Google Scholar]
- 16. Piralla A, et al. 2012. Human parechovirus infections in patients admitted to hospital in northern Italy, 2008–2010. J. Med. Virol. 84:686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Renaud C, et al. 2011. Introduction of a novel parechovirus RT-PCR clinical test in a regional medical center. J. Clin. Virol. 51:50–53 [DOI] [PubMed] [Google Scholar]
- 18. Selvarangan R, et al. 2011. Human parechovirus 3 causing sepsis-like illness in children from midwestern United States. Pediatr. Infect. Dis. 30:238–242 [DOI] [PubMed] [Google Scholar]
- 18a. Sharp J, et al. Characteristics of young infants in whom human parechovirus, enterovirus or neither were detected in cerebrospinal fluid during sepsis evaluations. Pediatr. Infect. Dis. J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tauriainen S, et al. 2007. Human parechovirus 1 infections in young children—no association with type 1 diabetes. J. Med. Virol. 79:457–462 [DOI] [PubMed] [Google Scholar]
- 20. van der Sanden S, et al. 2008. Prevalence of human parechovirus in the Netherlands in 2000 to 2007. J. Clin. Microbiol. 46:2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verboon-Maciolek MA, et al. 2008. Severe neonatal parechovirus infection and similarity with enterovirus infection. Pediatr. Infect. Dis. J. 27:241–245 [DOI] [PubMed] [Google Scholar]
- 22. Villarruel GR, Langley GE, Oberste MS, Pallansch M. 2010. Nonpolio enteroviruses and human parechovirus surveillance—United States, 2006–2008. MMWR Morb. Mort. Wkly. Rep. 59:1577–1580 [PubMed] [Google Scholar]
- 23. Yamamoto M, et al. 2009. Epidemic of human parechovirus type 3 in Hiroshima City, Japan in 2008. Jpn. J. Infect. Dis. 62:244–245 [PubMed] [Google Scholar]