Abstract
Acremonium species cause a variety of human infections, while Lecanicillium species have not been reported as human pathogens. We describe a pseudo-outbreak involving both organisms, highlighting the role and limitations of molecular methods in the characterization of rare fungal isolates. Repeated isolation of these fungi from patient tissue samples raises concerns about exogenous contamination in the hospital environment.
TEXT
Acremonium species are filamentous fungi ubiquitous in the environment. They are associated with a wide spectrum of clinical diseases, including both localized and disseminated infections in immunocompetent and immunocompromised hosts (4, 6, 12, 14, 15). Acremonium species are also pathogenic to other fungi (mycopathogenic) and have been used as biological controls for fungal plant pathogens (3, 13). To date, no known clusters or outbreaks of human cases involving Acremonium species have been described. Lecanicillium species are a group of fungi that are pathogenic both to insects (entomopathogenic) and to other fungi and have been recovered from a wide range of substrates (7, 8, 16). Some species of Lecanicillium have also been used as biocontrol agents (1, 7, 10, 13). Unlike Acremonium, however, this species has not been associated with disease in humans in spite of its abundance in the natural environment (10). Acremonium and Lecanicillium share many morphological features, including hyaline colony, cottony texture, and similar arrangement of conidia and conidiophores, that make morphological distinction between the two fungal taxa challenging (4, 7, 9). We investigated a cluster of Acremonium and Lecanicillium isolates recovered from tissues removed from patients during orthopedic surgery at hospital A. The present study describes the cluster, methods used for fungal species identification, and the limitations and challenges associated with currently employed methods for identification of fungal species.
Patient tissues were collected in the operating room from various body sites (hand, knee, and elbow) during routine irrigation and debridement procedures (Table 1). Fungus culture had been ordered on all samples. The protocol at hospital A for culturing tissue samples called for separate samples to be collected for bacterial and fungal testing. When separate samples were not available, tissue was divided in a biological safety cabinet in the hospital microbiology lab. Bacterial cultures were plated at hospital A. All tissue samples designated for fungus culture were sent to commercial laboratory B. Tissue specimens received at laboratory B were then sent unopened to either commercial laboratory C or university laboratory D for fungal culture and phenotypic identification.
TABLE 1.
Description of Acremonium/Lecanicillium cases at hospital Aa
| Case | Date of culture | Specimen site | Laboratory | Initial fungal identification | Bacterial organism coinfection | Histopathology done | Fungal isolate to CDC |
|---|---|---|---|---|---|---|---|
| 1 | 10/1/2008 | Left lumbar region | ULD | Acremonium | MRSA, Peptostreptococcus asaccharolyticus | No | |
| 2 | 11/22/2008 | Right hand | ULD | Acremonium | No | Yesc | |
| 3 | 2/12/2009 | Right elbow bursa | ULD | Acremonium | MRSA, Propionibacterium acnes | No | |
| 4 | 2/17/2009 | Left elbow allograft | ULD | Acremonium | Enterobacter cancerogenus, Staphyloccocus aureus, Candida parapsilosis | No | |
| 5 | 3/4/2009 | Right knee pretibial | CLC | Acremonium | No | Yesc | Lecanicilliumb |
| 3/4/2009 | Right knee anterior fat pad | CLC | Acremonium | No | Lecanicillium | ||
| 3/4/2009 | Right knee suprapatellar pouch | CLC | Acremonium | P. acnes, Mycobacterium chelonae | Acremonium | ||
| 3/11/2009 | Right knee tissue | ULD | Acremonium | P. acnes, M. chelonae | No | Lecanicillium | |
| 6 | 4/2/2009 | Synovium | ULD | Acremonium | MRSA | No | Lecanicillium |
ULD, university laboratory D; CLC, commercial laboratory C; MRSA, methicillin-resistant Staphylococcus aureus.
In case 5, the three isolates from the knee were sent to the CDC without labeling indicating the specific knee source. The three results cannot be directly correlated to an individual specimen.
Histopathology was performed on two samples, but no fungal elements were seen using Gomori silver staining.
Filamentous fungi were recovered from a cluster of patient tissues at hospital A between October 2008 and April 2009 (Table 1). The first case was a postoperative lumbar wound infection from which fungi resembling Acremonium species and several bacteria were recovered. Over the next 6 months, Acremonium was identified in orthopedic tissue samples from six additional patients. Histopathology was performed on cases 2 and 5 and was negative for fungal elements using Gomori methenamine silver staining. None of the patients received antifungal treatment.
The CDC was invited to assist in an investigation of the cases after the recovery of the fifth isolate identified as Acremonium species from one of these tissue samples. Environmental sampling was performed throughout hospital A, with a focus on the operating rooms and the laboratory. Spongesicle swabs were collected from the hospital A laboratory, facility storage room, and surgical suite, the commercial laboratory B transport vehicle, and the laboratory B processing room. Air samples (13) were collected from the hospital A surgical suite, air intake, rooftop, offsite storage center, storage room, microbiology laboratory, another laboratory (control), commercial laboratory B parking lot, and laboratory B processing room. Samples of water-damaged flooring (3), the used laboratory and operating suite HVAC air filters, and unused specimen collection cups (3) were also examined. Nine samples of soil and leaves were collected from plants in the hospital A laboratory and from a plant in the hospital atrium (control). All environmental samples were directly sent to the CDC for fungal culture and isolation.
Between January 2008 and April 2009, hospital A sent 320 samples for fungus culture, and 26 were positive for fungal growth at one of the two referral laboratories used by hospital A (commercial laboratory C or university laboratory D). Nine of these 26 samples were originally identified morphologically as Acremonium species: all 4 isolates from case 5 were identified at laboratory C, and 5 isolates from all the remaining cases were identified at laboratory D (Table 1). Five fungal isolates from cases 5 (four isolates) and 6 (one isolate) were sent to the CDC for species confirmation; isolates from the previous 4 cases had been discarded. Morphological examination and comparative DNA sequence analysis were used by the CDC to confirm the identity of the isolates. Genomic DNA was extracted and purified from the five clinical isolates and one environmental isolate (see below). The internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was PCR amplified and sequenced as described previously (2). The identity of the isolates was determined by comparison of approximately 550 bases of the contiguous sequence from the ITS region to sequences in the GenBank database.
Sequence-confirmed Lecanicillium isolates were further characterized by inter-simple sequence repeat (ISSR) PCR (18). Two epidemiologically unrelated Lecanicillium isolates (L. lecanii and L. attenuatum) from the CDC culture collection were used as controls for ISSR typing. Four 6-carboxyfluorescein (6-FAM)-labeled ISSR primers designed to flank tri- and tetranucleotide repeats were used for PCR amplification of the fragments constituting the ISSR fingerprint as described previously (11). An unweighted-pair group method using average linkages (UPGMA) tree, rooted with the outgroup Acremonium species (isolate 5c), was generated using PHYLIP (5).
Four of the five case isolates (5a, 5b, 5d, and 6) were reidentified as Lecanicillium lecanii at the CDC based on ITS sequencing. The ITS sequences for all four isolates were identical to one another and to the Lecanicillium lecanii sequence in the GenBank database (100% homology to GenBank accession number FJ515771.1, 90 to 99% query coverage). The fifth isolate (5c) was confirmed as Acremonium species, with 100% homology at 100% query coverage to the ITS regions of two species, Acremonium strictum (GenBank accession number GU595023.1) and Acremonium kiliense (GenBank accession number FR694874.1), and was identified as Acremonium species for the purposes of this study. Environmental surface swabs, materials, and air samples were negative for Lecanicillium and Acremonium species. Acremonium species was recovered from the soil of a potted plant in the hospital A laboratory. Comparative sequence analyses of the ITS region of this environmental isolate revealed that it was 99% homologous with 100% query coverage to Acremonium alternatum (GenBank accession number U57674.1) and Acremonium strictum (GenBank accession number AY138844.1). ITS sequence comparison of the environmental isolate to the clinical Acremonium isolate 5c demonstrated only 82% homology, suggesting that these isolates were not closely related.
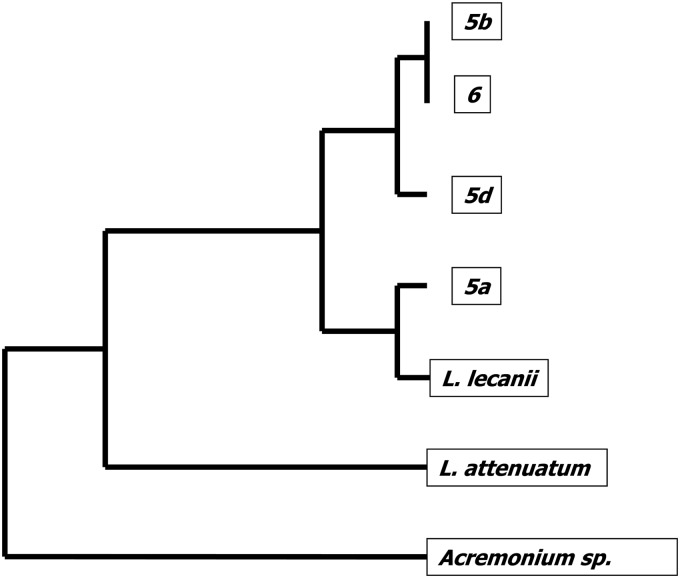
The four isolates identified as Lecanicillium lecanii by molecular methods were further characterized by ISSR PCR using four primers. Four ISSR primers yielded 467 reproducible markers, and the results demonstrated that the four Lecanicillium isolates in the cluster shared between 84 and 100% of their ISSR markers. Isolates 6 and 5b (from two different patients) were identical across all markers, suggesting that they may have a single source. Isolate 5a shared the least number of markers with other isolates. The UPGMA tree shows relatedness of the isolates based on ISSR data (Fig. 1).
Fig 1.
UPGMA tree based on ISSR data for Lecanicillium isolates. Phylogenetic relationship among Lecanicillium isolates recovered from patient tissues. L. lecanii and L. attenuatum are epidemiologically unrelated Lecanicillium species used as controls. The tree is rooted with the outgroup Acremonium species.
After a detailed epidemiological investigation of the cluster of infections, the outbreak was determined to be a pseudo-outbreak. There was no clear pattern associated with the recovery of fungi and the laboratories that processed the specimens. Histopathology performed on tissues from two cases did not reveal fungal elements. Visual observation of the tissue preparation procedures in both laboratories and the specimen transport system did not demonstrate breaches in sterile processes. Environmental sampling in these areas did not yield any fungal isolates morphologically similar to Acremonium or Lecanicillium beyond a genetically unrelated Acremonium in a potted plant. Personnel records for the operating rooms and laboratory specimen-processing area did not reveal any trend in personnel present in the rooms during these procedures. Furthermore, tissues for fungal culture were collected at the beginning of the surgery, before irrigation was begun, making the surgical procedure less likely to have been the source of contamination. We cannot rule out the possibility that contamination could have arisen in the operating room during collection of the tissues, but our retrospective investigation did not detect any breaches in sterile procedures or inappropriate instrument sterilization methods. We believe that the most likely source of these fungal isolates was contamination introduced in hospital A, because these fungal species were recovered at both commercial laboratory C and university laboratory D from samples originating at hospital A, where the tissues were collected and divided into two parts. True infection was considered unlikely based on full clinical recovery of all the patients without antifungal treatment. The specific source of Lecanicillium and Acremonium species in this cluster remains unknown. To our knowledge, no further cases involving these organisms have been detected in hospital A since the investigation.
Comparative sequence analysis allowed unambiguous identification of most of the isolates in the cluster as Lecanicillium lecanii and, in one case, confirmed the initial identification of Acremonium species. The identity of the fungus in four other cases is not known because the isolates were no longer available but may have been Lecanicillium species. Extensive environmental sampling from the hospital air intake, air samples in various locations in the hospital, and air filters in the operating room and laboratories did not yield Lecanicillium or Acremonium species. We were able to demonstrate the presence of Acremonium species in the hospital environment by recovering one isolate from the soil of a plant in the hospital A laboratory. However, molecular methods demonstrated that the environmental isolate was genetically distinct from the clinical isolate.
Both Lecanicillium lecanii and Acremonium species were recovered from case 5, and further genotyping of the Lecanicillium isolates revealed at least three unique genotypes. In contrast, one Lecanicillium species recovered from case 5 and another from case 6 were identical to each other by the ISSR PCR fingerprinting method. The results of our study demonstrate that there could have been multiple different sources of Lecanicillium/Acremonium seeding the original samples.
One of the challenges in this study was the lack of available robust sequences representing the species of Acremonium in the GenBank sequence database, which is most commonly used for DNA sequence comparison. Currently in GenBank, sequences of Acremonium are poorly characterized and annotated, with many sequences lacking a species designation, severely limiting the utility of this commonly available database for species level identification of Acremonium. Given this limitation, we could identify the isolates recovered in this study only as Acremonium species. Ongoing taxonomic studies of this genetically diverse genus might help to rectify this issue (17).
The recovery of soil fungi that are not traditionally thought to be human pathogens from multiple human samples over a long time period (6 months) raises concern about the handling and processing of human tissue samples in the hospital. This cluster also provides another example of the utility of DNA sequence-based identification. Because Lecanicillium is an unusual human pathogen, further scrutiny of the patients, the samples, and their handling process might have occurred sooner had Lecanicillium been correctly identified in the initial fungus cultures. DNA sequencing would have effectively determined the correct genus identification in these cases, thus changing the course of the investigation and allowing the hospital to initiate control measures more quickly. The reporting of fungus culture results that include molds not traditionally considered human pathogens is an area for which, to our knowledge, no guidelines currently exist. Some laboratories report such isolates with the comment of “possible contaminant.” We are reluctant to endorse this practice because isolates from immunosuppressed hosts that might reflect true disease might instead be disregarded. A comment such as “this organism has not been reported as a human pathogen: correlate with histopathology and clinical condition” might be useful to alert the clinician and infection control officer that an unusual mold of unclear clinical significance is being reported.
ACKNOWLEDGMENTS
Carolyn O. S. Neal was supported by an Emerging Infectious Diseases Fellowship sponsored by the Association of Public Health Laboratories and the Centers for Disease Control and Prevention.
We thank Judith Noble-Wang and Carol Rao of the CDC Division of Healthcare Quality Promotion, Anne Whitney and the CDC Genomics Unit for assistance with DNA sequencing and ISSR typing, and the laboratory staff at hospital A for their work in the investigation.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Ansari MA, Pope EC, Carpenter S, Scholte EJ, Butt TM. 2011. Entomopathogenic fungus as a biological control for an important vector of livestock disease: the Culicoides biting midge. PLoS One 6:e16108 doi:10.137/journal.pone.0016108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balajee SA, et al. 2009. Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot. Cell 8:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi GJ, Kim JC, Jang KS, Cho KY, Kim HT. 2008. Mycoparasitism of Acremonium strictum BCP on Botrytis cinerea, the gray mold pathogen. J. Microbiol. Biotechnol. 18:167–170 [PubMed] [Google Scholar]
- 4. Das S, Saha R, Dar SA, Ramachandran VG. 2010. Acremonium species: a review of the etiological agents of emerging hyalohyphomycosis. Mycopathologia 170:361–375 [DOI] [PubMed] [Google Scholar]
- 5. Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package) version 3.68. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 6. Guarro J, Gams W, Pujol I, Gene J. 1997. Acremonium species: new emerging fungal opportunists—in vitro antifungal susceptibilities and review. Clin. Infect. Dis. 25:1222–1229 [DOI] [PubMed] [Google Scholar]
- 7. Kim HY, Lee HB, Kim YC, Kim IS. 2008. Laboratory and field evaluations of entomopathogenic Lecanicillium attenuatum CNU-23 for control of green peach aphid (Myzus persicae). J. Microbiol. Biotechnol. 18:1915–1918 [PubMed] [Google Scholar]
- 8. Kouvelis VN, Sialakouma A, Typas MA. 2008. Mitochondrial gene sequences alone or combined with ITS region sequences provide firm molecular criteria for the classification of Lecanicillium species. Mycol. Res. 112:829–844 [DOI] [PubMed] [Google Scholar]
- 9. Liu K, Howell DN, Perfect JR, Schell WA. 1998. Morphologic criteria for the preliminary identification of Fusarium, Paecilomyces, and Acremonium species by histopathology. Am. J. Clin. Pathol. 109:45–54 [DOI] [PubMed] [Google Scholar]
- 10. Madsen AM, Hansen VM, Meyling NV, Eilenberg J. 2007. Human exposure to airborne fungi from genera used as biocontrol agents in plant production. Ann. Agric. Environ. Med. 14:5–24 [PubMed] [Google Scholar]
- 11. Neal CO, et al. 2011. Global population structure of Aspergillus terreus inferred by ISSR typing reveals geographical subclustering. BMC microbiology 11:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perdomo H, et al. 2011. Spectrum of clinically relevant Acremonium species in the United States. J. Clin. Microbiol. 49:243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romero D, Rivera ME, Cazorla FM, de Vicente A, Perez-Garcia A. 2003. Effect of mycoparasitic fungi on the development of Sphaerotheca fusca in melon leaves. Mycol. Res. 107:64–71 [DOI] [PubMed] [Google Scholar]
- 14. Sahin GO, Akova M. 2006. Treatment of invasive infections due to rare or emerging yeasts and moulds. Expert Opin. Pharmacother. 7:1181–1190 [DOI] [PubMed] [Google Scholar]
- 15. Saldarreaga A, et al. 2004. Antifungal susceptibility of Acremonium species using E-test and Sensititre. Rev. Esp. Quimioter. 17:44–47 (In Spanish.) [PubMed] [Google Scholar]
- 16. Sukarno N, et al. 2009. Lecanicillium and Verticillium species from Indonesia and Japan including three new species. Mycoscience 50:369–379 [Google Scholar]
- 17. Summerbell RC, et al. 2011. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 68:139–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zietkiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183 [DOI] [PubMed] [Google Scholar]