Abstract
We report the first case of deep-wound colonization by Dietzia cinnamea in a patient who had been bitten by a dog.
CASE REPORT
In January 2012, a 47-year-old otherwise healthy woman who was diagnosed with Liddle's disease was admitted to the emergency department at Vaasa Central Hospital (VCH) after being attacked by a Rottweiler dog on the same day. Several bite wounds, approximately 2 cm wide, were found in both arms on the left and right proximal antebrachia on the volar and medial surfaces. Additionally, there were extensive hematomas in both arms, but arterial or nerve damage was not observed. X-rays showed no fractures.
Microbiological and other laboratory tests were ordered according to the hospital routine practice in bite wound cases. A microbiological swab sample was taken deep in the wounds, and blood cultures were collected prior to the initiation of empirical antimicrobial treatment with amoxicillin-clavulanic acid (500 mg/125 mg every 8 h, orally [p.o.]). Laboratory tests revealed no signs of infection. The level of C-reactive protein (CRP) was 2 mg/liter (normal value, <10 mg/liter), the white blood cell count was 8.2 × 109 cells/liter (normal range, 3.4 × 109 to 8.2 × 109 cells/liter), and the platelet count was 244 × 109 cells/liter (normal range, 150 × 109 to 360 × 109 cells/liter). Wounds were not sutured but were covered with elastic bandages, and the patient was transferred to a hospital ward for observation. On the next day, the patient was discharged home, and follow-up controls were arranged at the wound polyclinic of VCH.
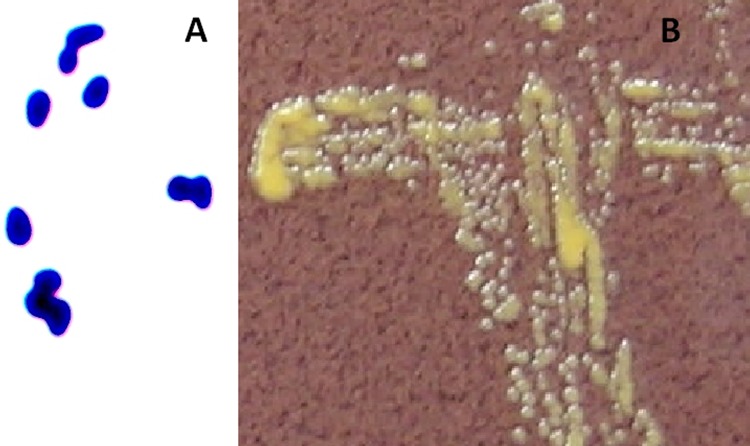
The wound sample was cultured on chocolate agar (CA) (Becton, Dickinson, Sparks, MD), fastidious anaerobe agar (FAA) (Lab M Ltd., Lancashire, United Kingdom), and on blood agar (BA) (Becton, Dickinson) with and without colistin and oxolinic acid, as well as in fastidious anaerobe broth (FAB) (Lab M Ltd.). After 18 h of incubation at 35°C in air supplemented with 5% CO2, a weak but abundant (3+) colorless aerobic growth was observed on CA and on nonselective BA plates. A colony Gram stain revealed short, drop-like Gram-positive bacilli exhibiting snapping division (Fig. 1A). Colonies were catalase positive and oxidase negative. After an additional 24 h of incubation at 35°C, smooth, deep-yellow-pigmented, and nonmucoid colonies (Fig. 1B) were seen as pure growth on all plates except FAA, which was incubated under an anaerobic atmosphere. The organism was not able to be identified with the RapID CB Plus identification system using an ERIC electronic compendium (Thermo Fisher Scientific, Lenexa, KS), although several enzymatic reactions were observed after 4 h of incubation (phenotypic identification profile was 0207053) (Table 1). Further identification with an API Coryne 3.0 (bioMérieux, Marcy l'Etoile, France) test strip gave Rhodococcus spp. (phenotypic identification profile was 1110004) at a good identification level using the apiweb database (Table 1). The possibility of Dietzia spp., Gordonia spp., and Nocardia spp. was mentioned as a notification. As the colonial and cell features of the isolate were slightly different from those of rhodococci (11), a further identification was performed. On modified Kinyoun acid-fast stain, the isolate proved to be non-acid fast. The optimal growth temperature was determined by incubating the strain on CA plates for 2 days at 25°C, 35°C, and 42°C. Although growth was evident at 25°C and 42°C, the best colony formation was observed at 35°C. In addition, molecular identification by PCR amplification and sequencing of the bacterial 16S rRNA gene was performed on the isolate. The primers used were as described previously (9). The consensus sequence was trimmed and submitted to GenBank for comparative analysis using the Basic Local Alignment Search Tool, available at the BLAST website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence of the 16S rRNA gene (GenBank accession number JX560162) showed 100.0% similarity (418 bp out of 418 bp) to that of the Dietzia cinnamea type strain (DSM 44904; GenBank accession number FJ468339.3).
Fig 1.
(A) Gram staining of the isolate identified as Dietzia cinnamea (oil immersion; magnification, ×1,000). (B) Colonial appearance of the isolate on chocolate agar after 48 h of incubation in air supplemented with 5% CO2.
TABLE 1.
Phenotypic characteristics of the isolate identified as Dietzia cinnamea
| Phenotypic characteristic | Test resulta |
|
|---|---|---|
| RapID CB Plus | API Coryne | |
| Acid production from: | ||
| Glucose | – | – |
| Sucrose | – | – |
| Ribose | – | – |
| Maltose | – | – |
| Xylose | NI | – |
| Mannitol | NI | – |
| Lactose | NI | – |
| Glycogen | NI | – |
| Activity of: | ||
| N-Acetyl-β-glucosaminidase | – | – |
| Glycosidase | – | NI |
| β-Galactosidase | – | – |
| α-Glucosidase | + | + |
| β-Glucosidase | – | – |
| Urease | – | – |
| Nitrate reductase | + | + |
| Pyrazinamidase | NI | – |
| Alkaline phosphatase | + | + |
| β-Glucuronidase | NI | – |
| Hydrolysis of: | ||
| Proline-β-naphtylamide | + | NI |
| Tryptophan-β-naphtylamide | –/w | NI |
| Pyrrolidine-β-naphtylamide | – | – |
| Leucyl-glycine-β-naphtylamide | –/w | NI |
| Leucine-β-naphtylamide | + | NI |
| Gelatin | NI | – |
| Fatty acid ester | + | NI |
NI, not included; +, positive; w, weak positive; –, negative.
Antimicrobial susceptibility of the isolate was determined by E-tests (bioMérieux, Marcy l'Etoile, France), according to the manufacturer's instructions, on Mueller-Hinton fastidious agar. The MIC values are shown in Table 2. The non-species-related breakpoints recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) were used for all drugs except erythromycin and clindamycin (5). As non-species-related breakpoints for erythromycin and clindamycin have not been established, EUCAST Staphylococcus species breakpoints were used instead. The isolate was interpreted as sensitive to all antimicrobial agents tested except erythromycin (resistant) and clindamycin (intermediate sensitivity).
TABLE 2.
MICs for the isolate identified as Dietzia cinnamea
| Antimicrobial agent | MIC (μg/ml) |
|---|---|
| Amoxicillin-clavulanic acid | 0.047 |
| Cefuroxime | 0.094 |
| Ceftriaxone | 0.094 |
| Ciprofloxacin | 0.094 |
| Clindamycin | 1.5 |
| Erythromycin | 16 |
| Gentamicin | 0.064 |
| Imipenem | 0.064 |
| Levofloxacin | 0.125 |
| Meropenem | 0.012 |
| Piperacillin-tazobactam | 0.023 |
| Tetracycline | 0.094 |
| Tobramycin | ≤0.064 |
| Trimethoprim-sulfamethoxazole | 0.75 |
Blood culture bottles (BacT/Alert FAN aerobic [n = 2] and standard anaerobic [n = 2]; bioMérieux, Marcy l'Etoile, France), incubated in the BacT/Alert 3D automated continuous growth monitoring system for 7 days, remained negative.
Two weeks after the injury, the patient's hematomas in the arms were drained, additional microbiological culture samples were collected, and the wounds were rinsed with disinfectant solution and covered with wound bandages. All additional culture samples were negative after 7 days of incubation. However, wound healing was slow, and in March 2012, a revision of necrotic subcutaneous tissue was performed. The last visit to the wound polyclinic was in April 2012, and there was still some swelling in the arms but the wounds had closed up. No signs of infection were seen during the follow-up period of the patient.
Dietzia cinnamea is a Gram-positive, rod-shaped, non-acid-fast bacterium that was first isolated in 2006 by Yassin and colleagues from a perianal swab of a patient with a bone marrow transplant (19). This aerobic, catalase-positive, and oxidase-negative bacterium was found to grow at temperatures in the range of 22 to 45°C, forming smooth and yellow-pigmented colonies. In addition, Yassin and colleagues found that the type strain, DSM 44904, hydrolyzed urea but was, e.g., unable to utilize citrate as the sole carbon source. In a recent publication by Niwa and colleagues, however, the D. cinnamea type strain proved to be negative in Christensen's urea hydrolysis test (11). Urea hydrolysis was also proven to be negative in commercial identification kits used in our study. However, despite the fact that Dietzia spp. form many enzymatic and biochemical reactions in these commercial identification tests, they fail to identify Dietzia species to the genus level and, e.g., API Coryne misidentifies Dietzia spp. as Rhodococcus spp. This may explain why only a few cases of Dietzia species isolations from human sources have been reported, although members of this genus are widely distributed in diverse habitats, such as marine, soil, and even hospital environments (2, 3, 8, 10, 11, 19). Recently, some additional phenotypic tests have been reported to help distinguish Dietzia spp. from Rhodococcus spp., particularly from Rhodococcus equi (11). These tests included, e.g., Christie-Atkins-Munch-Petersen (CAMP) and adenine hydrolyze tests in which Dietzia spp. were negative and R. equi was positive and acid production from d-fructose in Gordon's base medium in which Dietzia spp. were positive and R. equi was negative. Moreover, D. cinnamea may be distinguished from other yellow-pigmented Dietzia spp., including Dietzia maris, by its nonmucoid colonial morphology. The colonial difference is best seen after 3 days of incubation, as all other Dietzia spp. have been shown to form at least slightly mucoid colonies in this time period (11). Furthermore, in clinical samples, Dietzia spp. can be reliably differentiated from members of other mycolic acid-containing genera by using 16S rRNA gene sequencing analysis (8, 11, 13).
Although the potential of D. cinnamea to cause an infection was not able to be shown in our study, as an effective antimicrobial treatment was introduced without delay, previous reports by other authors suggest that Dietzia species have the potential to act as opportunistic pathogens (1, 7, 12, 13, 19). The cell structure of Dietzia spp. has proved to contain a component of lipoarabinomannan, which has been shown to be a powerful stimulator of tumor necrosis factor alpha (16, 18). Furthermore, in a recent study by Procópio and colleagues, D. cinnamea P4, a strain type isolated in tropical soil habitats, was found to contain the full metabolic pathway for the biosynthesis of l-rhamnose (14), the component of cell walls and capsular polysaccharides of pathogenic bacteria (6, 15). We do not believe, however, that Liddle's disease had an impact on colonization or the clinical picture since it is not known to predispose to infectious diseases.
A review of the veterinary literature revealed that Dietzia spp. are shown to be normally present in canine intestine and oral cavity, especially in a dog's tooth plaque (4, 17). Unfortunately, in our study, microbiological samples were not able to be collected from the Rottweiler's oral cavity. However, as no bacteria other than D. cinnamea were observed in the fresh wound sample, the fact that the isolate originated from the dog oral cavity was strongly supported.
In conclusion, this study demonstrated that Dietzia species should be considered possible infectious agents among other microbes in dog bite wounds. In addition, the need for particular phenotypic tests and 16S rRNA gene sequencing for reliable differentiation of Dietzia spp. from Rhodococcus spp. was highlighted.
Footnotes
Published ahead of print 3 October 2012
REFERENCES
- 1. Bemer-Melchior P, Haloun A, Riegel P, Drugeon HB. 1999. Bacteremia due to Dietzia maris in an immunocompromised patient. Clin. Infect. Dis. 29:1338–1340 [DOI] [PubMed] [Google Scholar]
- 2. Colquhoun JA, et al. 1998. Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie Van Leeuwenhoek 74:27–40 [DOI] [PubMed] [Google Scholar]
- 3. Duckworth AW, Grant S, Grant WD, Jones BE, Meijer D. 1998. Dietzia natronolimnaios sp. nov., a new member of the genus Dietzia isolated from an East African soda lake. Extremophiles 2:359–366 [DOI] [PubMed] [Google Scholar]
- 4. Elliott DR, Wilson M, Buckley CMF, Spratt DA. 2005. Cultivable oral microbiota of domestic dogs. J. Clin. Microbiol. 43:5470–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. European Committee on Antimicrobial Susceptibility Testing 2012. Breakpoint tables for interpretation of MICs and zone diameters, version 2.0. http://www.eucast.org/clinical_breakpoints/
- 6. Feng L, et al. 2005. Structural and genetic characterization of the Shigella boydii type 18 O antigen. Gene 355:79–86 [DOI] [PubMed] [Google Scholar]
- 7. Jones AL, Koerner RJ, Natarajan S, Perry JD, Goodfellow M. 2008. Dietzia papillomatosis sp. nov., a novel actinomycete isolated from the skin of an immunocompetent patient with confluent and reticulated papillomatosis. Int. J. Syst. Evol. Microbiol. 58:68–72 [DOI] [PubMed] [Google Scholar]
- 8. Koerner RJ, Goodfellow M, Jones AL. 2009. The genus Dietzia: a new home for some known and emerging opportunist pathogens. FEMS Immunol. Med. Microbiol. 55:296–305 [DOI] [PubMed] [Google Scholar]
- 9. Kommedal Ø, Simmon K, Karaca D, Langeland N, Wiker HG. 2012. Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens. J. Clin. Microbiol. 50:1289–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mayilraj S, Suresh K, Kroppenstedt RM, Saini HS. 2006. Dietzia kunjamensis sp. nov., isolated from the Indian Himalayas. Int. J. Syst. Evol. Microbiol. 56:1667–1671 [DOI] [PubMed] [Google Scholar]
- 11. Niwa H, et al. 2012. Characterization of human clinical isolates of Dietzia species previously misidentified as Rhodococcus equi. Eur. J. Clin. Microbiol. Infect. Dis. 31:811–820 [DOI] [PubMed] [Google Scholar]
- 12. Pidoux O, Argenson JN, Jacomo V, Drancourt M. 2001. Molecular identification of a Dietzia maris hip prosthesis infection isolate. J. Clin. Microbiol. 39:2634–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pilares L, Agüero J, Vázquez-Boland JA, Martínez-Martínez L, Navas J. 2010. Identification of atypical Rhodococcus-like clinical isolates as Dietzia spp. by 16S rRNA gene sequencing. J. Clin. Microbiol. 48:1904–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Procópio L, et al. 2012. Insight from the draft genome of Dietzia cinnamea P4 reveals mechanisms of survival in complex tropical soil habitats and biotechnology potential. Antonie Van Leeuwenhoek 101:289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramos PI, et al. 2012. Pyrosequencing-based analysis reveals a novel capsular gene cluster in a KPC-producing Klebsiella pneumoniae clinical isolate identified in Brazil. BMC Microbiol. 12:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhoades ER, Ullrich HJ. 2000. How to establish a lasting relationship with your host: lessons learned from Mycobacterium spp. Immunol. Cell Biol. 78:301–310 [DOI] [PubMed] [Google Scholar]
- 17. Suchodolski JS, et al. 2009. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sutcliffe IC. 2000. Characterisation of a lipomannan lipoglycan from the mycolic acid containing actinomycete Dietzia maris. Antonie Van Leeuwenhoek 78:195–201 [DOI] [PubMed] [Google Scholar]
- 19. Yassin AF, Hupfer H, Schaal KP. 2006. Dietzia cinnamea sp. nov., a novel species isolated from a perianal swab of a patient with a bone marrow transplant. Int. J. Syst. Evol. Microbiol. 56:641–645 [DOI] [PubMed] [Google Scholar]