Abstract
Recurrent Clostridium difficile infection (CDI) occurs in up to 35% of patients. Recurrences can be due to either relapse with the same strain or reinfection with another strain. In this study, multilocus variable-number tandem-repeat analysis (MLVA) was performed on C. difficile isolates from patients with recurrent CDI to distinguish relapse from reinfection. In addition, univariate and multivariate analyses were performed to identify risk factors associated with relapse. Among patients with a single recurrence, relapse due to the original infecting strain was more prevalent than reinfection and the interval between episodes was shorter than among patients who had reinfections. Among patients with >1 recurrence, equal distributions of relapse and reinfection or a combination of the two episode types were observed. Initial infection with the BI/NAP1/027 epidemic clone was found to be a significant risk factor for relapse. This finding may have important implications for patient therapy. Classification of recurrent CDI episodes by MLVA can be utilized to make informed patient care decisions and to accurately define new CDI cases for infection control and reimbursement purposes.
INTRODUCTION
One of the most problematic aspects of Clostridium difficile infections (CDI) is the propensity of recurrence in 15% to 35% of patients who initially respond to antimicrobial therapy. In addition, recurrent CDI is difficult to treat and contributes to significant morbidity and mortality and increased health care expenditures (10, 11, 27).
The molecular epidemiology of CDI has changed since 2000 with the global spread of an epidemic clone, designated BI/NAP1/027 by restriction endonuclease analysis (REA), pulsed-field gel electrophoresis, and PCR ribotyping. However, recent studies have raised doubts regarding the role of BI/NAP1/027 in increased CDI incidence, severity, and recurrence rates (4, 21).
Recurrent CDI can caused by either relapse due to the original infecting strain or reinfection with a new strain. Previous studies have demonstrated that continued non-CDI antibiotic treatment and a failed immune response to C. difficile toxins A and B are risk factors for recurrent CDI (15, 20, 25). Most recently, lower cure rates and higher rates of recurrence due to BI/NAP1/027 infection were reported in phase 3 clinical trials of fidaxomicin (26). Recent estimates suggest that 65% to 88% of recurrent CDI is attributable to relapse with the original infecting strain (2, 3, 12). However, some of the molecular typing methods used in these studies such as PCR-ribotyping and random amplification of polymorphic DNAs (RAPD) lack sufficient discriminatory power and may have misclassified reinfections as relapses. In a study using restriction endonuclease analysis (REA), 83.3% of recurrences were due to relapse (8). In this study, multilocus variable-number tandem-repeat analysis (MLVA), a highly discriminatory C. difficile genotyping method, was used to define relapse in patients with recurrent CDI and to identify risk factors associated with relapse.
MATERIALS AND METHODS
Setting.
The University of Pittsburgh Medical Center—Presbyterian (UPMC) is a 724-bed, tertiary-care teaching facility with a previously described CDI epidemic and enacted control measures (23, 24). Considering only first infections per person, the BI/NAP1/027 prevalence rates at UPMC were 46.7%, 40.0%, and 50.7% in 2001, 2005, and 2009, respectively.
Study patients.
This study was approved by the University of Pittsburgh Institutional Review Board. Cell culture cytotoxicity assay of stool (Diagnostic Hybrids, Athens, OH) was performed for laboratory diagnosis of CDI throughout the study period. Patients with recurrent CDI were defined as having diarrhea (≥2 bowel movements per day) and a C. difficile-positive stool culture for both initial and subsequent episodes (interval of at least 14 days). C. difficile isolates were obtained from stool samples submitted for toxin testing by selective culture (22). A total of 149 patients with multiple positive stool cultures seen at UPMC between March 2001 and November 2009 were available for inclusion. For logistical purposes, a systematic sampling of these 149 patients was performed to limit the number of patients to 100.
Chart review.
On retrospective chart review of the 100 patients, 18 did not meet the recurrent CDI definition. Therefore, 82 patients with recurrent CDI were available for analysis. Data representing patient demographics, comorbidities, clinical symptoms (diarrhea, emesis, abdominal pain, abdominal distension), laboratory parameters (maximum white blood cell count, maximum band percentage, minimum albumin, lactate, maximum creatinine) from −2 to +5 days from the day of stool toxin testing, length of hospital stay, length of intensive care unit (ICU) stay, concurrent infection, duration and class of non-CDI antimicrobials administered (−5 to +10 days from day of testing), discharge to home versus another health care facility, and survival to hospital discharge were collected. Radiology records were reviewed for evidence of pan-colitis, bowel wall thickening or edema, pneumatosis, ileus, and ascites. Exposures to other hospitals or health care institutions, dialysis centers, outpatient clinics, and infants as well as outpatient and inpatient antibiotics, H2 blocker/proton pump inhibitor (PPI) use, tube feeds, enemas, antimotility agents, and immunosuppressive drugs were determined for the 12-week period prior to CDI recurrence for each recurrent episode.
Genotyping.
MLVA and tcdC genotyping were performed on 199 isolates (82 original and 117 recurrent) from 82 patients as previously described (18). Isolates with the tcdC-1 genotype were defined as belonging to the BI/NAP1/027 epidemic clone (6). The sum of the tandem-repeat differences (STRD) between MLVA genotypes in consecutive C. difficile isolates from each individual was used to define each recurrent episode as either relapse or reinfection. Consecutive episodes with STRD ≤ 2 were defined as relapses, while consecutive episodes with STRD ≥ 3 were defined as reinfections. This cutoff was selected based on previous data from consecutive patient isolates demonstrating only 1 or 2 tandem-repeat changes over as many as 90 days (17).
For comparison with MLVA, all recurrent CDI episodes for the 82 patients (n = 117) were also classified according to the Centers for Disease Control and Prevention Ad Hoc C. difficile Surveillance Working Group recommendations (19). Under these guidelines, episodes that occur 2 to 8 weeks after resolution of the last episode represent recurrent CDI and episodes that occur >8 weeks after the onset of a previous episode represent new infections (19). The genetic diversity of BI/NAP1/027 was assessed by MLVA genotyping of 856 BI/NAP1/027 isolates collected from 2001 to 2009 at our institution.
Statistics.
Univariate logistic regression analysis was performed to determine risk factors for relapses versus reinfections using SAS (v9.2; SAS Institute). Stepwise multivariable logistic regression analysis was performed to identify independent risk factors for relapse. All variables that were significant in the univariate analysis at P = 0.2 were eligible for inclusion in the multivariable model. The stay criterion for the model was P < 0.05. The odds ratio (OR) was expressed as the odds for relapse.
Patients with >1 recurrence (≥3 episodes) were examined with respect to the interval between the first and second episode only. An interval was defined as the number of days between the stool specimen collection dates from consecutive episodes. Given the potential for misclassification of isolates with STRD 3 to 9, analyses were performed that both included and excluded recurrent CDI episodes within this STRD range.
RESULTS
Of the 82 patients that met the clinical definition of recurrent CDI, 51 had relapse and 31 had reinfection by MLVA. Patients with relapse or reinfection were similar in age, gender, race, and Charlson index data (Table 1). In univariate analysis of comorbidities, malignancy was more common in patients with relapse than in patients with reinfection and the opposite was true for neurologic disorders. Patients with relapse were more likely to have received an opiate than patients with reinfection (Table 1). In univariate analysis of characteristics associated with the index case, there was a strong association between prior BI/NAP1/027 infection and the risk of relapse observed (OR, 3.8; 95% confidence interval [CI], 1.4, 10.1; P = 0.008).
TABLE 1.
Comparison of characteristics of patients (n = 82) with a second episode of CDI
| Demographic | No. of casesa |
OR (95% CI) | P value | |
|---|---|---|---|---|
| Relapse (n = 51) | Reinfection (n = 31) | |||
| Age (median, range) | 64 (29–87) | 64 (18–93) | 1.0 (0.97, 1.02) | 0.57 |
| Male gender | 24 (47.1) | 14 (45.2) | 1.1 (0.4, 2.6) | 0.87 |
| Black race | 13 (25.5) | 9 (29.0) | 0.8 (0.3, 2.3) | 0.73 |
| Comorbidity | ||||
| Modified Charlson index (median, range) | 5 (0–11) | 4 (0–16) | 1.0 (0.9, 1.1) | 0.92 |
| Chronic renal insufficiency | 17 (33.3) | 9 (29.0) | 1.2 (0.5, 3.2) | 0.69 |
| Diabetes | 20 (39.2) | 12 (38.7) | 1.0 (0.4, 2.6) | 0.96 |
| Diverticulosis | 9 (17.6) | 5 (16.1) | 1.1 (0.3, 3.7) | 0.86 |
| Infection | 32 (62.7) | 17 (54.8) | 1.4 (0.6, 3.4) | 0.48 |
| Ischemic vascular disease | 21 (41.2) | 17 (54.8) | 0.6 (0.2, 1.4) | 0.23 |
| Inflammatory bowel disease | 2 (3.9) | 4 (12.9) | 0.3 (<0.1, 1.6) | 0.15 |
| Immunocompromised host | 23 (45.1) | 16 (51.6) | 0.8 (0.3, 1.9) | 0.57 |
| Lung disease | 13 (25.5) | 14 (45.2) | 0.4 (0.2, 1.1) | 0.07 |
| Malignancy | 20 (39.2) | 5 (16.1) | 3.4 (1.1, 10.2) | 0.03 |
| Neurologic disorder | 3 (5.9) | 7 (22.6) | 0.2 (<0.1, 0.9) | 0.04 |
| Obesity | 6 (11.8) | 1 (3.2) | 4.0 (0.5, 34.9) | 0.21 |
| Abdominal surgery | 24 (47.1) | 14 (45.2) | 1.1 (0.4, 2.6) | 0.87 |
| Medication use (during 1st episode) | ||||
| Metronidazole as CDI treatment | 34 (69.4) | 22 (71.0) | 0.9 (0.3, 2.5) | 0.88 |
| Non-CDI antibiotic | 34 (66.7) | 26 (83.9) | 0.4 (0.1, 1.2) | 0.09 |
| Antimotility (excluding opiates) | 8 (16.3) | 2 (6.5) | 2.8 (0.6, 14.3) | 0.21 |
| Antacid | 17 (34.7) | 6 (19.4) | 2.2 (0.8, 6.4) | 0.14 |
| PPI | 28 (57.1) | 16 (51.6) | 1.2 (0.5, 3.1) | 0.63 |
| Opiate(s) | 30 (61.2) | 10 (32.3) | 3.3 (1.3, 8.6) | 0.01 |
| Probiotic(s) | 17 (34.7) | 16 (51.6) | 0.5 (0.2, 1.2) | 0.14 |
| Tube feeding (12 wk prior to 2nd episode) | 19 (37.3) | 15 (48.4) | 0.6 (0.3, 1.6) | 0.32 |
| Exposures (following 1st episode) | ||||
| Healthcare facility since discharge for previous episode | 44 (86.3) | 27 (90.0) | 0.7 (0.2, 2.9) | 0.62 |
| Homeb | 13 (25.5) | 5 (16.7) | 1.7 (0.5, 5.4) | 0.36 |
| Antibiotic(s) (outpatient/outside hospital) | 25 (50.0) | 22 (73.3) | 0.4 (0.1, 1.0) | 0.04 |
| Antimicrobial(s) (12 wk prior to 2nd episode) | 40 (81.6) | 29 (93.5) | 0.3 (<0.1, 1.5) | 0.15 |
| Characteristics of index case | ||||
| Infection with BI/NAP1/027 strain | 29 (56.9) | 8 (25.8) | 3.8 (1.4, 10.1) | 0.008 |
| High (>3) CD prognosis score | 9 (17.6) | 6 (19.4) | 0.9 (0.3, 2.8) | 0.85 |
| Radiologic abnormality | 25 (49.0) | 10 (32.3) | 2.0 (0.8, 5.1) | 0.14 |
| No. (range) of days between positive cultures | 40 (17–402) | 90 (18–1,260) | 1.0 (0.99, 1.00) | 0.01 |
| Median (range) no. of days of hospitalization | 15 (0–257) | 10 (0–122) | 1.0 (0.99, 1.02) | 0.45 |
Data in parentheses represent percentages except where otherwise indicated.
Data exclude patients exposed to rehabilitation and skilled-nursing facilities or other hospitals prior to discharge.
In the multivariate analysis, prior infection with a BI/NAP1/027 strain and use of opiates were also associated with relapse (Table 2). In contrast, the use of non-CDI antibiotics during the initial episode, use of any antimicrobial in the 12 weeks prior to the second episode, and inflammatory bowel disease (IBD) were associated with a second episode of CDI attributable to reinfection. Excluding the 7 patients with STRD 3 to 9 did not substantially change these results (data not shown).
TABLE 2.
Multivariate analysis of factors associated with relapse or reinfection among 82 patients with a second episode of CDI
| Factor associated with indicated disease category | OR (95% CI) | P value |
|---|---|---|
| Relapse | ||
| Infection with BI/NAP1/027 strain | 6.9 (1.7, 28.2) | 0.007 |
| Opiate use during previous episode | 13.1 (3.2, 54.0) | <0.001 |
| Reinfection | ||
| Non-CDI antibiotic (previous episode) | 0.1 (0.02, 0.5) | 0.007 |
| Inflammatory bowel disease | 0.04 (0.0, 0.5) | 0.011 |
| Antimicrobial(s) (12 wk prior prior to 2nd episode) | 0.1 (0.01, 0.8) | 0.033 |
Among the 52 patients with a single CDI recurrence, 36 (69.2%) were classified as having a relapse by MLVA and 16 (30.8%) patients had reinfections (Table 3). The median interval to relapse (48 days) was significantly shorter than the median interval to reinfection (108 days; P = 0.01) in patients with a single recurrence. Among 30 patients with multiple recurrences, 11 patients had relapses, 8 had recurrences due solely to reinfections, and 11 had recurrences attributed to both relapse and reinfection (Table 3). The median intervals to relapse or reinfection in patients with multiple recurrences were not significantly different (30 and 40 days, respectively). The median interval to recurrence among the 11 patients with both relapse and reinfection (78 days) was significantly (P = 0.03) longer than the interval to recurrence in patients with either relapse or reinfection alone. When all 117 episodes were stratified by the prior episode strain type (BI/NAP1/027 versus non-BI/NAP1/027), the interval to recurrence tended to be shorter for episodes due to BI/NAP1/027 strains than for those due to non-BI/NAP1/027 strains (43 versus 62 days) but this difference was not significant (P = 0.64). Similarly, among second episodes only (n = 82), the interval to recurrence was shorter for episodes due to BI/NAP1/027 strains (40 versus 79 days) but not significantly so (P = 0.11).
TABLE 3.
Median time interval between 117 recurrent CDI episodes for 82 patients
| Episode type | No. (%) of patients | Interval (days) | Range (days) | P value |
|---|---|---|---|---|
| Single recurrence | ||||
| Relapse (n = 36) | 36 (69.2) | 48 | 19–402 | 0.01 |
| Reinfection (n = 16) | 16 (30.8) | 108 | 23–1,260 | |
| Multiple recurrence | ||||
| Relapse (n = 23) | 11 (36.7) | 30 | 15–319 | 0.03 |
| Reinfection (n = 17) | 8 (26.6) | 40 | 17–325 | |
| Relapse + reinfection (n = 25) | 11 (36.7) | 78 | 17–778 |
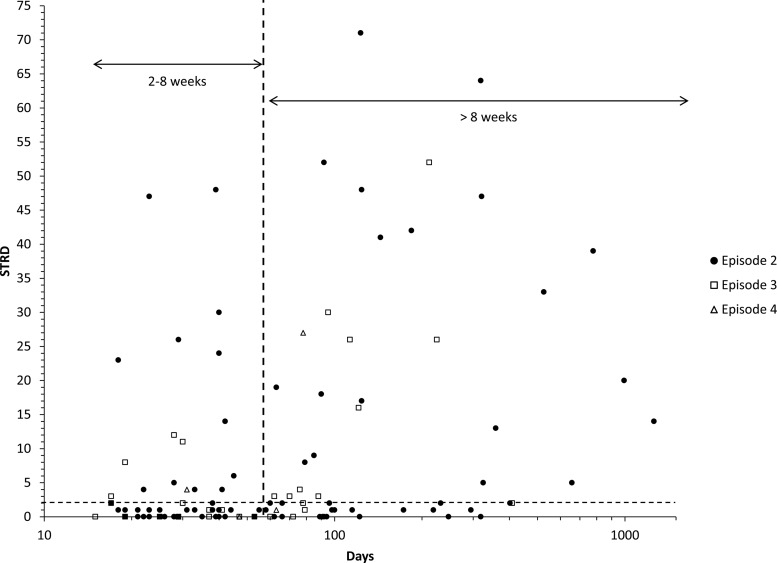
A comparison of all CDI recurrences defined by either MLVA or CDC recommendations (interval between episodes) was performed. There were 61 recurrent CDI episodes that occurred within 2 to 8 weeks (representing relapses as defined by CDC recommendations) of the previous episode. Of these, 17 (28%) were considered reinfections as determined by MLVA (STRD ≥ 3) (Fig. 1). There were 56 recurrent CDI episodes that occurred more than 8 weeks after the previous episode that would be considered reinfections by CDC recommendations. Of these, 27 (48%) were classified as relapses by MLVA (STRD ≤ 2) (Fig. 1). Thus, the 8-week cutoff misclassified 44/117 (38%) recurrent CDI episodes.
Fig 1.
Recurrent CDI episodes (n = 117) plotted as a function of STRD and interval (days). Episode 2, n = 82; episode 3, n = 30; episode 4, n = 5. Vertical and horizontal dashed lines indicate interval and MLVA cutoffs, respectively, for defining relapse and reinfection. STRD, sum of the tandem-repeat differences.
DISCUSSION
In this study, prior infection with the BI/NAP1/027 epidemic strain was a significant risk factor for the development of recurrent CDI due to relapse. Two very recent studies have described increased recurrence rates and a trend toward a higher incidence of relapse among patients infected with BI strains as defined by REA typing (8, 26). These data could have major implications for treatment and development of new therapeutics that specifically target infection with BI/NAP1/027 strains. Treatment of recurrent CDI is more difficult, as no single therapy has been proven to prevent recurrence in all patients (11). While the recurrence rate observed in patients treated with fidaxomicin was lower than was seen with vancomycin, this effect was limited to patients with non-BI/NAP1/027 infections (15.4% versus 25.3%; P = 0.005) (16). Similar results were observed in recent phase 3 clinical trials, where recurrence rates were significantly reduced in patients treated with fidaxomicin versus vancomycin for non-BI/NAP1/027 infections but no difference in rates of recurrence was observed in BI/NAP1/027 infections (26). Moreover, the same study demonstrated that patients with BI/NAP1/027 infections have a reduced overall cure rate (26). Together, these results demonstrate the need for further characterization of BI/NAP1/027 to assess the biological basis for relapse among these strains.
The strong association of opiates with CDI relapse may be due to the antimotility effect of these agents. The clinical utility of antimotility drugs for adjunctive therapy of CDI has recently been questioned for several reasons, including the finding that resolution of CDI symptoms correlates with a decrease in detectable C. difficile levels in patient stool (1, 9). Slowing the transit of bowel contents may impede effective elimination of the organism, lead to spore accumulation, and contribute to future recurrent CDI episodes due to the original infecting strain.
This study demonstrated an association of IBD with CDI reinfection. This finding is consistent with a recent point-prevalence investigation of C. difficile environmental contamination in a hospital-based outpatient GI clinic (S. R. Curry, N. T. Brown, J. W. Marsh, C. A. Muto, L. H. Harrison, and D. Binion, presented at the Society for Healthcare Epidemiology of America, Dallas, TX, 2 April 2011). In that study, 3/6 GI examination rooms were found to be contaminated with toxigenic C. difficile (Curry et al., 2 April 2011). This finding not only highlights the need for improved infection control practices in outpatient clinics but also suggests that recurrent CDI in IBD patients could be due to reinfection from the clinic environment. Further molecular epidemiologic investigations of IBD reinfections are required to validate this hypothesis.
Understanding the relative rates of relapse and reinfection in recurrent CDI is important from a number of perspectives. This information can help elucidate the pathology of the organism and may reveal host immune deficiencies or genetic predispositions for relapse that have yet to be explored. In addition, rates of relapse and reinfection are important from a health care provider standpoint. Increased rates of reinfection indicate a need for enhanced infection control measures.
MLVA is an objective, highly discriminatory molecular genotyping tool that can be used to define relapse versus reinfection in recurrent CDI cases. We report rates of relapse in our study that were lower than the rates in recent reports that used PCR-ribotyping, multilocus sequence typing (MLST), and REA to classify recurrent CDI (7, 8, 13). These methods may overestimate relapse and underestimate reinfection rates because they are less discriminatory than MLVA (14). The MLVA data presented in this study suggest that definitions of recurrent CDI based on the interval between episodes alone do not provide an accurate measurement of the relative frequencies of relapse and reinfection. MLVA, on the other hand, provides a more reliable estimate of the contribution of relapse and reinfection to recurrent CDI.
This study used a strict MLVA cutoff STRD ≤ 2 to define relapse. This cutoff was selected based upon a previous study which demonstrated that MLVA genotypes obtained from consecutive isolates from individual patients varied by 1 or 2 tandem repeats at one or two loci over as many as 90 days (5). There were 16 isolates in the current study with STRD 3 to 9 compared to the previous episode's isolate. Seven of these isolates were single-locus variants with STRD 3 to 5 and shared the same tcdC genotype as the previous isolate. Further investigation of the mutation rates at MLVA loci are required to determine whether these recurrent isolates can be classified as representing relapses. Thus, the definition of relapse in this study is conservative.
While some patient samples in this retrospective study were collected during a BI/NAP1/027 epidemic at our institution, it is unlikely that the association of BI/NAP1/027 with relapse was a consequence of misclassification bias resulting from hospital transmission of circulating clones classified as identical by MLVA. Substantial genetic diversity of BI/NAP1/027 strains at our institution was observed by MLVA, and a conservative STRD cutoff ≤ 2 was used to define relapse. Among 856 BI/NAP1/027 isolates at our institution, 439 different MLVA genotypes were identified. Only 10 MLVA genotypes included ≥10 isolates, and no genotype included more than 20 isolates. This study was limited by the fact that only patients with recurrent CDI from a single institution were included. Prospective studies of CDI patients are required to validate the role of BI/NAP1/027 in recurrent CDI due to relapse.
In summary, this study used MLVA to define recurrent CDI as either relapse due to the original infecting strain or reinfection with a new strain. Based upon this analysis, we demonstrate that an initial infection with BI/NAP1/027 is a significant risk factor for relapse in patients with recurrent disease. Thus, strain-specific characteristics may play a role in recurrent CDI. This finding may have important implications for the rational design of future therapeutics targeting BI/NAP1/027 infections.
ACKNOWLEDGMENTS
This work was supported in part by a research grant from the Investigator-Initiated Studies Program of Merck Sharp & Dohme Corp. J.W.M. and L.H.H. have received research funding from ViroPharma Inc.
The opinions expressed in this paper are ours and do not necessarily represent those of Merck Sharp & Dohme Corp.
We gratefully acknowledge the assistance of Lloyd Clarke and Melissa Saul for data retrieval and management of patient medical records.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Al-Nassir WN, et al. 2008. Comparison of clinical and microbiological response to treatment of Clostridium difficile-associated disease with metronidazole and vancomycin. Clin. Infect. Dis. 47:56–62 [DOI] [PubMed] [Google Scholar]
- 2. Alonso R, et al. 2001. Molecular analysis of relapse vs re-infection in HIV-positive patients suffering from recurrent Clostridium difficile associated diarrhoea. J. Hosp. Infect. 48:86–92 [DOI] [PubMed] [Google Scholar]
- 3. Barbut F, et al. 2000. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2386–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cloud J, Noddin L, Pressman A, Hu M, Kelly C. 2009. Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin. Gastroenterol. Hepatol. 7:868–873.e2 [DOI] [PubMed] [Google Scholar]
- 5. Curry SR, et al. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45:215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dingle KE, et al. 2011. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 6:e19993 doi:10.1371/journal.pone.0019993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eyre DW, et al. 2012. Clostridium difficile mixed infection and reinfection. J. Clin. Microbiol. 50:142–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figueroa I, et al. 2012. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin. Infect. Dis. 55(Suppl 2):S104–S109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerding DN. 2009. Antimotility agents for the treatment of Clostridium difficile infection: is the juice worth the squeeze? Clin. Infect. Dis. 48:606–608 [DOI] [PubMed] [Google Scholar]
- 10. Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW. 2010. Economic healthcare costs of Clostridium difficile infection: a systematic review. J. Hosp. Infect. 74:309–318 [DOI] [PubMed] [Google Scholar]
- 11. Johnson S. 2009. Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J. Infect. 58:403–410 doi:10.1016/j.jinf.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 12. Johnson S, Adelmann A, Clabots CR, Peterson LR, Gerding DN. 1989. Recurrences of Clostridium difficile diarrhea not caused by the original infecting organism. J. Infect. Dis. 159:340–343 [DOI] [PubMed] [Google Scholar]
- 13. Kamboj M, et al. 2011. Relapse versus reinfection: surveillance of Clostridium difficile infection. Clin. Infect. Dis. 53:1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Killgore G, et al. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kyne L, Warny M, Qamar A, Kelly CP. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357:189–193 doi:10.1016/S0140-6736(00)03592-3 [DOI] [PubMed] [Google Scholar]
- 16. Louie TJ, et al. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364:422–431 [DOI] [PubMed] [Google Scholar]
- 17. Marsh JW, et al. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J. Clin. Microbiol. 44:2558–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsh JW, et al. 2010. Multilocus variable-number tandem-repeat analysis and multilocus sequence typing reveal genetic relationships among Clostridium difficile isolates genotyped by restriction endonuclease analysis. J. Clin. Microbiol. 48:412–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDonald LC, et al. 2007. Recommendations for surveillance of Clostridium difficile-associated disease. Infect. Control Hosp. Epidemiol. 28:140–145 [DOI] [PubMed] [Google Scholar]
- 20. McFarland LV, et al. 1999. Recurrent Clostridium difficile disease: epidemiology and clinical characteristics. Infect. Control Hosp. Epidemiol. 20:43–50 [DOI] [PubMed] [Google Scholar]
- 21. Morgan OW, et al. 2008. Clinical severity of Clostridium difficile PCR ribotype 027: a case-case study. PLoS One 3:e1812 doi:10.1371/journal.pone.0001812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mundy LS, Shanholtzer CJ, Willard KE, Gerding DN, Peterson LR. 1995. Laboratory detection of Clostridium difficile. A comparison of media and incubation systems. Am. J. Clin. Pathol. 103:52–56 [DOI] [PubMed] [Google Scholar]
- 23. Muto CA, et al. 2007. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin. Infect. Dis. 45:1266–1273 [DOI] [PubMed] [Google Scholar]
- 24. Muto CA, et al. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26:273–280 [DOI] [PubMed] [Google Scholar]
- 25. Nair S, Yadav D, Corpuz M, Pitchumoni CS. 1998. Clostridium difficile colitis: factors influencing treatment failure and relapse—a prospective evaluation. Am. J. Gastroenterol. 93:1873–1876 [DOI] [PubMed] [Google Scholar]
- 26. Petrella LA, et al. 2012. Decreased cure and increased recurrence rates for Clostridium difficile infection caused by the epidemic C. difficile BI strain. Clin. Infect. Dis. 55:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Surawicz CM, Alexander J. 2011. Treatment of refractory and recurrent Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 8:330–339 [DOI] [PubMed] [Google Scholar]