Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a relatively new addition to the clinical microbiology laboratory. The performance of the MALDI Biotyper system (Bruker Daltonics) was compared to those of phenotypic and genotypic identification methods for 690 routine and referred clinical isolates representing 102 genera and 225 unique species. We systematically compared direct-smear and extraction methods on a taxonomically diverse collection of isolates. The optimal score thresholds for bacterial identification were determined, and an approach to address multiple divergent results above these thresholds was evaluated. Analysis of identification scores revealed optimal species- and genus-level identification thresholds of 1.9 and 1.7, with 91.9% and 97.0% of isolates correctly identified to species and genus levels, respectively. Not surprisingly, routinely encountered isolates showed higher concordance than did uncommon isolates. The extraction method yielded higher scores than the direct-smear method for 78.3% of isolates. Incorrect species were reported in the top 10 results for 19.4% of isolates, and although there was no obvious cutoff to eliminate all of these ambiguities, a 10% score differential between the top match and additional species may be useful to limit the need for additional testing to reach single-species-level identifications.
INTRODUCTION
Recent decades have seen advances in automation of traditional phenotypic and biochemical methods for microbial identification (ID), and advances in sequencing and the proliferation of genomic data hold great promise for further improvements. The development of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has brought microbial diagnostics to another cusp of rapid development. The speed and low cost of bacterial identification by MALDI-TOF MS make it an attractive technology in the clinical microbiology laboratory, and it has shown promise for identification of Gram-positive cocci (2, 6, 8), enteric and nonfermenting Gram-negative rods (11, 21, 24), HACEK organisms (10), anaerobes (14, 17, 19, 20, 31), and broad cohorts of clinically relevant bacteria (3, 4, 22, 27, 30).
Commercial MALDI-TOF systems identify a broad range of microorganisms based on analysis of unique “fingerprints” of abundant proteins from whole cells or cellular extracts (15, 23, 26, 28). These profiles are searched against databases of reference spectra, and similarity scores for the top database matches are used to determine the identification of unknown isolates. As observed previously, a systematic evaluation of scoring criteria on diverse isolates could improve results (2, 10, 25, 27, 29). Identification may be complicated when multiple species- or genus-level matches are among the top 10 results. Most current publications on the MALDI Biotyper system (Bruker Daltonics, Billerica, MA) do not address these complicated situations; however, one example where this problem is addressed is the use of the “10% rule,” which states that any species scoring >10% below the top-scoring match may be excluded (24). Another approach is a system introduced in the MALDI Biotyper software (v3.0) that categorizes results based on the identification consistency among the top 10 matches. In the current study, we evaluated the performance of the Biotyper system on a diverse set of routine and unusual isolates and determined optimal thresholds for species- and genus-level identifications. We also used a custom computational approach to search for optimal values for exclusion of additional species in the context of the newly introduced Biotyper consistency categories.
MATERIALS AND METHODS
Bacterial isolates.
Routine and referred clinical isolates (n = 690) representing 102 genera and 225 unique species of broad phylogenetic distribution were analyzed by MALDI-TOF MS between January 2010 and January 2012. Isolates were analyzed prospectively, although to maintain diversity, very common organisms were limited by randomly including only a portion of those encountered. Of the 690 isolates, 50 were selected from archives to expand diversity of the cohort and were analyzed retrospectively. Among this cohort were 577 isolates (93 genera and 225 species) that were identified to the species level by one or more standard laboratory methods. These fully identified isolates served as the core set for quantitative analyses to allow direct comparison of species-level performance. Isolates were identified by the following standard methods: (i) sequencing of the first 500 bp of the 16S rRNA gene (n = 388; 304 to the species level) (18), (ii) the BD Phoenix (BD Diagnostics, Sparks, MD) automated identification system (n = 179; 168 to the species level), and (iii) traditional phenotypic methods (n = 101; 83 to the species level) (33), submitting client identification (n = 4; 4 to the species level), or quality control strains (n = 18; 18 to the species level) (Table 1).
TABLE 1.
Distribution of study isolates by organism category
| Organism category | All study isolates (no.) | Isolates identified to species (no.)a |
|---|---|---|
| Gram-positive cocci | 165 | 133 |
| Gram-positive rods | 142 | 102 |
| Gram-negative cocci | 27 | 23 |
| Gram-negative rods | ||
| Enteric | 170 | 164 |
| Nonfermenting | 103 | 77 |
| Fastidious | 60 | 57 |
| Other | 23 | 21 |
| Total | 690 | 577 |
Isolates identified to the species level by standard methods.
MALDI-TOF MS.
Isolates were cultivated in pure culture on Columbia sheep blood, chocolate, or brucella agar plates (Hardy Diagnostics, Santa Maria, CA) at 35°C under anaerobic (90% N2, 5% CO2, 5% H2) or aerobic (5% CO2) atmosphere as required for optimal growth. Organisms were harvested at 24 to 48 h depending on growth rate and available cellular mass.
The direct-smear method was evaluated on a taxonomically diverse set of 183 isolates. A thin film of bacterial cells was spread evenly on a polished steel target (Bruker Daltonics), overlaid with 1.75 μl matrix (saturated alpha-cyano-4-hydroxycinnamic acid [HCCA] in 50% acetonitrile–2.5% trifluoroacetic acid) and allowed to air dry.
The extraction method was performed on all 690 isolates as previously described (10). Bacterial cells (∼5 to 10 mg) were suspended in 300 μl distilled water (dH2O) and mixed by inversion with 900 μl absolute ethyl alcohol (EtOH). Cells were pelleted (16,000 × g, 2 min), and EtOH was completely removed after a second centrifugation (16,000 × g, 2 min). Cells were resuspended in 50 μl of 70% formic acid, vortexed for 1 min, and mixed by pipetting with 50 μl pure acetonitrile. Samples were centrifuged (16,000 × g, 2 min), and 75 μl of supernatant (bacterial extract) was transferred to fresh tubes. Bacterial extract (1.25 μl) was spotted onto polished steel targets, air dried, and overlaid by 1.75 μl of HCCA matrix, which was allowed to air dry.
Mass spectra were acquired as previously described (10) using a single spot for each isolate. Data were collected between 2,000 and 20,000 m/z in linear positive ionization mode (microflex; Bruker Daltonics). Each spectrum was a sum of 500 shots collected in increments of 100. If scores from the initial automated data collection and analysis were <1.9, new spectra were collected in manual acquisition mode. If the score remained <1.9, the isolate was recultivated, reextracted, and reanalyzed. If scores did not improve after the second extraction, the higher score of the two attempts was recorded. Spectra that repeatedly scored <1.7 were manually reviewed. Spectra were analyzed with the MALDI Biotyper 3.0 software (Bruker Daltonics) using the MALDI Biotyper library (version 3.0; 3,995 spectra). Each spectrum was assigned a similarity score (0 to 3) to the best 10 database matches, which were recorded for further analysis. Results were also assigned a consistency category based on the manufacturer's criteria as follows: A, species consistency (all matches scoring ≥1.9 are of the same species, and all matches scoring ≥1.7 are of the same genus); B, genus consistency (top match score is 1.899 to 1.7, or matches scoring ≥1.9 are not of the same species, but all matches scoring ≥1.7 are of the same genus); C, no consistency (top match score is <1.7, or matches scoring ≥1.7 are not of the same genus).
The term “mismatch” indicates results in the top 10 MALDI Biotyper report that differ at the species or genus level from the top identification. “Significant mismatch” indicates mismatches, as defined above, that score ≥1.9 (species mismatch) or ≥1.7 (genus mismatch). “Threshold,” as used by the manufacturer, indicates MALDI-TOF MS scores used to differentiate between species, genus, or unreliable identifications (IDs). “Cutoff” indicates the percentage below the top score used to eliminate additional species from the top 10 results. Discrepant results (incorrect genus or species matches with scores of ≥1.9 or incorrect genus matches scoring between 1.7 and 1.899) were reanalyzed by MALDI-TOF, and those that remained unresolved were subjected to complementary testing (e.g., testing with a different method than used for original identification) and/or reanalysis by the original identification method. Concordance was calculated after resolving discrepant results and applying scoring thresholds (≥1.9 for species-level and ≥1.7 for genus-level identification).
Data analysis.
Due to the limitations of current standard methods to identify some isolates to the species level, 70 isolates in this study were identified only to the genus level, 30 to the group or complex level, and 13 to two possible species (slash calls; see Table S1 in the supplemental material). When species-level Biotyper identifications matched a member of a group or complex or one of the species of a slash call, the identification was considered correct for concordance calculations (2). Isolates identified only to the genus level by a standard method were considered correct only to the genus level regardless of the level of Biotyper identification.
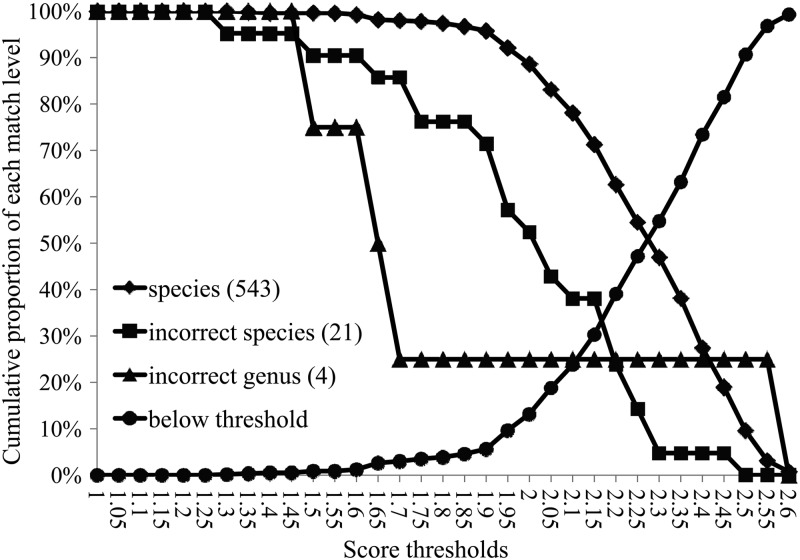
To find the optimum score thresholds for genus- and species-level identification, isolates were assigned to a match level (species, incorrect species, or incorrect genus) at a range of identification thresholds (1 to 2.6, the maximum score in this data set). The cumulative proportion of isolates in each category was plotted as a function of the identification score thresholds to detect scores that maximized the ratio of species to incorrect identifications.
To attempt to identify the optimal value for excluding mismatches among the 10 results generated per isolate, we wrote a custom MATLAB (Mathworks, Natick, MA) script (available upon request) to calculate the percent difference between the top result and subsequent mismatches and then plotted the cumulative proportion of isolates with significant mismatches over a range of percent difference cutoff scores (0 to 35%). The distribution of significant mismatches by species was evaluated with the statistical computing software R (v.2.15.0; http://www.R-project.org) using the R Commander package (v.1.7.0; http://socserv.socsci.mcmaster.ca/jfox/Misc/Rcmdr). Box and whisker plots showing the minimum, lower quartile, median, upper quartile, and maximum percent difference from the top-scoring matches were generated using the R Commander default boxplot command.
Associations between categorical variables were analyzed by Fisher's exact or chi-square tests using R, and means were compared using the Student t test in Microsoft Excel (Redmond, WA). P values <0.05 were considered statistically significant.
RESULTS
Score threshold optimization for species and genus identification.
To determine the optimal score thresholds for genus- and species-level identifications, the 568 isolates (82.3%) identified to the species level by standard methods and represented in the Biotyper library were assigned to each match level (species, incorrect species, or incorrect genus) over a wide range of MALDI-TOF MS score thresholds (1 to 2.6) (Fig. 1). When the species threshold was moved from the default of 2.0 to 1.9, there was a significant increase in isolates correct to the species level (39 of 568; 6.9%; P = 0.0004) at the cost of only 4 isolates (0.7% of 568; P = 0.43) incorrect to the species level but still correct to the genus or group level. This change also resulted in a lower false-negative rate (11.2% at 2.0 to 4% at 1.9). It is interesting to note that the 4 species-level discrepant results that arose due to lowering the threshold were a Burkholderia cepacia isolate identified as Burkholderia cenocepacia, 2 isolates of Streptococcus intermedius identified as Streptococcus constellatus, and a Fusobacterium nucleatum isolate identified as Fusobacterium naviforme. All would have been called only to the genus level at the manufacturer's cutoff. Three of these four are routinely identified only to the complex or group level (B. cepacia complex and S. anginosus group); thus, lowering the threshold would effectively result in correct complex/group-level identification for three-fourths of these “discrepants” and a single reporting difference (a Fusobacterium sp. reported as F. naviforme) that would be unlikely to have treatment implications (16). Moreover, by changing from 2.0 to 1.9, the number of isolates called to category A (high-confidence identifications [IDs]) increased by 14, with a corresponding loss of 14 isolates from category B (lower-confidence IDs). Thus, changing the species threshold to 1.9 resulted in a nearly 7% increase in correct species-level IDs and 14 fewer isolates requiring review due to the shift from category B to A. In contrast, the default genus threshold of 1.7 provided an optimal balance between correct, incorrect, and unreliable identifications (Fig. 1).
Fig 1.
Cumulative proportion of isolates above a chosen MALDI-TOF MS threshold (1 to 2.6) reaching species (diamonds), reaching genus only (squares), or showing no agreement (triangles) between results of MALDI-TOF MS and standard methods.
Direct-smear versus extraction methods.
A very taxonomically diverse set of 183 isolates spanning 72 genera and 146 unique species was chosen for comparison of direct-smear and cell extraction sample preparation methods (see Table S2 in the supplemental material). The extraction method yielded significantly higher average scores than the direct-smear method across this diverse set of isolates (P < 0.001), with 78.7% of isolates scoring higher by extraction. In addition, significantly more extracted samples were identified correctly to the species level (164 versus 140, P = 0.0001) and fewer gave unreliable identifications than with the direct-smear method (Table 2). Because of the superior results, the remainder of the study utilized extracted samples.
TABLE 2.
Concordance of direct smear and extraction sample preparation methods with standard identification methods on a taxonomically diverse set of organismsa
| Match category | Result (% concordance [95% confidence interval]) |
|
|---|---|---|
| Direct smear | Extraction | |
| Speciesb | 140/142 (98.6% [95.0–99.8]) | 164/167 (98.2% [94.8–99.6]) |
| Genusc | 167/168 (99.4% [96.7–100]) | 175/176 (99.4% [96.9–100]) |
| No reliable identificationd | 9/15 (60% [32.3–83.7]) | 4/7 (57.1% [18.4–90.1]) |
| Total | 183 | 183 |
The sample preparation method had a significant effect on distribution across match categories (P = 0.004, Fisher's exact test).
Number of isolates concordant to species level with standard method identification/total number of isolates scoring ≥1.9.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring ≥1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring <1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Overall concordance.
Of the 611 isolates characterized to the species, group, or complex level by a standard method, 557 (91.2%) were correctly identified by MALDI-TOF to the species level with scores of ≥1.9. Overall, 637 of 690 (92.3%) were correctly identified to the genus level with scores of ≥1.7 and 43 (6.2%) failed to achieve a reliable identification (see Table S1 in the supplemental material). Of the 690 isolates evaluated, 9 were not represented in the MALDI Biotyper database (see Tables S1 and S3 in the supplemental material). Of these 9 isolates, 5 scored ≥1.9: 2 Shigella (S. sonnei and S. flexneri) isolates were predictably misidentified as Escherichia coli (4, 20, 22, 27); a Citrobacter werkmanii isolate was identified as Citrobacter freundii, which belongs to the same complex; a Clostridium bolteae isolate was identified as Clostridium clostridioforme, which is phenotypically very similar to C. bolteae but was recently described as genetically distinct (12); and a Vibrio cholerae isolate was identified as Vibrio albensis, which is considered by many as a biovar of V. cholerae. Only 1 of the remaining 4 isolates scored between 1.7 and 1.899, and it was correct at the genus level (isolate 451, a Nocardia sp.). Of the remaining 568 isolates identified by standard methods to the species level, 522 of 537 (97.2%) with scores of ≥1.9 were correct to the species level and 551 of 552 (99.8%) with scores of ≥1.7 were correct to the genus level (Table 3). The remaining 15 species-level-discrepant results belonged primarily to the genera Citrobacter, Enterobacter, Streptococcus, Burkholderia, and Corynebacterium. The single genus-level discrepant was a Klebsiella oxytoca isolate identified as Raoultella ornithinolytica (see Table S3 in the supplemental material).
TABLE 3.
Concordance between MALDI-TOF MS and standard identification methodsa
| Match category | Result (% concordance [95% confidence interval]) |
|||
|---|---|---|---|---|
| 16S | Phoenix | Traditional phenotypic methods | Overall | |
| Speciesb | 263/269 (97.8% [95.2–99.2]) | 160/167 (95.8% [91.6–98.3]) | 99/101 (98.0% [93.0–99.8]) | 522/537 (97.2% [95.4–98.4]) |
| Genusc | 282/282 (100% [98.7–100]) | 166/167 (99.4% [96.6–100]) | 103/103 (100% [96.5–100]) | 551/552 (99.8% [99.0–100]) |
| No reliable identificationd | 13/16 (81.3% [54.4–96]) | 0/0 | 0/0 | 13/16 (81.3% [54.4–96]) |
| Total | 298 | 167 | 103 | 568 |
The standard identification method had a significant effect on distribution across match categories (P = 0.0008, Fisher's exact test).
Number of isolates concordant to species level with standard method identification/total number of isolates scoring ≥1.9.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring ≥1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring <1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Concordance by method of identification.
Sequencing the first 500 bp of the 16S rRNA gene (16S sequencing) was used on isolates that are difficult to identify using routine phenotypic methods. The remaining isolates were identified by the Phoenix automated bacterial identification system or traditional phenotypic methods and were composed primarily of routinely encountered organisms such as staphylococci, enterococci, and enteric and common nonfermenting Gram-negative rods. Species-level concordance between isolates identified by MALDI-TOF MS and the more challenging subset identified by 16S sequencing was 88.3% (263 of 298). Not surprisingly, 5.4% (16 of 298) of these isolates could not be reliably identified due to scores of <1.7 (Table 3; see Table S1 in the supplemental material). More commonly encountered isolates identified by the Phoenix system and traditional phenotypic methods had good concordance with MALDI-TOF MS, with >95.8% correctly identified to the species level and >99% of remaining isolates identified correctly to the genus level (Table 3).
Concordance by major organism categories.
Identification of enteric Gram-negative rods was good by MALDI-TOF MS, with concordance of 96.3% (154 of 160) to the species level and 99.4% (159 to 160) to the genus level (Table 4). Among nonfermenting and fastidious Gram-negative rods, 90.9% (70 of 77) and 91.2% (52 of 57), respectively, were correctly identified to the species level (Table 4). Species-level concordance among Gram-positive cocci, Gram-negative cocci, and Gram-positive rods was 93.2% (123 of 132), 87% (20 of 23), and 87.9% (87 of 99), respectively (Table 4). Anaerobes showed slightly lower concordance with standard methods, with 66 of 74 (89.2%) correctly identified to the species level, 70 of 74 (94.6%) to the genus level, and 4 of 74 (5.4%) producing unreliable identifications (see Table S1 in the supplemental material). Overall, 16 of 568 (2.8%) isolates identified to the species level by standard methods failed to give reliable identifications by MALDI-TOF MS (i.e., they scored <1.7): Aggregatibacter segnis (2 of 2), Cardiobacterium hominis (1 of 2), Clostridium bifermentans (1 of 2), Corynebacterium tuberculostearicum (1 of 1), Fusobacterium nucleatum (1 of 2), Inquilinus limosus (1 of 1), Neisseria elongata (1 of 3), Nocardia farcinica (2 of 2), Paracoccus yeei (1 of 3), Parvimonas micra (1 of 3), Propionibacterium acnes (1 of 6), Rhodococcus corynebacterioides (1 of 1), Sphingomonas mucosissima (1 of 1), and Variovorax paradoxus (1 of 1) (see Table S1).
TABLE 4.
Concordance between MALDI-TOF MS and standard identification methods across major organism categoriesa
| Match category | Result (% concordance [95% confidence interval]) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gram-positive cocci | Gram-positive rods | Gram-negative cocci | Gram-negative rods |
Overall | ||||
| Enteric | Nonfermenting | Fastidious | Overall | |||||
| Speciesb | 123/126 (97.6% [93.2–99.5]) | 87/89 (97.8% [92.1–99.7]) | 20/20 (100% [83.2–100]) | 154/160 (96.3% [92.0–98.6]) | 70/72 (97.2% [90.3–99.7]) | 52/53 (98.1% [89.9–100]) | 292/302 (96.7% [94.0–98.4]) | 522/537 (97.2% [95.4–98.4]) |
| Genusc | 131/131 (100% [97.2–100]) | 93/93 (100% [96.1–100]) | 22/22 (100% [84.6–100]) | 159/160 (99.4% [96.6–100]) | 74/74 (100% [95.1–100]) | 53/53 (100% [93.3–100]) | 305/306 (99.7% [98.2–100]) | 551/552 (99.8% [99.0–100]) |
| No reliable identificationd | 1/1 (100% [25–100]) | 6/6 (100% [54.1–100]) | 1/1 (100% [25–100]) | 0/0 | 2/3 (66.7% [9.4–99.2]) | 3/4 (75% [19.4–99.4]) | 5/8 (62.5% [24.5–91.5]) | 13/16 (81.3% [54.4–96]) |
| Total | 132 | 99 | 23 | 160 | 77 | 57 | 314 | 568 |
The distribution of identifications by match category was not significantly different across the major organism categories (P = 0.247, chi-square test).
Number of isolates concordant to species level with standard method identification/total number of isolates scoring ≥1.9.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring ≥1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring <1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Concordance by consistency categories.
MALDI-TOF results for isolates in consistency category A showed excellent species-level concordance with routine methods (421 of 427, 98.6%), and the remaining isolates were correctly identified to the genus level (Table 5). Consistency category B isolates showed only 68.1% (47 of 69) species-level concordance with standard methods, yet all were correctly identified to the genus level. Within consistency category C, 75% (54 of 72) were correct to the species level, 76.4% (55 of 72) were correct to the genus level, and 16 of 72 (22.2%) of isolates produced unreliable scores (<1.7). Overall, within our diverse core collection of 568 isolates, over 80% of isolates (427 with species-level scores categorized as A, 15 scoring between 1.7 and 1.899 categorized as B, and 16 with unreliable identification scores of <1.7 categorized as C) could be reported confidently using the consistency categories and identification scores, while the remaining 20% (110 of 568) of isolates would require further analysis of the top 10 results prior to reporting.
TABLE 5.
Concordance between MALDI-TOF MS and standard identification methods across Biotyper consistency categoriesa
| Match category | Result (% concordance [95% confidence interval]) |
|||
|---|---|---|---|---|
| A | B | C | Overall | |
| Speciesb | 421/427 (98.6% [97.0–99.5]) | 47/54 (87.0% [75.1–94.6]) | 54/56 (96.4% [87.7–99.6]) | 522/537 (97.2% [95.4–98.4]) |
| Genusc | 427/427 (100% [99.1–100]) | 69/69 (100% [94.8–100]) | 55/56 (98.2% [90.4–100]) | 551/552 (99.8% [99.0–100]) |
| No reliable identificationd | 0/0 | 0/0 | 13/16 (81.3% [54.4–96]) | 13/16 (81.3% [54.4–96]) |
| Total | 427 | 69 | 72 | 568 |
The distribution of identifications by match category was significantly different across Biotyper consistency categories (P < 0.0001, Fisher's exact test).
Number of isolates concordant to species level with standard method identification/total number of isolates scoring ≥1.9.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring ≥1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Number of isolates concordant to genus level with standard method identification/total number of isolates scoring <1.7. Isolates scoring <1.7 were considered unreliable for identification by MALDI regardless of their concordance with standard method identification.
Variability among top 10 database matches.
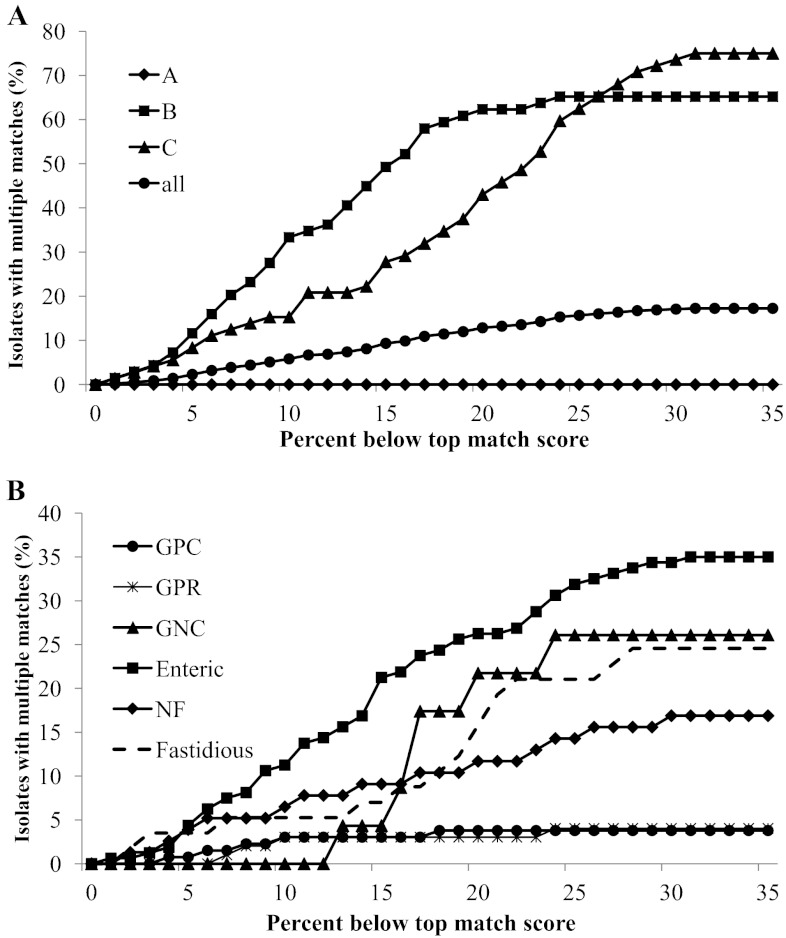
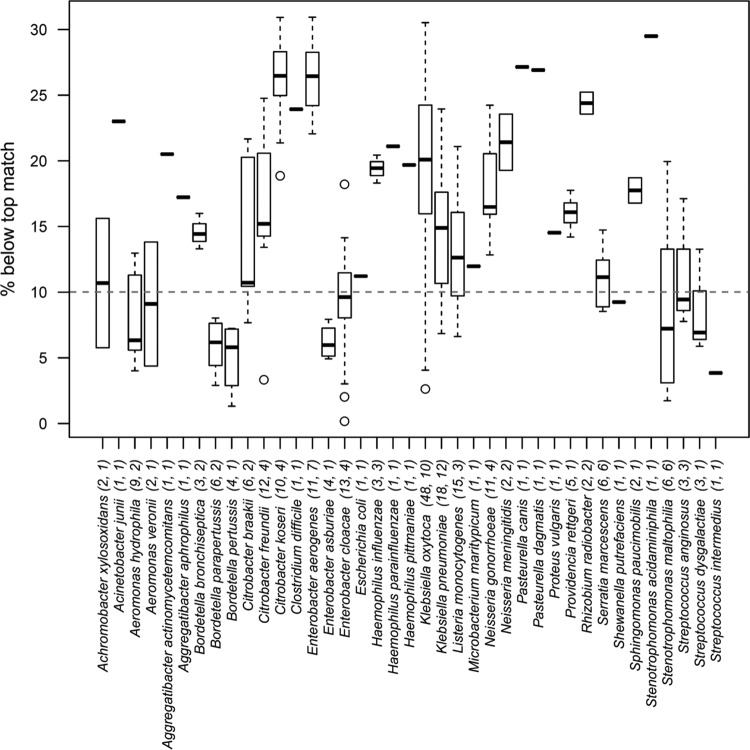
By default, the 10 best database matches are reported by the Biotyper software for each isolate. For 99 of the 568 (17.4%) concordant isolates, the top 10 results contained at least one significant mismatch. In an effort to develop a cutoff score for exclusion of additional incorrect matches, we plotted the cumulative proportion of isolates with significant mismatches versus the percent difference below the top database match score using cutoffs ranging from 0 to 35% (Fig. 2A and B). When all 568 core isolates were considered, there was a linear relationship observed between the cutoff score and the fraction of isolates with a significant mismatch up to a cutoff of ∼27% (R2 = 0.997; Fig. 2A), suggesting that the 10% rule used in previous studies (11, 24) may not be applicable to all isolates. To investigate if this rule might apply to subsets of our isolates, we plotted significant mismatches by cutoff percentage for Biotyper consistency categories as well as major organism categories (Fig. 2A and B). As expected, isolates in consistency category A did not have significant species or genus mismatches (Fig. 2A). Category B isolates showed essentially a linear relationship to a cutoff of ∼17% (R2 = 0.995), whereas category C isolates showed a slightly higher rate of increase in mismatches beyond a 10% cutoff (Fig. 2A). At a 10% cutoff, 23 of 69 (33%) and 11 of 72 (15%) isolates categorized as B and C, respectively, had at least one significant genus- or species-level mismatch among the top 10 results (Fig. 2A). Among the major organism categories, enteric Gram-negative rods showed the highest proportion of significant mismatches, followed by Gram-negative cocci and fastidious Gram-negative rods. The relationship between cutoff score and mismatches was essentially linear for enteric Gram-negative rods up to a cutoff of ∼26% (R2 = 0.988) (Fig. 2B) but showed an increased rate of mismatches above 12 to 13% for fastidious Gram-negative rods and Gram-negative cocci. At a 10% cutoff, nearly 11% of enteric and ∼6% of nonfermenting and fastidious Gram-negative rods, 3% of Gram-positive cocci and rods, and none of the Gram-negative cocci had significant mismatches (Fig. 2B). Interestingly, Gram-negative organisms had much higher proportions of mismatches overall than was seen with Gram-positive organisms (Fig. 2B). When mismatch score distributions were evaluated at higher resolution (Fig. 3), it became clear that certain genera, namely, Aeromonas, Bordetella, Enterobacter, Stenotrophomonas, and Streptococcus, had higher proportions of mismatches that scored very close to the top match.
Fig 2.
Cumulative proportion of the 568 core isolates with a significant species or genus mismatch among the top 10 MALDI-TOF MS results that scored within cutoff values ranging from 1 to 35% below the top match score. (A) Proportion of mismatches within each Biotyper consistency category (A, 427 isolates; B, 69 isolates; C, 72 isolates). (B) Proportion of mismatches within major organism categories. GPC, Gram-positive cocci (n = 132); GPR, Gram-positive rods (n = 99); GNC, Gram-negative cocci (n = 23); Enteric, enteric Gram-negative rods (n = 160); NF, nonfermenting Gram-negative rods (n = 77); Fastidious, fastidious Gram-negative rods (n = 57).
Fig 3.
Box and whisker plot showing the median (solid horizontal lines), upper and lower quartiles (boxes), upper and lower 1.5 interquartile ranges (whiskers), and outliers (open circles) of the percentage differences from the top-scoring match for all significant mismatches by species. In parentheses are the numbers of all significant mismatches followed by the number of isolates contributing to those mismatches. The area below the dashed horizontal line illustrates the relative proportion of isolates that would include mismatches using the 10% cutoff rule.
DISCUSSION
Recent studies have demonstrated the tremendous potential of MALDI-TOF MS for bacterial identification in the clinical microbiology laboratory, primarily because of its speed and cost-effectiveness. However, as this technology is more widely adopted, several important issues remain. Among these is the need for validation of the existing databases across broad collections of microorganisms. The range of organisms not readily identified by direct-smear approaches needs further clarification to allow development of efficient testing algorithms that incorporate extraction only when necessary. Finally, scoring algorithms should be critically evaluated to optimize identification of clinically relevant organisms and to resolve reporting issues that arise when multiple database matches meet genus- or species-level identification criteria. This study was designed to address these issues with the goal of improving the performance of MALDI-TOF MS in the clinical microbiology laboratory.
Although the direct-smear sample preparation method is simpler than extraction, it is also more susceptible to overloading, which can lead to poor results (reference 4 and data not shown). The majority of published studies using MALDI-TOF MS for bacterial identification used the direct-smear method for most isolates and reserved extraction for isolates that were not initially identified, yet extraction has been shown to improve identification (2, 4, 30). A recent study comparing these methods for Gram-positive cocci showed that only 56% and 20% of the isolates could be identified to genus and species levels, respectively, by direct smear, whereas extraction yielded genus and species identification for 95% and 69% of isolates, respectively (2). Among our diverse group of isolates, nearly 76% (140 of 183) were correctly identified by the direct-smear method, indicating that this is a viable first approach. However, the fact that the extraction method yielded significantly higher scores with more species-level identifications and fewer unreliable identifications reinforces the notion that it is more robust.
Most studies to date for routine bacterial identification use the Biotyper's recommended thresholds of ≥2.0 for species-level, 1.7 to 1.999 for genus-level, and <1.7 for unreliable identifications. However, alternative scores have been suggested (1, 11, 29), and the optimal thresholds are subject to debate. To address this question empirically, we evaluated a range of score thresholds on the achievable level of identification (Fig. 1). Although a simple receiver operating characteristic analysis suggested a higher species threshold (2.03), this type of analysis does not always yield the optimal value (13), and in this case, additional criteria, including a reduced false-negativity rate, increased yield of true positives, and improved consistency category distribution, resulted in substantial gains in efficiency at very modest cost to specificity at a threshold of 1.9. A recent study exploring species-specific thresholds for routine clinical isolates found that a wide variety of thresholds could be derived depending on the type of organism being evaluated (29). Their method resulted in thresholds that were dependent on the number of isolates tested for a given species. Although species-specific thresholds may allow fine-tuning of the identification algorithm, they could be cumbersome in the clinical laboratory without direct integration in identification software, and database updates would likely necessitate significant reanalysis to establish new thresholds. Broadly applicable thresholds determined empirically as described here can outperform default parameters and are a reasonable approach until automation of potentially higher-resolution analysis methods is widely available.
Numerous studies evaluating MALDI-TOF MS for identification of routine isolates such as staphylococci, enterococci, and enteric and common nonfermenting Gram-negative rods have reported >90% concordance with phenotypic methods (3, 4, 24, 30). Similarly, our study showed high concordance rates of 95.8% and 96.1% relative to the Phoenix and traditional phenotypic methods. As a reference laboratory, we encounter many isolates that are difficult to identify by phenotypic methods. Such isolates are often identified by 16S rRNA gene sequencing. Not unexpectedly, species-level identification rates were significantly lower for this category compared to those for routine isolates (P < 0.0001), and all of the isolates that MALDI-TOF MS could not identify due to low scores belonged to this category (Table 3; see Table S1 in the supplemental material). Among these 16 isolates were Corynebacterium tuberculostearicum, Nocardia farcinica, and Fusobacterium nucleatum, which have all been noted as difficult to identify by MALDI-TOF (1, 19, 31).
Several incorrect identifications seen here have been noted in other recent studies. Such examples include misidentification of Enterococcus casseliflavus as E. phoeniculicola (2, 5, 30), K. oxytoca as R. ornithinolytica (4), and Stenotrophomonas maltophilia as Pseudomonas hibiscicola (4). Some of these errors appear to be associated with previous versions of the Biotyper database, as 7 initially discrepant isolates in our study were resolved after upgrading from the 3740 to the 3995 database (Bordetella bronchiseptica to B. parapertussis [n = 2], Enterococcus phoeniculicola to E. casseliflavus, Enterococcus cecorum to E. casseliflavus, R. ornithinolytica to K. oxytoca, and P. hibiscicola to S. maltophilia [n = 2]). Multiple studies have described the poor resolution of MALDI-TOF among the S. mitis group (2, 7, 20, 22). Only 2 of our 9 (22%) non-pneumoniae S. mitis group isolates were misidentified as S. pneumoniae, suggesting that newer databases may potentially overcome this significant problem.
Additional factors could lead to unreliable identifications with MALDI-TOF MS, including database composition and depth. There is a demonstrated positive relationship between increased numbers of database spectra and improved MALDI-TOF MS scores (10, 27, 31, 32). Our data using a recent Biotyper database showed a similar trend (see Table S1 in the supplemental material). Isolates scoring ≥1.9 matched species with significantly more reference spectra than isolates scoring <1.9 (P < 0.0001). Another potential factor in unreliable identification is the inherent resolution of the spectra in the database. Biotyper results may have more than one species or genus scoring above the respective thresholds for a given isolate. Few studies have addressed this issue, and those that have done so resolved it by excluding any result scoring >10% below the top-scoring match (11, 24). Unfortunately, this 10% rule was developed using a custom database with different analysis parameters than the Biotyper (11). The newest version of the Biotyper software (3.0) attempts to address this problem by assigning consistency categories to the results. However, nearly 20% (110 of 568) of our isolates with species-level scores (≥1.9) were placed into categories B or C, indicating lower-confidence identifications. When the proportion of isolates with significant mismatches was plotted against the difference from the top-scoring match (cutoff score) for each consistency category, or all categories combined, there was no obvious inflection, suggesting that there is no universal score cutoff that applies to all species (Fig. 2A). For category C isolates, there was a small inflection at 10 to 14%, suggesting that the 10% cutoff rule may be somewhat more applicable to isolates in this category. This analysis is particularly useful in evaluating the proportion of isolates that would yield ambiguous results at a given cutoff (e.g., 33% of category B isolates at a 10% cutoff). Importantly, this analysis confirmed the reliability of category A results, which are defined as having unambiguous species-level consistency. A similar analysis of major organism categories revealed that Gram-negative rods (primarily enteric and fastidious organisms) and Gram-negative cocci had the highest proportions of significant mismatches (Fig. 2B). Of the 39 species resulting in significant mismatches, 10 enteric Gram-negative rods account for nearly 62% (137 of 220) of all ambiguities (Fig. 3). In general, Gram-positive organisms had substantially lower proportions of isolates with significant mismatches (Fig. 2B). The profiles of these curves trended toward linearity well beyond a 20% cutoff for enteric Gram-negative rods, but there were inflection points for fastidious Gram-negative rods and Gram-negative cocci at 13% and 12%, respectively, perhaps justifying a 10% cutoff for the latter organisms. Since a single universal cutoff may not be ideal for all classes of organisms, particularly Gram-negative rods, we evaluated the distribution of mismatch scores by species (Fig. 3). As observed in this analysis, a 10% cutoff would eliminate a substantial fraction of significant mismatches, but some species from genera like Achromobacter, Aeromonas, Bordetella, Citrobacter, Enterobacter, Klebsiella, Listeria, Serratia, Shewanella, Stenotrophomonas, and Streptococcus may require tighter cutoffs or additional testing to resolve ambiguous results (Fig. 3).
An important aspect of a cutoff score is whether it excludes correct results. In our core set of 568 isolates, there were 11 (1.9%) isolates scoring ≥1.9 with an incorrect top result above a correct but lower-scoring match (see Table S4 in the supplemental material). Most of these isolates were in consistency category B or C and belonged to the same complex or were very closely related to the top match. The only genus mismatch among this group was a K. oxytoca isolate called R. ornithinolytica by the Biotyper. This could be a significant error for laboratories that rely on the CLSI extended-spectrum β-lactamase (ESBL) screen/confirm algorithm because K. oxytoca, but not R. ornithinolytica, is among the recommended organisms to test for ESBL production (9). Although it does not appear to be a universal solution, some aspects of our data appear to support the use of a 10% cutoff, and it may be a pragmatic solution for handling mismatches among Biotyper results in the busy clinical laboratory. Ultimately, a species-specific cutoff score, tempered by the clinical relevance of species-level identification and the laboratory's testing capability, will likely be the optimal approach.
There are several limitations of our study. First, the inclusion of retrospective samples (50 of 690) to expand diversity could bias results in favor of higher concordance. Second, although our cohort was diverse, not all spectra in the database were tested; therefore the thresholds we propose may not be applicable to all species. Further, we chose a species-level threshold that increases sensitivity at the cost of specificity, which was advantageous among these isolates but may not always be optimal. Finally, we used extraction for all isolates (n = 690) and compared direct smear for a diverse yet smaller subset of isolates (n = 183), while routine use is largely focused on direct-smear analysis.
Although MALDI-TOF MS has several limitations, such as database diversity and resolution of closely related species (2, 4, 27, 30), many other microbial identification systems suffer from similar problems. Overall, we demonstrated the capabilities of the technology in correctly identifying vastly divergent groups of bacteria, including both routine and unusual species. Although the extraction sample preparation method was used throughout the study, we showed that the direct-smear method is broadly applicable and can be successfully applied as a first approach for the majority of routinely encountered organisms. Finally, we illustrated limitations in the current state of MALDI-TOF MS data analysis. There is still debate on the optimal score thresholds for identification, but we illustrated that they can be optimized to provide more species-level identifications than the conservative Biotyper settings. There is yet no consensus on how to handle multiple species or genera among the top 10 results, but the newly introduced consistency categories at least highlight problematic isolates. Additional cutoff algorithms could be developed in each laboratory based on the most frequently encountered organisms. Overall, MALDI-TOF MS is a powerful tool for the clinical microbiology laboratory with tremendous potential to improve patient care through rapid and accurate bacterial identification.
Supplementary Material
ACKNOWLEDGMENT
We thank the members of the ARUP Bacteriology laboratory for assistance in completion of this study.
Footnotes
Published ahead of print 19 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alatoom AA, Cazanave CJ, Cunningham SA, Ihde SM, Patel R. 2012. Identification of non-diphtheriae Corynebacterium by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:160–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424 doi:10.1371/journal.pone.0016424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bizzini A, et al. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cherkaoui A, Emonet S, Fernandez J, Schorderet D, Schrenzel J. 2011. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J. Clin. Microbiol. 49:3004–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherkaoui A, et al. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christensen JJ, et al. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis of Gram-positive, catalase-negative cocci not belonging to the Streptococcus or Enterococcus genus and benefits of database extension. J. Clin. Microbiol. 50:1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CLSI 2011. Performance standards for antimicrobial susceptibility testing. Twenty-first informational supplement. CLSI document M100–S21. Clinical and Laboratory Standards Institute; Wayne, PA [Google Scholar]
- 10. Couturier MR, Mehinovic E, Croft AC, Fisher MA. 2011. Identification of HACEK clinical isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1104–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Degand N, et al. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finegold SM, et al. 2005. Clostridium clostridioforme: a mixture of three clinically important species. Eur. J. Clin. Microbiol. Infect. Dis. 24:319–324 [DOI] [PubMed] [Google Scholar]
- 13. Fluss R, Faraggi D, Reiser B. 2005. Estimation of the Youden Index and its associated cutoff point. Biom. J. 47:458–472 [DOI] [PubMed] [Google Scholar]
- 14. Fournier R, et al. 2012. Chemical extraction versus direct smear for MALDI-TOF mass spectrometry identification of anaerobic bacteria. Anaerobe 18:294–297 [DOI] [PubMed] [Google Scholar]
- 15. Freiwald A, Sauer S. 2009. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 4:732–742 [DOI] [PubMed] [Google Scholar]
- 16. George WL, Kirby BD, Sutter VL, Citron DM, Finegold SM. 1981. Gram-negative anaerobic bacilli: their role in infection and patterns of susceptibility to antimicrobial agents. II. Little-known Fusobacterium species and miscellaneous genera. Rev. Infect. Dis. 3:599–626 [DOI] [PubMed] [Google Scholar]
- 17. Justesen US, et al. 2011. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 49:4314–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kallstrom G, et al. 2011. Recovery of a catalase-negative Staphylococcus epidermidis strain in blood and urine cultures from a patient with pyelonephritis. J. Clin. Microbiol. 49:4018–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knoester M, van Veen SQ, Claas EC, Kuijper EJ. 2012. Routine identification of clinical isolates of anaerobic bacteria: matrix-assisted laser desorption ionization–time of flight mass spectrometry performs better than conventional identification methods. J. Clin. Microbiol. 50:1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martiny D, et al. 2012. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mellmann A, et al. 2008. Evaluation of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neville SA, et al. 2011. Utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryzhov V, Fenselau C. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746–750 [DOI] [PubMed] [Google Scholar]
- 24. Saffert RT, et al. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saffert RT, Cunningham SA, Mandrekar J, Patel R. 2012. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagn. Microbiol. Infect. Dis. 73:21–26 [DOI] [PubMed] [Google Scholar]
- 26. Sauer S, et al. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843 doi:10.1371/journal.pone.0002843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seng P, et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 28. Simmon KE, et al. 2011. Isolation and characterization of “Pseudomonas andersonii” from four cases of pulmonary granulomas and emended species description. J. Clin. Microbiol. 49:1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szabados F, et al. 2012. Evaluation of species-specific score cutoff values of routinely isolated clinically relevant bacteria using a direct smear preparation for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based bacterial identification. Eur. J. Clin. Microbiol. Infect. Dis. 31:1109–1119 [DOI] [PubMed] [Google Scholar]
- 30. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veloo AC, Welling GW, Degener JE. 2011. The identification of anaerobic bacteria using MALDI-TOF MS. Anaerobe 17:211–212 [DOI] [PubMed] [Google Scholar]
- 32. Verroken A, et al. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Versalovic J, et al. 2011. Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.