Abstract
To be acceptable for use in cervical cancer screening, a new assay that detects DNA of high-risk human papillomavirus (hrHPV) types must demonstrate high reproducibility and performance not inferior to that of a clinically validated HPV test. In the present study, a real-time quantitative PCR (qPCR) assay targeting the E6 and E7 genes of hrHPV was compared with Hybrid Capture 2 (hc2) in a Belgian cervical cancer screening setting. In women >30 years old, the sensitivity and specificity for intraepithelial neoplasias of grade 2 or worse (93 cases of cervical intraepithelial neoplasias of grade 2 or worse (CIN2+) and 1,207 cases of no CIN or CIN1) were 93.6% and 95.6%, respectively, and those of hc2 were 83.9% and 94.5%, respectively {relative sensitivity of qPCR/hc2 = 1.12 [95% confidence interval (CI), 1.01 to 1.23]; relative specificity = 1.01 [95% CI, 0.99 to 1.03]}. A score test showed that the sensitivity (P < 0.0001) and specificity (P < 0.0001) of the qPCR assay were not inferior to those of hc2 at the required thresholds of 90% and 98%, respectively. The overall agreement of hrHPV positivity between the two runs of the qPCR tests was 98.7% (95% CI, 97.5 to 99.4%), with a kappa value of 0.96 (95% CI, 0.83 to 1.00). The qPCR assay used in this study can be considered a reliable HPV assay that fulfills the clinical validation criteria defined for use in cervical cancer screening.
INTRODUCTION
Momentum is building toward the understanding and awareness that persistent infection with high-risk human papillomavirus (hrHPV) is the primary risk factor for the development of cervical cancer and its precursor lesions (7, 19, 27). Today, evidence is available from randomized trials that screening with Hybrid Capture 2 (hc2) or with a GP5+/6+ PCR-enzyme immunoassay (EIA) results in a reduced incidence of cervical intraepithelial neoplasia of grade 3 or worse (CIN3+) lesions and even of invasive cervical cancer in the second or subsequent screening rounds (3, 4). Therefore, these two tests are considered clinically validated for use in screening for cervical cancer.
Recently, the cross-sectional equivalency criteria that a candidate HPV assay has to fulfill were outlined by an international consortium based on a comparison of the new assay with hc2 or a GP5+/6+ PCR-EIA (17, 21).
In the present study, we evaluated a type-specific real-time quantitative PCR (qPCR) assay targeting the viral E6/E7 genes included as high-risk types in the hc2 according to this validation paradigm (17).
MATERIALS AND METHODS
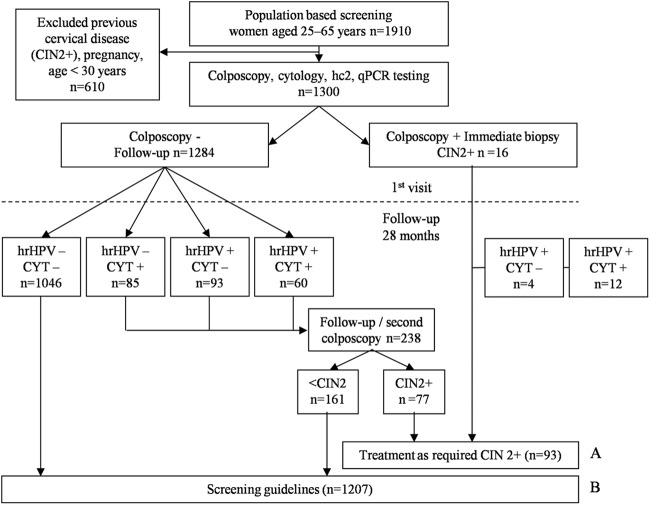
Women coming for routine cervical cancer screening from August 2008 until August 2009 were asked to participate in a controlled colposcopy trial (DRKS00000408). After giving written informed consent, all of the women underwent a colposcopic examination immediately after the collection of a cervical cell sample. All cervical cell specimens were tested by cytology, hc2, and qPCR assay (Fig. 1). Positivity by one of the HPV tests and/or abnormal cytology prompted a second colposcopy. Colposcopists and histologists were unaware of the cytology or HPV test results. This study was approved by the local ethical committee (Ziekenhuis Oost-Limburg, Genk, Belgium).
Fig 1.
Study algorithm. hrHPV: hc2 and/or qPCR positive for 1 of the 13 hrHPV types. (A) Samples selected for clinical sensitivity analysis (n = 93). (B) Samples selected for clinical specificity analysis (n = 1,207).
Cervical cells were collected by using the Cervex-Brush Combi (Rovers, Oss, The Netherlands) as recommended in the European Union guidelines (2). After collection, the head of the brush was left in a vial containing ethanol-based BD SurePath preservative fluid (BD SurePath; BD Diagnostics-TriPath, Burlington, NC). The vial was then transported to RIATOL, Department of Molecular Diagnostics, Sonic Healthcare Benelux, Antwerp, Belgium, where all samples were prepared. A density sedimentation method (BD PrepMate; BD Diagnostics-Tripath, Burlington, NC) was used to enrich the cell samples by removing obscuring elements such as blood, inflammatory cells, necrotic debris, and mucus. DNA was isolated from the cellular pellet remaining after cytologic processing as previously described (10). The qPCR assay involves automated sample preparation and DNA extraction combined with real-time PCR technology to detect and quantify 17 different HPV types, including the 13 HPV types considered high-risk types in hc2, i.e., HPV type 6 (HPV6), HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV53, HPV56, HPV58, HPV59, HPV66, HPV67, and HPV68, as previously described by Micalessi et al. (18). A β-globin real-time qPCR assay was used to assess DNA quality and to estimate the number of cells in the test sample (18). This β-globin control PCR was considered positive when at least 1,000 cells could be measured. The analytical sensitivities of the different HPV type-specific qPCR assays vary between 1 and 100 HPV copies/reaction (11). The number of HPV copies was divided by the number of cells to calculate the viral load (the number of HPV copies/cell). The threshold of positivity was 0.0001 HPV copy/cell.
Since June 2006, more than 600,000 liquid-based cytology samples have been tested with this assay (1).
The clinical performance of the qPCR assay for the 13 types included in hc2 was compared with that of the hc2 test, which detects HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, and HPV68 (15, 20). hc2 testing was performed with BD-SurePath specimens in a central lab (Laborverbund für Medizinische Diagnostik, Heidelberg, Germany) throughout the trial in accordance with the manufacturer's instructions. A ratio of relative light units (RLU) to a standard positive control of >1 was considered positive. The qPCR assay was considered positive if at least 1 of the 13 high-risk types targeted by hc2 was present. The cumulative hrHPV load measured by the qPCR assay was defined as the sum of the type-specific loads of the 13 high-risk types.
Sensitivity was assessed with samples with histologically confirmed CIN2+, whereas specificity was evaluated for women with an outcome of CIN1 or absence of CIN. The clinical sensitivity and specificity of the qPCR assay for CIN2+ were compared to those of hc2 by using a noninferiority score test considering a relative sensitivity threshold of at least 0.90 and a specificity threshold of at least 98% (17). P values for noninferiority were computed as described previously (26). Moreover, differences in sensitivity and specificity between hc2 and the qPCR assay were assessed by using McNemar's exact χ2 test.
To determine the intrasystem reproducibility of the quantitative HPV genotyping assay, two portions (half of the extracted DNA of the original sample) of a set of 633 samples were retested by two independent laboratory technicians using the same reagent lot numbers after 6 weeks (RIATOL, Sonic Healthcare Benelux, Antwerp, Belgium). The two runs were performed with different PCR machines at two different locations (Antwerp and Hoboken). The concordance for the presence of high-risk types was assessed by the percent agreement and kappa values (13). The type-specific viral load agreement between the two runs was assessed by using Bland-Altman graphs, which plot the paired differences against the pairwise load means for each sample that is positive for a given hrHPV type by the qPCR assay (6). The plot further contains the limits of agreement, which correspond to ±1.96 standard deviations of the pairwise load differences. A horizontal line through the mean of the differences near the line of perfect agreement (through zero on the y axis) indicates good agreement. The change in sensitivity and specificity with different viral load cutoffs was assessed by receiver operating characteristic (ROC) curve analysis. Fitted maximum-likelihood ROC curves were estimated by assuming a binomial distribution of the underlying sensitivity and specificity using the MedCalc program (MedCalc Software, Mariakerke, Belgium) (22, 23). All other statistical analyses were performed with Stata version 10 0.1 (StataCorp, College Station, TX).
RESULTS
Out of the 1,300 liquid-based cervical smears from the controlled colposcopy trial, two sets were selected. The first set was for clinical sensitivity analysis (Fig. 1A) and included 93 smears from women (median age of 36 [range, 30 to 65] years) who had histologically confirmed CIN2+ lesions (i.e., 42 with CIN2, 45 with CIN3, 1 with adenocarcinoma, and 5 with squamous cell carcinoma). The median follow-up time was 3.3 (range, 0 to 33) months. Women were identified through HPV testing (hc2 and/or qPCR assay), by cytology, or by the baseline colposcopy. A second set for clinical specificity analysis (Fig. 1B) included 1,207 representative smears from women (median age of 45 [range, 30 to 65] years) without a CIN2+ diagnosis within a follow-up time of 28 months.
The sensitivity and specificity values of the qPCR assay for CIN2+ were 93.6% (87/93; 95% confidence interval [CI], 86.5 to 97.6%) and 95.6% (95% CI, 94.3 to 96.7%), respectively. The sensitivity and specificity values of hc2 for CIN2+ were 83.9% (78/93; 95% CI, 74.8 to 90.7%) and 94.5% (95% CI, 93.0 to 95.7%), respectively. Both the clinical sensitivity and specificity for CIN2+ of the quantitative HPV genotyping assay were not inferior to those of the hc2 (P < 0.0001 and P < 0.0001, respectively). Moreover, the quantitative HPV genotyping assay was not only superior to hc2 with respect to the sensitivity of CIN2+ detection, with a sensitivity difference of 9.7% (CI, 0.1 to 19.2%) (P = 0.029; McNemar's test), but also more specific than hc2, with a specificity difference of 1.6% (CI, 1.3 to 1.6%) (P = 0.016; McNemar's test). An overview of the real-time qualitative HPV assay results stratified by CIN2+ disease status is given in Table 1.
TABLE 1.
Real-time qualitative HPV assay results of 1,300 liquid-based cervical smears stratified by CIN2+ disease status
| Status and qPCR result | No. of smears with hc2 result of: |
Total no. of smears | |
|---|---|---|---|
| hrHPV− | hrHPV+ | ||
| <CIN2 | |||
| hrHPV− | 1,130 | 24a | 1,154 |
| hrHPV+ | 10 | 43 | 53 |
| Total | 1,140 | 67 | 1,207 |
| CIN2+ | |||
| hrHPV− | 2 | 4b | 6 |
| hrHPV+ | 13c | 74 | 87 |
| Total | 15 | 78 | 93 |
Of the 24 hc2 hrHPV-positive/qPCR hrHPV-negative specimens, 5 were positive by qPCR for genotypes that are not included in the 13 HPV types, i.e., HPV53 (n = 1), HPV66 (n = 2), and HPV67 (n = 2).
All four patients were CIN2, three patients were positive for HPV types not included in the 13 HPV types with the qPCR, namely, two HPV53-positive patients and one HPV66 patient, and all three patients had a high viral load levels.
All of the 13 hc2 hrHPV-negative/qPCR hrHPV-positive patients (3 HPV16; HPV16 and HPV18; HPV16, HPV31, and HPV39; HPV31; 2 HPV33; HPV51; HPV51 and HPV52; HPV52; and 2 HPV56) had a low mean viral load level (2.11 log HPV copies/cell, which was significantly lower than the mean viral load level of the 74 hc2 hrHPV-positive/qPCR hrHPV-positive patients (3.43 log HPV copies/cell; P = 0.0003).
The reproducibility was very high for the 13 pooled high-risk types considered together (98.7% [95% CI, 97.5 to 99.4%]), as well as for each HPV type separately, between 98 and 100%. The positive agreement for results that were hrHPV positive in the first or second run was 93% (95% CI, 87 to 97%). The kappa value was 0.96 (95% CI, 0.83 to 1.00) for hrHPV and >0.87 for separate hrHPV types.
The average number of hrHPV infections per sample did not differ in samples where both runs yielded a positive hrHPV result.
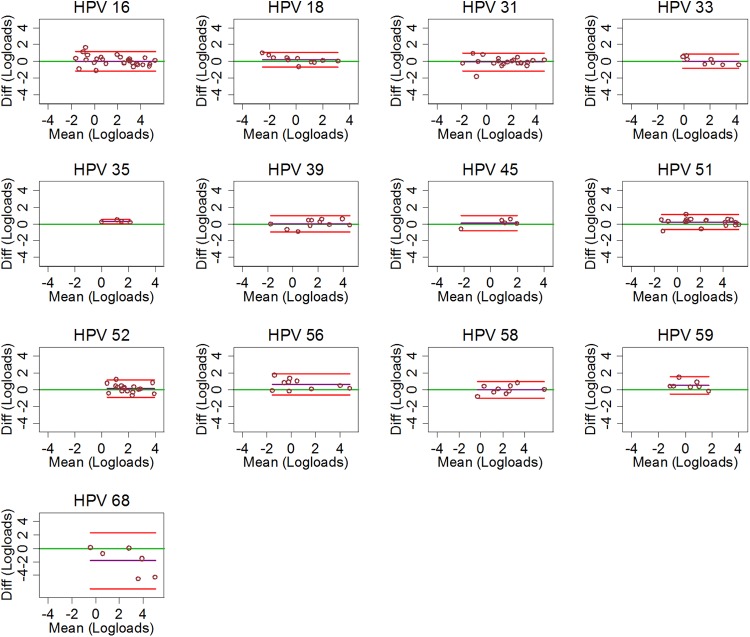
For the reproducibility of viral load measurements (log10 of viral load), the analysis was restricted to samples were both measurements were positive. The Bland-Altman plots of the 13 hrHPV types are displayed in Fig. 2. The purple horizontal line through the average difference of log loads was always located near the green line of perfect agreement (Fig. 2).
Fig 2.
Bland-Altman plots displaying the viral load agreement of each of the 13 hrHPV types between the first and second qPCR assay assessments according to the HPV type. The green horizontal line corresponds to perfect agreement, the purple line corresponds to the average difference (Diff) in type-specific log loads, and the red lines correspond to the 95% limits of agreement.
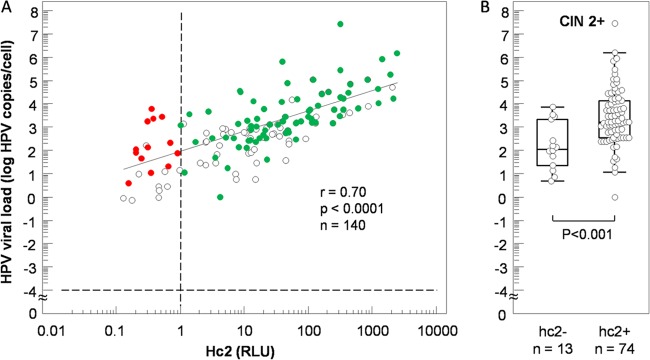
The correlation between the viral loads measured by hc2 (expressed in RLU) and the qPCR assay (log of the sum of viral loads of 13 hrHPV types) is shown in Fig. 3. The correlation was r = 0.70 (95% CI, 0.60 to 0.78; P < 0.0001) for samples with a positive hrHPV qPCR assay results (n = 140) (Fig. 3A). The distribution of the cumulative viral load in all of the CIN2+ cases is also shown for hc2-negative (n = 13) and hc2-positive cases (n = 78) in Fig. 3B. ROC curve analysis for these hc2 cases showed a cutoff for CIN2+ at 2.35 log hrHPV copies/cell with a sensitivity of 90.5% and a specificity of 69.2% with an area under the ROC curve of 0.791 and a 95% CI between 0.691 and 0.871.
Fig 3.
(A) Scatterplot showing the cumulative loads of all 13 hrHPV types included in hc2 as a function of the hc2 RLU value (circles). Red circles, hc2− CIN2+ cases. Green circles, hc2+ CIN2+ cases. (B) Cumulative hrHPV loads of CIN2+ cases according to hc2 status.
The ROC curves for both tests are displayed in Fig. 4. The sensitivities of hc2 and the qPCR assay for CIN2+ jointly rise with a lower viral load cutoff up to approximately 73% at a specificity of approximately 97%. The sensitivity of the qPCR assay continues to rise steeply to a maximum of 94% at a specificity of 96%. However, the hc2 ROC curve shows a less steep slope and any further sensitivity increase is accompanied by a substantial loss of specificity.
Fig 4.
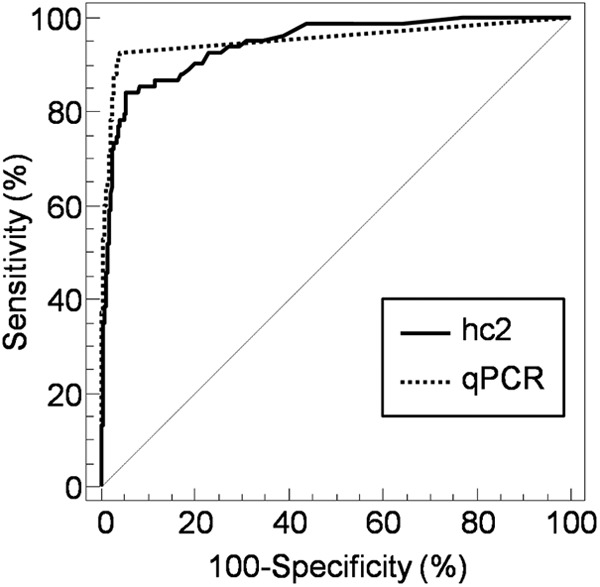
ROC curves for hc2 (RLU) and the qPCR assay (log number of hrHPV copies/cell) in discriminating between samples from women without (n = 1,207) or with (n = 93) CIN2+.
An overview of the ROC curve analysis of both hc2 and the qPCR assay for the detection of CIN2+ is given in Table 2. Comparison of ROC curves for a given sensitivity of 95%, however, showed that the qPCR assay was significantly more specific (25.9%). Also for a given specificity of 95%, the qPCR assay showed significantly better sensitivity (15.2%) than hc2 in the detection of CIN2+ (Table 2).
TABLE 2.
Comparison of ROC curve analyses of hc2 and qPCR for detection of CIN2+
| Test | For 95% sensitivity |
For 95% specificity |
||||
|---|---|---|---|---|---|---|
| % Specificity | 95% CI | Threshold value | % Sensitivity | 95% CI | Threshold value | |
| hc2a | 69.2 | 55.7–81.0 | >0.2015 | 78.3 | 63.9–85.5 | >1.477 |
| qPCRb | 95.2c | 94.2–96.6 | >−4 | 93.6c | 87.1–96.8 | >−4 |
Results are expressed in RLU.
Results are expressed in log number of hrHPV copies per cell.
P < 0.05 compared to hc2.
DISCUSSION
The aim of the present study was to compare the clinical accuracy of a type-specific real-time qPCR assay with the clinically validated reference HPV test, hc2, with samples from women coming for routine cervical cancer screening and included in a controlled colposcopy trial. The results show higher clinical sensitivity (93.6% versus 83.9%) and specificity (95.6% versus 94.5%) of the qPCR assay (0.0001 HPV copies/cell) than of hc2 (>1 RLU) for CIN2+. Indeed, at a sensitivity of 95%, hc2 was approximately 25% less specific than the qPCR assay, and at a specificity of 95%, hc2 was also 15% less specific. These differences could be explained, on the one hand, by the nature of the assay. Indeed, the qPCR assay is more sensitive than liquid hybridization and also more specific because there is no cross-reaction among the different type-specific E6/E7 qPCR assays, in contrast to the known cross-reactivity with low-risk HPV types exhibited by the high-risk probes of hc2 (8). Furthermore, on the other hand, during recent years, we improved the clinical sensitivity for detection of CIN2+. This improvement in sensitivity may have been the result of modification of the sampling device (Cervex-Brush Combi, yielding a 50-fold greater viral load) left in the transport liquid (10); this brush could be better at sampling squamo-columnar junction cells at the transition zone, where neoplastic lesions preferentially occur (14); and the enrichment of the cell suspension used to make a liquid-based cytology specimen and the BD FocalPoint assisted cytological interpretation with knowledge of the HPV status. We demonstrated that prior knowledge of the HPV status improves the sensitivity of cytology for CIN2+ detection (5). The efficacy of using a clinically more sensitive assay has recently been well illustrated by four randomized controlled trials comparing primary HPV screening with cytology screening (4). These randomized trials consistently showed, in the second screening round, a significant reduction in the incidence of CIN3+ and even of invasive cancer (CIN3 precedes cervical cancer) by HPV screening, reducing the number of women dying from cervical cancer.
In the present study, ROC curve analysis showed a 3-log-lower clinical threshold for the qPCR assay (−0.04 log HPV copies/cell) than hc2 (2.35 log HPV copies/cell) for the detection of CIN2+. Further, ROC curve analysis also showed that the clinical threshold for hc2 was lower (0.77 RLU) than that given in the product insert (>1 RLU), corroborating the improvement of our clinical sensitivity.
Concerning transferability to other laboratories, we recommend the use of a cell enrichment method and correction for the number of cells before performing DNA extraction, instead of taking a fixed volume from the original vial. Furthermore, we encourage the use of primers targeting the E6 or E7 region and not L1, as L1-negative cancers have been described (27). And finally, we recommend the use of individual type-specific and normalized (for the number of cells) HPV tests which allow monitoring of individual patients over time (12). Such serial measurements without clinical threshold constraints (9, 12, 16), in comparison to a fixed single viral load threshold (24, 25), could allow earlier detection of CIN2+ and improve clinical sensitivity.
Whether the use of the qPCR assay would result in less cancer than that of hc2 and whether it would allow the safe extension of screening intervals cannot be derived from this cross-sectional validation study. However, this study clearly indicates that the qPCR assay fulfills and even exceeds the requirements defined in the international guidelines for HPV DNA assays that can be used for cervical cancer screening (17).
ACKNOWLEDGMENTS
This work was supported in part by Biokring II vzw (R10-023). M.A. received financial support from the International Agency for Research on Cancer (IARC), Lyon, France; the 7th Framework Programme of DG Research of the European Commission through the PREHDICT project (grant 242061, coordinated by the Vrije Universiteit Amsterdam, Amsterdam, The Netherlands); and the Belgian Foundation Against Cancer (Brussels, Belgium). J.J.B. received financial support from the 7th Framework Programme of DG Research of the European Commission through the HPV-AHEAD project coordinated by IARC and from the Belgian Foundation Against Cancer (Brussels, Belgium).
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Arbyn M, et al. 2009. Prevaccination distribution of human papillomavirus types in women attending at cervical cancer screening in Belgium. Cancer Epidemiol. Biomarkers Prev. 18:321–330 [DOI] [PubMed] [Google Scholar]
- 2. Arbyn M, et al. 2007. European guidelines for quality assurance in cervical cancer screening: recommendations for collecting samples for conventional and liquid-based cytology. Cytopathology 18:133–139 [DOI] [PubMed] [Google Scholar]
- 3. Arbyn M, Ronco G, Meijer CJ, Naucler P. 2009. Trials comparing cytology with human papillomavirus screening. Lancet Oncol. 10:935–936 [DOI] [PubMed] [Google Scholar]
- 4. Arbyn M, et al. 2012. EUROGIN 2011 roadmap on prevention and treatment of HPV-related disease. Int. J. Cancer 131:1969–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benoy IH, et al. 2011. Prior knowledge of HPV status improves detection of CIN2+ by cytology screening. Am. J. Obstet. Gynecol. 205:569.e1–569.e7 [DOI] [PubMed] [Google Scholar]
- 6. Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307–310 [PubMed] [Google Scholar]
- 7. Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castle PE, et al. 2008. Human papillomavirus genotype specificity of hybrid capture 2. J. Clin. Microbiol. 46:2595–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Constandinou-Williams C, et al. 2010. Is human papillomavirus viral load a clinically useful predictive marker? A longitudinal study. Cancer Epidemiol. Biomarkers Prev. 19:832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Depuydt CE, et al. 2006. Improved endocervical sampling and HPV viral load detection by Cervex-Brush Combi. Cytopathology 17:374–381 [DOI] [PubMed] [Google Scholar]
- 11. Depuydt CE, et al. 2007. Comparison of MY09/11 consensus PCR and type-specific PCRs in the detection of oncogenic HPV types. J. Cell Mol. Med. 11:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Depuydt CE, et al. 14 September 2012. Changes in type-specific human papillomavirus load predict progression to cervical cancer. J. Cell Mol. Med. (Epub ahead of print.) doi:10.1111/j.1582-4934.2012.01631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleiss JL. 1981. Statistical methods for rates and proportions. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 14. Herfs M, et al. 2012. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc. Natl. Acad. Sci. U. S. A. 109:10516–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lörincz AT. 1996. Hybrid capture method for detection of human papillomavirus DNA in clinical specimens: a tool for clinical management of equivocal Pap smears and for population screening. J. Obstet. Gynaecol. Res. 22:629–636 [DOI] [PubMed] [Google Scholar]
- 16. Marks M, et al. 2011. Kinetics of DNA load predict HPV 16 viral clearance. J. Clin. Virol. 51:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meijer CJ, et al. 2009. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int. J. Cancer 124:516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Micalessi IM, Boulet GA, Bogers JJ, Benoy IH, Depuydt CE. 2012. High-throughput detection, genotyping and quantification of the human papillomavirus using real-time PCR. Clin. Chem. Lab. Med. 50:655–661 [DOI] [PubMed] [Google Scholar]
- 19. Nobbenhuis MA, et al. 1999. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet 354:20–25 [DOI] [PubMed] [Google Scholar]
- 20. Poljak M, Brencic A, Seme K, Vince A, Marin IJ. 1999. Comparative evaluation of first- and second-generation digene hybrid capture assays for detection of human papillomaviruses associated with high or intermediate risk for cervical cancer. J. Clin. Microbiol. 37:796–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rijkaart DC, et al. 2012. Evaluation of 14 triage strategies for HPV DNA-positive women in population-based cervical screening. Int. J. Cancer 130:602–610 [DOI] [PubMed] [Google Scholar]
- 22. Schoonjans F, Depuydt C, Comhaire F. 1996. Presentation of receiver-operating characteristics (ROC) plots. Clin. Chem. 42:986–987 [PubMed] [Google Scholar]
- 23. Schoonjans F, Zalata A, Depuydt CE, Comhaire FH. 1995. MedCalc: a new computer program for medical statistics. Comput. Methods Programs Biomed. 48:257–262 [DOI] [PubMed] [Google Scholar]
- 24. Snijders PJ, et al. 2006. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int. J. Cancer 119:1102–1107 [DOI] [PubMed] [Google Scholar]
- 25. Snijders PJ, van den Brule AJ, Meijer CJ. 2003. The clinical relevance of human papillomavirus testing: relationship between analytical and clinical sensitivity. J. Pathol. 201:1–6 [DOI] [PubMed] [Google Scholar]
- 26. Tang NS, Tang ML, Chan IS. 2003. On tests of equivalence via non-unity relative risk for matched-pair design. Stat. Med. 22:1217–1233 [DOI] [PubMed] [Google Scholar]
- 27. Walboomers JM, et al. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19 [DOI] [PubMed] [Google Scholar]