Abstract
Catheter-associated urinary tract infection is caused by bacteria, which ascend the catheter along its external or internal surface to the bladder and subsequently develop into biofilms on the catheter and uroepithelium. Antibiotic-treated bacteria and bacteria residing in biofilm can be difficult to culture. In this study we used culture-based and 16S rRNA gene-based culture-independent methods (fingerprinting, cloning, and pyrosequencing) to determine the microbial diversity of biofilms on 24 urinary catheters. Most of the patients were catheterized for <30 days and had undergone recent antibiotic treatment. In addition, the corresponding urine samples for 16 patients were cultured. We found that gene analyses of the catheters were consistent with cultures of the corresponding urine samples for the presence of bacteria but sometimes discordant for the identity of the species. Cultures of catheter tips detected bacteria more frequently than urine cultures and gene analyses; coagulase-negative staphylococci were, in particular, cultured much more often from catheter tips, indicating potential contamination of the catheter tips during sampling. The external and internal surfaces of 19 catheters were separately analyzed by molecular methods, and discordant results were found in six catheters, suggesting that bacterial colonization intra- and extraluminally may be different. Molecular analyses showed that most of the species identified in this study were known uropathogens, and infected catheters were generally colonized by one to two species, probably due to antibiotic usage and short-term catheterization. In conclusion, our data showed that culture-independent molecular methods did not detect bacteria from urinary catheters more frequently than culture-based methods.
INTRODUCTION
Urinary catheters are commonly used to manage urinary incontinence and retentions, to measure urine output, or to help patients who had surgery or other medical conditions. Up to 25% of hospitalized patients need urinary catheters; however, catheterization is associated with increased risk of infection in the urinary tract, and >40% of all nosocomial infections in hospitals and other health care units are attributed to catheter-associated urinary tract infections (CAUTIs) (21). The presence of a catheter predisposes the patient to acquire infections by providing access to uropathogens into the bladder along its external and internal surfaces, which develop into biofilms on the catheters and uroepithelium (41). The risk of UTI is associated with the duration of catheterization. The daily increase in prevalence of bacteriuria after catheterization is estimated to be 3 to 10%, and bacteriuria occurs for almost all patients with an indwelling catheter over 30 days (48). Exposure to antibiotics can delay the onset of bacteriuria in catheterized patients; however, the effect is relatively short. The prevalence of bacteriuria will eventually reach the same level in patients with or without antibiotic treatment after 14 days (19).
Traditionally, the diagnosis of CAUTI has relied on clinical signs and symptoms and microbiological culture of urine, which are still the recommended methods (21). When the suspected catheter is removed, the catheter can be used for microbiological culture. However, on the surfaces of catheters bacteria grow in biofilms, which are structured communities of microorganisms surrounded by their extracellular products. Biofilm bacteria differ in their behavior and in phenotypic form from their planktonic counterparts. They grow poorly on agar plates when recovered from their natural or pathogenic ecosystems, and generally only planktonic cells can be cultured (7). Antibiotic usage, inappropriate culture conditions, and viable but nonculturable bacteria may also cause false-negative culture (51). To improve the recovery of the biofilm attached to the catheters, swabbing (32) and sonication (20) of the catheter have been used, and greater sensitivity was achieved by both approaches. In addition, pathogens can gain access to catheters extra- or intraluminally (42), and the microbial population on the two sides of the catheter can be different (15); hence, analyzing one side of the catheter or the catheter tip alone may be insufficient to detect all of the microbes on the catheter.
Many infecting organisms have been isolated from CAUTIs. Depending on the patient population, prior antimicrobial exposure, and the organisms unique to each hospital, the spectrum of microorganisms varies. The common CAUTI microorganisms detected by conventional culture are members of the fecal microbial communities, such as Escherichia coli, Pseudomonas aeruginosa, Proteus mirabilis, enterococci, Klebsiella species, and Citrobacter species, while occasionally coagulase-negative staphylococci (CoNS), Candida, and other species are also involved. CoNS and Enterococcus spp. are considered relatively avirulent and are more frequently recovered from asymptomatic infections than from symptomatic infections. Moreover, short-term catheters are frequently colonized by a single species, whereas CAUTIs in long-term catheterized patients are usually polymicrobial (21, 34).
In recent years culture-independent methods are increasingly applied to study infections, which often detect previously undescribed levels of bacterial diversity. For instance, to investigate chronic wound microbiota several studies (9, 13, 38, 44, 45) have applied denaturant gradient gel electrophoresis (DGGE), cloning, and pyrosequencing, which have revealed a substantially higher level of bacterial diversity than found by conventional culture. Another major advantage of molecular methods compared to culture-based methods is that detection of bacterial DNA is unaffected after antimicrobial exposure; however, positive molecular detection may originate from DNA from dead bacteria. This issue can be overcome by only extracting DNA from intact bacterial cells (22, 35, 36). Notably most of the hospitalized patients with an indwelling urinary catheter (up to 80%) receive antimicrobial therapy due to indication of infection (21, 23), and this may greatly influence the outcome of culture.
In this study, we examined the microbial diversity on the extra- and intraluminal surfaces of the urinary catheter by culture-independent molecular methods, including 16S rRNA gene PCR, DGGE, cloning, and next-generation sequencing. The molecular findings were subsequently compared to data obtained by routine microbiological culture of urine and catheter tips. The purpose was to investigate whether culture-independent molecular methods could detect more bacterial species than the culture-based methods from urinary catheters removed from patients.
MATERIALS AND METHODS
Patients and catheters.
Urinary catheters and urine were routinely sent to the Department of Clinical Microbiology, Rigshospitalet, Denmark, for culture. Samples were collected from 24 patients undergoing removal of the urinary catheter at Rigshospitalet. The catheters were randomly selected irrespective of the underlying disease. Fourteen patient journals (patients 1 to 14) were retrieved, whereas the medical information was not available for 10 patients (patients 15 to 24). Corresponding urine samples were available from 16 patients (patients 1 to 3, 7 to 9, 11, 13, 15, and 17 to 23).
For the 14 patients whose journals were available, the catheters' indwelling period varied from 3 days to long-term (≥30 days) catheterization, with only one catheter placed for less than a week. Except for patient 12, all of the patients had prior or ongoing antibiotic treatment either because of UTI or infection at another site. Four patients (7, 8, 10, and 11) were suspected of having UTI. The decision to analyze the samples was made by the medical doctors in different departments, and the Department of Clinical Microbiology processed the samples according to the established procedure without differentiating bacteriuria and asymptomatic UTI. Details for these patients can be found in Table S1 in the supplemental material.
Urine collection and culture.
Urine samples obtained from the catheters were stored in sterile 10-ml tubes at 4°C and processed within 24 h at the Department of Clinical Microbiology. A wet smear was performed, and phase-contrast microscopy was done. The presence of rods, cocci in chains or clusters, and yeast was noted. If there were visible microorganisms in every field, it was considered “positive”. If there were visible microorganisms in some fields, it was considered “maybe positive.” In addition, samples were plated using a 1- and a 10-μl eyelet onto 5% blood agar (Statens Seruminstitut [SSI], Copenhagen, Denmark) and on a modified Conradi-Drigalski (SSI) medium selective for Gram-negative rods and a Chromagar (SSI) using a 1-μl eyelet. If microscopy was positive, an additional tellurite-containing and α-glucuronidase-detecting agar medium (PGUA agar [SSI]) was plated. Cultured bacteria were identified using an Api kit (bioMérieux) of automatic systems for identification and analyzed on an ATB Expression 1550 Vitek system (bioMérieux). Identification of Enterobacteriaceae was performed with ID32E, and Enterococcus species were identified based on hemolysis, motility, and the fermentation of arabinose, raffinose, sorbitol, and arginine. Streptococci were identified by using a Rapid ID32 STREP, and hemolysis grouping was performed using Partho DX (Remel, Santa Fe, NM). Staphylococcus identification was primarily based on novobiocin sensitivity and subsequently analyzed by ID32 STAPH. Corynebacteria, especially Corynebacterium urealyticum, were identified with Api Coryne. Yeasts were identified with ID32C and/or Chromagar Candida (Becton Dickinson, Franklin Lakes, NJ). Culture results were provided in a quantified manner as the CFU ml−1. Cultivation and identification were always performed by the same trained individuals.
Catheter collection and culture.
Immediately after removal from the patient, the catheter tip was cut off (typically 5 to 7 cm) and transferred aseptically to a sterile tube and transported to the Department of Clinical Microbiology within 24 h. Catheter tips were rolled onto a 5% blood agar (SSI) and modified Conradi-Drigalski agar (SSI), followed by stepwise spreading. All species were identified using the Api kit of automatic systems for identification and analyzed on an ATB expression 1550 Vitek system as described above. Cultivation and identification were always performed by the same trained individuals. After the catheter samples for microbiological culture were obtained, the catheters were kept on ice during the 5 to 6 h of transportation to Aalborg University. The catheters were processed immediately upon arrival.
Isolation of biofilm and DNA extraction.
Collection of biomass from the internal and external surfaces of urinary catheters was done by forceful scraping with a sterile scalpel (29). However, in five catheters (i.e., catheters 20 to 24), it was impossible to separate the internal and external samples; thus, the internal and external samples were mixed before DNA extraction for these catheters. It resulted in a total of 43 samples for molecular analysis. DNA extraction was performed according to the method of Larsen et al. (29).
Partial 16S rRNA gene amplification, DGGE, and Sanger sequencing.
To obtain the PCR amplicon for DGGE analysis, the 16S rRNA gene was first amplified by PCR using Taq DNA polymerase with the primers 8F (24) and 1390R (28) according to the method of Thomsen et al. (43). Then, 1 μl of each PCR product was used as a template to prepare amplicons for DGGE using the primer 341F-GC (33) and the primer 907R (28). Negative controls including water and PCR mix were included for every five samples and were always negative, indicating that there was no contamination of the reagents. DGGE of the amplicons was performed according to the method of Larsen et al. (29). The purified PCR products were sequenced by Macrogen, Inc. (Seoul, South Korea), using the 341F primer.
Nearly full-length 16S rRNA gene amplification, cloning, and Sanger sequencing.
Nearly full-length 16S rRNA gene was amplified by PCR using Taq DNA polymerase with the primer pair 8F (24) and 1390R (28) and the primer pair 8F (24) and 1492R (28) according to the method of Thomsen et al. (43). The PCR products for each sample were pooled and purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and used for cloning. Cloning was performed using a TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). Following blue and white screening according to standard methods, more than 48 clones from each library were isolated and subcultured. The plasmids were amplified by rolling-cycle reaction using an Illustra TempliPhi kit (GE Healthcare, United Kingdom) and sequenced by Macrogen unidirectionally or bidirectionally using M13 primers.
Sanger sequencing data analysis.
Vector trimming, quality analysis, consensus assembly, and BLAST search of sequencing data were performed by using CLC Main Workbench (CLC Bio). The consensus sequence for each clone was compiled by assembling the forward and reverse sequences and trimming vector sequences. Each clone sequence and DGGE sequence was manually curated, and ambiguous nucleotides were resolved by inspecting the sequencing chromatogram. If the sequence contained many ambiguous nucleotides, indicating the presence of mixed species, the sequence was discarded. The primer sequences and vector sequence regions beyond the primers were trimmed. Checks for chimeric sequences were conducted using the program Mallard (4). Initial microbial species identification was performed by using the integrated blastn search in the NCBI database (http://www.ncbi.nlm.nih.gov/) with standard parameters except that “Nucleotide Collection“ was the chosen database and the “Entrez Query” was limited to “Bacteria [ORGN].” All sequences were aligned to SILVA rRNA database using the SINA Web Aligner (39). The aligned sequences were imported into the ARB software package for taxonomic lineage assignment (30) by calculating neighbor-joining, maximum-parsimony, and maximum-likelihood trees. The coverage of the clone library was assessed by Good's coverage estimator (18).
Pyrosequencing and data analysis.
A 466-bp fragment of 16S rRNA gene flanking V3 and V4 regions was amplified using the modified primers PRK341F (5′-CCTAYGGGRBGCASCAG-3′) and PRK806R (5′-GGACTACNNGGGTATCTAAT-3′) (50). The PCR incorporated an Illustra PuReTaq Ready-To-Go PCR beads kit (GE Healthcare), 2 μl of each primer (10 μM) (TAG, Copenhagen, Denmark), 1 μl of diluted template, and water to a total volume of 25 μl. The PCR incubation conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 20 s, 56°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. Reactions were analyzed by 1% agarose gel electrophoresis. A band of the approximately correct size was sliced from the gel and purified using a QIAEX II gel extraction kit (Qiagen). A second PCR amplification (25 μl) was performed with the fusion primers 341F and 806R with adapters and tags. The PCR mix of second amplification contained 5 μl of 5× Phusion HF buffer (7.5 mM MgCl2; Finnzymes, Finland), 0.5 μl of a 10 mM deoxynucleoside triphosphate mixture, 0.25 μl of Phusion Hot Start DNA polymerase (1 U/μl; Finnzymes), 1.25 μl of each primer (10 μM), 2 μl of PCR product amplified from the first step, and water to a total of 25 μl. The PCR incubation conditions of the second PCR were as follows: initial activation of the hot start polymerase at 98°C for 30 s, followed by 15 cycles of 98°C for 5 s, 56°C for 20 s, and 72°C for 20 s, with final extension at 72°C for 5 min, and the number of cycles was reduced to 15. Bands of ∼500 bp were cut out and purified as described above. The amplified fragments with adapters and tags were quantified using a Qubit fluorometer (Invitrogen). Tagged samples were mixed in approximately equal concentrations (4 × 106 copies/μl) to ensure equal representation of each sample. DNA samples were sequenced on the 75×75 Titanium GS PicoTiterPlate (PTP) by using a GS FLX pyrosequencing system according to the manufacturer's instructions (Roche).
Analysis of 16S rRNA gene amplicon sequences was performed by using the Quantitative Insights Into Microbial Ecology (QIIME; v1.3.0) pipeline (6). The sequencing data were processed initially with AmpliconNoise (40) to remove noises. Then, QIIME pipeline separates the sequences into individual communities by barcode and utilizes a suite of external programs to make taxonomic assignments and estimate the phylogenetic diversity. These data were used to generate taxonomic summaries. The default settings in QIIME were used for analysis, except that the sequences were clustered into operational taxonomic units by 99% sequence similarity, and taxonomy assignment was done by using BLAST and Greengenes taxonomy (12).
FISH.
Fluorescence in situ hybridization (FISH) was performed on fixed biofilms (2, 10). Visualization of the hybridized samples was carried out on a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Germany). The following oligonucleotide probes were used: Ent targeting Enterobacteriaceae except for Proteus spp. (25) and EUBmix (1, 8). Oligonucleotide probes were 5′ labeled with 5(6)-carboxyfluorescein-N-hydroxy-succinimide ester (FLUOS) or with sulfoindocyanine dyes (Cy3; Thermo Hybaid, Germany).
Statistics.
The Fisher exact probability test was used to determine statistically significant differences of positive rates between urine culture, catheter tip culture, and 16S rRNA gene PCRs of catheter surfaces.
Nucleotide accession numbers.
The GenBank accession numbers for the 16S rRNA gene sequences determined in the present study are JQ820171 to JQ820200.
RESULTS
Catheter and urine results.
All 24 removed catheters were examined by culture and broad-range 16S rRNA gene PCR (i.e., partial 16S rRNA gene PCR and DGGE), of which 16 had corresponding urine culture. The results for these 16 catheters showed that cultures of the catheter tip and urine were concordant for eight cases (seven positive and one negative) and discordant for the other eight cases (all were catheter tip culture positive and urine culture negative); the difference was statistically significant (Fisher exact test, two-tailed, P = 0.006). In comparison, the correlation between PCR and urine culture was greater with disagreement only observed in 2 of the 16 cases (7 positive, 7 negative, and 2 PCR positive but urine culture negative); the difference was statistically insignificant (Fisher exact test, two-tailed, P = 0.72). Comparison of the 24 catheters showed that all catheter tip cultures were positive except for one (catheter 15), while PCR was only positive for 14 catheters (Fisher exact test, two-tailed, P = 0.004).
Overall, culture of catheter tips resulted in more frequent positive identifications compared to urine culture and DGGE analysis (Table 1). Candida species were not found by DGGE because eukaryotic PCR primers were not used. In total, culture identified 19 species, including 2 fungal species, all of which were common pathogens involved in CAUTIs or normal skin microbes, such as CoNS and corynebacteria except for Achromobacter denitrificans. In comparison, DGGE and sequencing analysis identified 13 species, all of which were also found by culture except Actinomyces neuii, uncultured Corynebacterium sp., and Corynebacterium jeikeium. However, cultures of catheter tip and urine and DGGE analysis frequently disagreed on the species detected from the same patient.
TABLE 1.
Culture and DGGE results
| Parameter | Culture |
DGGE |
||||
|---|---|---|---|---|---|---|
| Catheter | Urine | Total | External | Internal | Mixed | |
| Total no. of analyzed catheters | 24 | 16 | 24 | 19 | 19 | 5 |
| No. positive/no. negativea | 23/1 | 7/9 | 14/10 | 10/9 | 10/9 | 1/4 |
| No. polymicrobial/no. monomicrobialb | 10/13 | 3/4 | 4/10 | 2/8 | 2/8 | 0/1 |
| Total no. of species | 16 | >7 | 13 | 9 | 11 | 1 |
| Catheter no. | ||||||
| Gram-negative bacteria | ||||||
| Achromobacter denitrificans | 11 | 11 | 11 | |||
| Citrobacter koseri | 9, 14 | 9 | 14 | 14 | ||
| Escherichia coli | 13 | |||||
| Enterobacter cloacae | 1 | |||||
| Enterobacter sp. | 1 | 1 | ||||
| Klebsiella oxytoca | 7 | 7 | 7 | 7 | ||
| Klebsiella pneumoniae | 10 | 10 | 10 | |||
| Morganella morganii | 13 | |||||
| Pseudomonas aeruginosa | 12 | 12 | ||||
| Stenotrophomonas maltophilia | 9 | 9 | ||||
| Gram-positive bacteria | ||||||
| Actinomyces neuii | 12 | 12 | ||||
| CoNS | 2, 8, 9, 11, 17–19, 20–23 | |||||
| Corynebacterium jeikeium | 19 | |||||
| Corynebacterium sp. | 6, 8 | |||||
| Uncultured Corynebacterium sp. | 2 | |||||
| Enterococcus faecalis | 3, 4, 13, 19, 21, 24 | 19 | 10, 13, 19 | 4, 19 | 21 | |
| Enterococcus faecium | 17, 24 | |||||
| Micrococcus luteus | 9 | |||||
| Nonhemolytic streptococci | 8 | 13 | ||||
| Staphylococcus epidermidis | 5, 16 | 6 | ||||
| Staphylococcus haemolyticus | 2, 8 | 2 | ||||
| Unidentified mixed bacterial species | 8, 9 | |||||
| Fungi | ||||||
| Candida albicans | 17 | |||||
| Candida glabrata | 20 | |||||
That is, the number of positive identifications/the number of negative identifications.
That is, the number of polymicrobial identifications/the number of monomicrobial identifications.
Table 1 shows also that polymicrobial community was more frequently found by catheter tip culture than by DGGE. The highest number of bacterial species identified from a single sample was four (culture of catheter 9). In addition, all of the patients, who were suspected of having CAUTIs, had bacteria detected by all three methods except the corresponding urine of catheter 10, which was not cultured (Table 1).
Extraluminal versus intraluminal identifications by molecular methods.
The insides and outsides of 19 catheters were analyzed separately by PCR, which showed concordant results in 13 catheters (6 negative and 7 positive) and discordant results in 6 catheters (Table 1). Sequencing of the DGGE bands from these samples identified 13 bacterial species in total (Table 1 and Fig. 1). Among the seven catheters with positive PCR from both sides of the catheter, three catheters (catheters 7, 11, and 14) had the same species on both sides (all were monomicrobial), while the remaining four catheters (catheters 2, 10, 12, and 19) had one common species on both sides and an additional species on one side of the catheter. These four catheters were also the only ones from which two species were identified, whereas all the other PCR-positive catheters were colonized with a single species according to DGGE analysis. Of the six discordant catheters, three catheters (catheters 8, 9, and 13) were PCR positive on the external surface, while the remaining three catheters (catheters 1, 4, and 6) were PCR positive on the internal surface. Hence, there was no difference in the positive rate between the two sides.
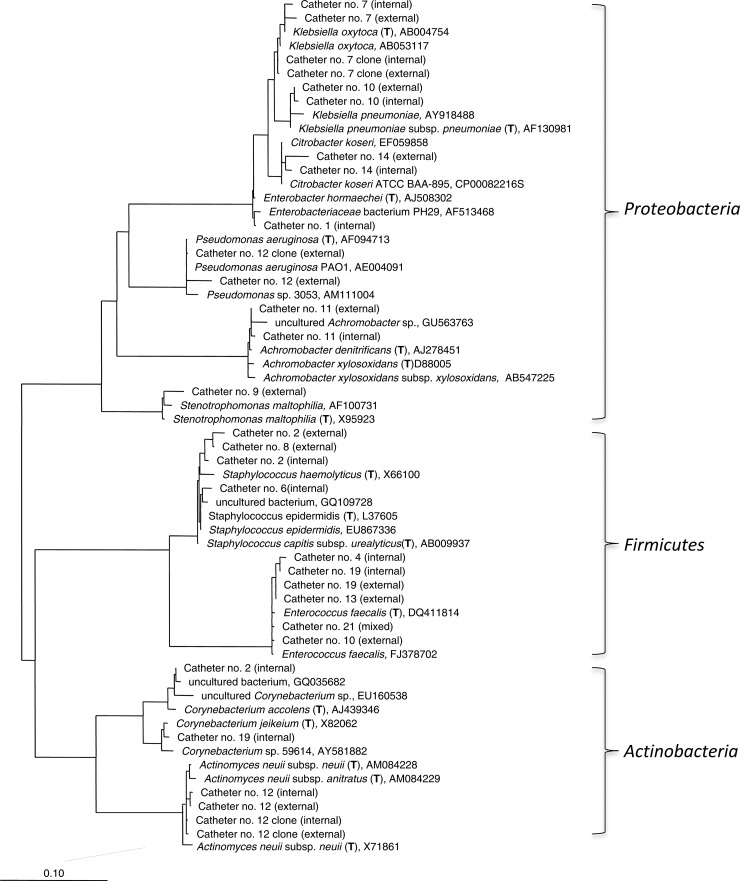
Fig 1.
Phylogenetic tree based on DGGE and cloning sequences obtained from the catheter samples. The tree was calculated using the maximum-likelihood method. The scale bar represents the 10% estimated sequence derivation. (T), type strains.
To determine whether DGGE was an adequate method for the investigation of bacterial diversity in the present study, two catheters (four DNA samples) were randomly selected for cloning and 454 pyrosequencing of the 16S rRNA genes. From each clone library, 60 to 190 clones were sequenced, and the coverage was 100%. The species identified by cloning were exactly the same as detected by DGGE for all four samples (Table 2). For each sample, the number of pyrosequencing reads after quality filtering by AmpliconNoise was >5,000, which provides higher sampling-depth than DGGE and cloning. From the external surface of catheter 12, Actinomyces and Pseudomonas were detected by all three molecular methods. However, pyrosequencing identified one more genus than DGGE and cloning in the other three samples. Enterobacter hormaechei was found on both the external and the internal surfaces of catheter 7, in addition to Klebsiella and, in addition to Actinomyces, Pseudomonas was detected on the interior of catheter 12. However, due to the relatively short length of the amplicons and the high degree of sequence similarity within Klebsiella and Pseudomonas genera, species-level assignment was not achieved.
TABLE 2.
Cloning and pyrosequencing results
| Catheter no. and position | DGGE | Cloning |
Pyrosequencing |
||||
|---|---|---|---|---|---|---|---|
| No. of clones | Species (% of cloning library) | No. of sequences | Family (% of total sequences) | Genus (% of total sequences) | Species (% of total sequences) | ||
| 7 | |||||||
| Exterior | K. oxytoca | 176 | K. oxytoca (100) | 7,085 | Enterobacteriaceae (100) | Enterobacter (68.0) | E. hormaechei (68.0) |
| Klebsiella (31.9) | Undetermined | ||||||
| Interior | K. oxytoca | 190 | K. oxytoca (100) | 5,683 | Enterobacteriaceae (100) | Enterobacter (66.3) | E. hormaechei (66.3) |
| Klebsiella (33.3) | Undetermined | ||||||
| 12 | |||||||
| Exterior | A. neuii | 73 | A. neuii (82) | 28,579 | Actinomycetaceae (50.8) | Actinomyces (50.8) | A. neuii (50.8) |
| P. aeruginosa | P. aeruginosa (18) | Pseudomonadaceae (49.1) | Pseudomonas (49.1) | Undetermined | |||
| Interior | A. neuii | 60 | A. neuii (100) | 5,795 | Actinomycetaceae (90.6) | Actinomyces (90.6) | A. neuii (90.6) |
| Pseudomonadaceae (9.4) | Pseudomonas (9.4) | Undetermined | |||||
Visualization of bacteria.
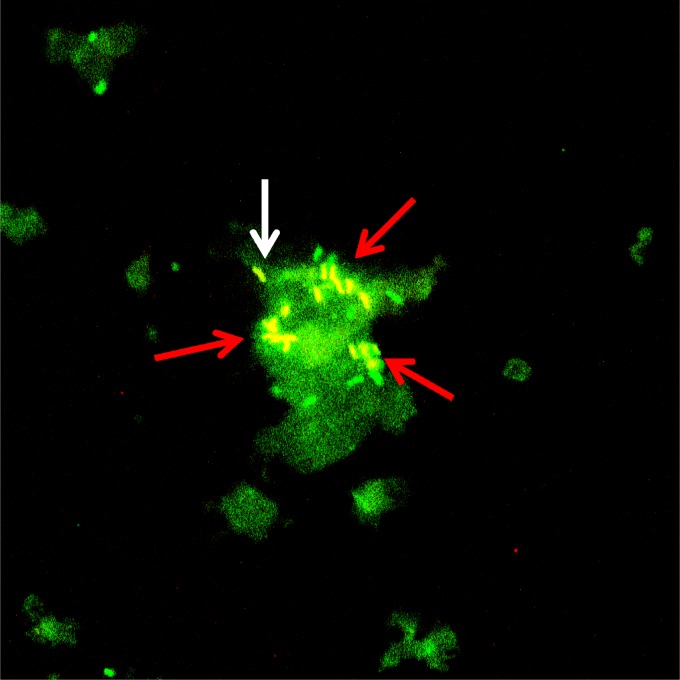
To visualize the presence and organization of bacteria, an additional removed urinary catheter was used for FISH. Both single cells and clusters of bacteria were detected in the material scraped from the catheter (Fig. 2) by the universal probe and the Ent probe, which correlates with the finding of E. faecalis and E. coli by culturing of the catheter tip. No large aggregates of bacteria were observed, indicating that only a small number of bacterial cells were unevenly distributed on the catheter.
Fig 2.
FISH micrograph of catheter biofilm. Material scraped from a catheter was hybridized with a universal bacterial probe (EUBmix) and an Enterobacteriaceae probe. Single cells (white arrow) and clusters of enterobacteria (red arrow) are visible in the biofilm. The large green background indicates unspecific binding of EUBmix probes to the biofilm. E. faecalis and E. coli were isolated from the catheter tip by culture.
DISCUSSION
To our knowledge, only one study (15) has been published using culture-independent molecular methods to investigate microbial diversity on the removed urinary catheters. The authors of that study examined 14 catheters removed after 14 days of catheterization using 16S rRNA gene PCR and cloning. They found that all eight catheters from patients without systemic antimicrobial treatment were colonized by multiple species (estimated average of >20 species), with extensive variation between the external and internal surfaces, whereas only one of the six catheters removed from patients receiving systemic antibiotics was PCR positive, and only a single species was identified on both sides of the catheter. Thus, antimicrobial agents significantly reduce the microbial diversity on the catheters, which was also observed in our study. In the present study, 13 of the 14 patients with available medical records were treated with antibiotics during catheterization, and PCR was positive in 12 of them. All of the PCR positive catheters were colonized by one to two species (1.3 on average), as shown by DGGE. Even when pyrosequencing with >5,000 sequences per sample was used, only one or two bacterial genera dominated 99.9 to 100% of the sequences for each sample (Table 2). The low diversity of the biofilm indicates that antibiotics or antibiotic prophylaxis kill some species but selected for a few and possibly resistant ones. Other likely factors contributing to the observed low diversity that we observed here are the relatively short catheter indwelling period (generally <30 days) and the microbial competition for resources resulting in the exclusion of some species.
Our results, overall, showed no apparent difference in positive rate and total number of identified bacterial species between the inside and outside of the catheters; nonetheless, it is likely that colonization of the bacteria was heterogeneous on the catheters (15). This is suggested by the FISH micrographs (Fig. 2) and indicated by the finding that six catheters had discordant PCR results between the inside and outside, and four of the seven concordant PCR-positive catheters had different species on the two sides of the catheters. This heterogeneous distribution of bacteria has also been observed in a number of biofilm-related infections, such as chronic wounds (26, 44) and the respiratory tracts of cystic fibrosis patients (5).
All microorganisms identified here (Table 1) are common pathogens involved in CAUTIs except for A. denitrificans, A. neuii, the uncultured Corynebacterium sp., and C. jeikeium. A. denitrificans has been isolated from various clinical specimens, such as urine, eye swabs, and pleural fluid, but no detailed reports about its clinical significance are available (47). A. neuii is a rare pathogen, which has mainly been reported to cause abscesses, but it has also been found in UTIs, skin lesions, bacteremia, endocarditis, etc. (16, 46). The two corynebacteria (uncultured Corynebacterium sp. and C. jeikeium) were only identified by DGGE. Corynebacteria constitute part of the normal skin flora, which makes it difficult to distinguish between infection, colonization, and contamination with these organisms. However, they are now recognized with increasing frequency as opportunistic human pathogens (17). Both corynebacteria were found on the inside of the catheters; hence, they were more likely colonizers than contaminants. We found E. coli, the most commonly isolated Gram-negative organism in CAUTIs, only once, and other common species such as Proteus mirabilis, Serratia spp. and Providencia stuartii were not detected at all. This was expected because these species are normally only isolated when infection is suspected. Only 24 catheters were analyzed; therefore, the species identified are not representative for our facility and should not be compared to other studies.
We provide here the first comparison of molecular and culture investigations of microbes on urinary catheters. Overall, culture of catheter tips by the rolling-plate method yielded significantly higher positive rates, which was unexpected since culture-independent molecular methods are generally more sensitive than culture for detecting bacteria in biofilm-related infections (11, 44, 49). However, the applied semiquantitative Maki's roll-plate method for catheter culture was originally developed for intravenous-catheter-related infections with a cutoff of 15 colonies per culture plate for distinguishing contamination (31). Its application on urinary catheters has never been studied. The urethra, which is normally colonized by many different bacteria from the vagina, skin, or feces, contains more microbes than the insertion site for intravenous catheters. Thus, using Maki's roll-plate method for urinary catheters may lead to overestimation of infection. Potential contamination is indicated by the finding that CoNS, including S. epidermidis, were isolated from 13 of the 24 catheter tips, whereas only three catheters were positive for Staphylococcus species by DGGE analysis and none of the urine cultures (Table 1). CoNS are previously reported to account for 20 to 30% of isolates from short-term catheters and less than 10% from long-term catheters (34). Since CoNS are frequently found on skin and mucosal membranes as part of the normal human microbes, the risk of urethral contamination during catheter removal is considerable. Rinsing the catheter tip with sterile phosphate-buffered saline before culturing may help to remove the loosely attached contaminating bacteria (15). Similarly, after the catheters are used for roll-plate culture, contaminants are significantly reduced; hence, the samples used for molecular studies here were less affected by contaminants. On the other hand, roll-plate culture probably also removed significant amounts of bacteria from the catheter surfaces in some cases, and the applied molecular methods were not sensitive enough to detect the remaining ones. Consequently, PCR had a lower positive rate than catheter tip culture.
Urine culture and PCR only showed disagreement in two catheters; however, the species identified by urine culture and DGGE were at least different in three of the seven concordant positive cases (catheters 1, 9, and 13). The differences could be reasoned by the presence of bacteria in biofilms colonizing only the catheter without a detectable level of planktonic bacteria in the urine (32), or the loosely attached bacteria were removed from the catheter by roll-plate method. Despite of this potential limitation, the overall findings in the present study suggest that culture of urine is still recommended for the routine diagnosis of CAUTI, whereas the catheter tips may overestimate microbial presence and diversity. This is in agreement with the most recent recommendations from the U.S. Centers for Disease Control and Prevention (www.cdc.gov/nhsn/pdfs/pscManual/7pscCAUTIcurrent.pdf), which state that urinary catheter tips should not be cultured and are not acceptable for the diagnosis of a UTI.
We used DGGE as the main molecular method to identify the bacterial species. DGGE is a method based on separating different DNA sequences according to their melting behavior (33), which has a relatively low resolution compared to cloning and next-generation sequencing technologies. DGGE and cloning identified the same species in all four samples. However, 454 pyrosequencing identified one more genus in three samples. Hence, it was likely that some of the other 16S rRNA gene PCR-positive samples contained more species than were revealed by DGGE. To detect the additional bacteria in the samples, all samples could have been analyzed by pyrosequencing; nevertheless, due to the cost of pyrosequencing, it was only used to evaluate DGGE results in the four samples.
It is also noteworthy that in the case of the exterior of catheter 12, the proportional read abundance of A. neuii and Pseudomonas differed between cloning and pyrosequencing, likely because of primer bias since different primer pairs were used for cloning and pyrosequencing (27). There are a few widely used broad-range 16S rRNA gene primers. Although “universal,” these primers differ in their taxonomic coverage, the degree of specificity of primers for different organisms, and the extent of phylogenetic information generated by the target 16S rRNA gene fragment (27). Cloning bias may also occur due to a different ligation efficiency of TA cloning system when cloning diverse insert pools (37). Similarly, 454 pyrosequencing is associated with various biases, such as those caused by emulsion PCR, systematic base calling errors, etc. Even so, the 454 pyrosequencing reads may be used to compare the proportional read abundance of a given species with itself across samples (3). Thus, the pyrosequencing data indicated that the proportions of Enterobacter and Klebsiella in the population were nearly the same on the two sides of catheter 7, whereas the proportions of Actinomyces and Pseudomonas were unequal on the two sides of catheter 12. For the purpose of absolute quantification, quantitative PCR with primers specific for an organism or an organism group is more suitable.
To study the microbial community composition, DGGE, cloning, and next-generation sequencing such as 454 pyrosequencing can provide detailed information; however, these approaches are too time-consuming to be applicable for routine clinical diagnosis. In comparison, quantitative PCR is a fast culture-independent molecular method and has been used routinely in many clinical microbiological departments. Choosing the appropriate quantitative PCR assay requires a priori supposition of the pathogens involved in contrast to these other methods. To detect and identify the causative agents quickly and without a priori supposition of the involved bacteria, the new PLEX-ID system may be a good candidate. This technology can in theory identify all present microbes in a sample within 6 h by using multiple primer sets to amplify multiple regions of the bacterial genomes, followed by mass spectrometry analysis (14).
In the present study, power calculation was not performed since the difference in sensitivity of the different methods was not known beforehand. In addition, it was not possible to calculate the correlation between the insertion time and the positivity of detection of organisms with catheter tip culture, urine culture, and PCR, since these three methods showed no difference in the positive rate for the catheters with known insertion times.
Conclusions.
This study suggests that prophylaxis and antibiotics reduce the diversity of microbial species associated with urinary catheters, which may be effective for the prevention of bacterial colonization in short-term catheterized patients. Surprisingly, culture-independent molecular methods did not detect bacteria more frequently than culture-based methods. Thus, based on this pilot study, urine cultures seem to be sufficient for the routine diagnosis of CAUTI, which, however, should be confirmed by other studies. Caution should be taken when using catheter tips for diagnosis to avoid contamination, and loosely attached bacteria should be removed by rinsing the catheter with, e.g., phosphate-buffered saline. 16S rRNA gene fingerprinting did not detect more bacterial species than culture-based methods from the urinary catheters, whereas deep sequencing using next-generation sequencing tools may uncover more species. Finally, the species identified are not representative for our facility and should not be compared to other studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jane Ildal, Lars Christophersen, and Masumeh Chavoshi for valuable technical assistance.
Footnotes
Published ahead of print 26 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amend AS, Seifert KA, Bruns TD. 2010. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Ecol. 19:5555–5565 [DOI] [PubMed] [Google Scholar]
- 4. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjarnsholt T, et al. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558 [DOI] [PubMed] [Google Scholar]
- 6. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costerton JW. 2007. The biofilm primer. Springer, Berlin, Germany [Google Scholar]
- 8. Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 9. Davies CE, et al. 2004. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J. Clin. Microbiol. 42:3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeLong EF, Wickham GS, Pace NR. 1989. Phylogenetic stains: rRNA-based probes for the identification of single cells. Science 243:1360–1363 [DOI] [PubMed] [Google Scholar]
- 11. Dempsey KE, et al. 2007. Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res. Ther. 9:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dowd SE, et al. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43 doi:10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ecker DJ, et al. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank DN, et al. 2009. Culture-independent microbiological analysis of Foley urinary catheter biofilms. PLoS One 4:e7811 doi:10.1371/journal.pone.0007811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Funke G, von Graevenitz A. 1995. Infections due to Actinomyces neuii (formerly “CDC coryneform group 1” bacteria). Infection 23:73–75 [DOI] [PubMed] [Google Scholar]
- 17. Funke G, Von Graevenitz A, Clarridge JE, III, Bernard KA. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 19. Hartstein AI, Garber SB, Ward TT, Jones SR, Morthland VH. 1981. Nosocomial urinary tract infection: a prospective evaluation of 108 catheterized patients. Infect. Control 2:380–386 [DOI] [PubMed] [Google Scholar]
- 20. Holá V, Ruzicka F, Horka M. 2010. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol. Med. Microbiol. 59:525–528 [DOI] [PubMed] [Google Scholar]
- 21. Hooton TM, et al. 2010. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin. Infect. Dis. 50:625–663 [DOI] [PubMed] [Google Scholar]
- 22. Horz H-P, Scheer S, Vianna ME, Conrads G. 2010. New methods for selective isolation of bacterial DNA from human clinical specimens. Anaerobe 16:47–53 [DOI] [PubMed] [Google Scholar]
- 23. Hustinx WN, Mintjes-de Groot AJ, Verkooyen RP, Verbrugh HA. 1991. Impact of concurrent antimicrobial therapy on catheter-associated urinary tract infection. J. Hosp. Infect. 18:45–56 [DOI] [PubMed] [Google Scholar]
- 24. Juretschko S, et al. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kempf VA, Trebesius K, Autenrieth IB. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirketerp-Møller K, Jensen PØ, et al. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuczynski J, et al. 2011. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 13:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lane D. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 29. Larsen MKS, Thomsen TR, Moser C, Hoiby N, Nielsen PH. 2008. Use of cultivation-dependent and -independent techniques to assess contamination of central venous catheters: a pilot study. BMC Clin. Pathol. 8:10 doi:10.1186/1472-6890-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maki DG, Weise CE, Sarafin HW. 1977. A semiquantitative culture method for identifying intravenous-catheter-related infection. N. Engl. J. Med. 296:1305–1309 [DOI] [PubMed] [Google Scholar]
- 32. Matsukawa M, Kunishima Y, Takahashi S, Takeyama K, Tsukamoto T. 2005. Bacterial colonization on intraluminal surface of urethral catheter. Urology 65:440–444 [DOI] [PubMed] [Google Scholar]
- 33. Muyzer G, De Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicolle L. 2005. Catheter-related urinary tract infection. Drugs Aging 22:627–639 [DOI] [PubMed] [Google Scholar]
- 35. Nocker A, Camper AK. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nocker A, Richter-Heitmann T, Montijn R, Schuren F, Kort R. 2010. Discrimination between live and dead cells in bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int. Microbiol. 13:59–65 [DOI] [PubMed] [Google Scholar]
- 37. Palatinszky M, Nikolausz M, Sváb D, Márialigeti K. 2011. Preferential ligation during TA-cloning of multitemplate PCR products: a factor causing bias in microbial community structure analysis. J. Microbiol. Methods 85:131–136 [DOI] [PubMed] [Google Scholar]
- 38. Price LB, et al. 2009. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One 4:e6462 doi:10.1371/journal.pone.0006462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned rRNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quince C, Lanzen A, Davenport R, Turnbaugh P. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38 doi:10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stickler DJ. 2008. Bacterial biofilms in patients with indwelling urinary catheters. Nat. Clin. Pract Urol. 5:598–608 [DOI] [PubMed] [Google Scholar]
- 42. Tambyah PA, Halvorson KT, Maki DG. 1999. A prospective study of pathogenesis of catheter-associated urinary tract infections. Mayo Clin. Proc. 74:131–136 [DOI] [PubMed] [Google Scholar]
- 43. Thomsen TR, Finster K, Ramsing NB. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomsen TR, et al. 2010. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen. 18:38–49 [DOI] [PubMed] [Google Scholar]
- 45. Tuttle MS, et al. 2011. Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J. Clin. Microbiol. 49:3812–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. von Graevenitz A. 2011. Actinomyces neuii: review of an unusual infectious agent. Infection 39:97–100 [DOI] [PubMed] [Google Scholar]
- 47. von König C-HW, Riffelmann M, Coenye T. 2011. Bordetella and related genera, p 739–750 In Versalovic JM, et al. (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 48. Warren JW. 2001. Catheter-associated urinary tract infections. Int. J. Antimicrob. Agents 17:299–303 [DOI] [PubMed] [Google Scholar]
- 49. Wolff TY, et al. 2011. Detection of microbial diversity in endocarditis using cultivation-independent molecular techniques. Scand. J. Infect. Dis. 43:857–869 [DOI] [PubMed] [Google Scholar]
- 50. Yu Y, Lee C, Kim J, Hwang S. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89:670–679 [DOI] [PubMed] [Google Scholar]
- 51. Zimmerli W, Trampuz A, Ochsner PE. 2004. Prosthetic-joint infections. N. Engl. J. Med. 351:1645–1654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.