Abstract
Reported methods for the detection of the yellow fever viral genome are beset by limitations in sensitivity, specificity, strain detection spectra, and suitability to laboratories with simple infrastructure in areas of endemicity. We describe the development of two different approaches affording sensitive and specific detection of the yellow fever genome: a real-time reverse transcription-quantitative PCR (RT-qPCR) and an isothermal protocol employing the same primer-probe set but based on helicase-dependent amplification technology (RT-tHDA). Both assays were evaluated using yellow fever cell culture supernatants as well as spiked and clinical samples. We demonstrate reliable detection by both assays of different strains of yellow fever virus with improved sensitivity and specificity. The RT-qPCR assay is a powerful tool for reference or diagnostic laboratories with real-time PCR capability, while the isothermal RT-tHDA assay represents a useful alternative to earlier amplification techniques for the molecular diagnosis of yellow fever by field or point-of-care laboratories.
INTRODUCTION
Yellow fever (YF) is a zoonotic flaviviral disease endemic and epidemic in tropical regions of South America and Africa. According to the World Health Organization (WHO), YF remains an important public health problem in Africa and is considered an emerging disease (9). The true incidence of YF infection is unknown due to insufficient reporting and ground surveillance but has been estimated at over 200,000 cases per year worldwide, causing 30,000 deaths (23).
The clinical diagnosis of YF is particularly difficult because the symptoms are quite similar to those of a wide range of diseases, including dengue fever, other hemorrhagic viral diseases, leptospirosis, viral hepatitis, and malaria; laboratory confirmation, therefore, is essential. The WHO recommends as criteria for laboratory YF diagnosis the detection of YF-specific IgM or a 4-fold or greater rise in specific serum IgG levels in the absence of recent yellow fever vaccination and negative detection results for other flaviviruses. Formal confirmation of yellow fever virus (YFV) infection, however, requires immunohistochemical detection of YFV antigen, amplification of YFV genomic sequences from blood or solid tissues by PCR, or a test for viremia involving cultivation of YFV infectious particles. In general, these assays are performed only in a few national or international reference laboratories.
Molecular methods for the detection of the viral genome offer a rapid, sensitive, and highly specific alternative to serological assays for early diagnosis during the viremic phase of infection or in postmortem tissues. A number of molecular assays have been reported in recent years for the detection of YFV RNA, with various levels of sensitivity and specificity (6). Conventional reverse transcription-PCR (RT-PCR) and RT-nested PCR assays are the most frequent methods for YFV diagnosis currently used by reference laboratories in areas of endemicity. The more sensitive nested PCR protocols, however, involve an increased risk of sample cross-contamination and the need to confirm the identity of the viruses by sequencing the PCR amplicons. Real-time PCR is a valuable technical alternative, but the high cost of instrumentation purchase and maintenance and the need for skilled personnel make it very challenging to implement where YF diagnosis is most needed, i.e., at the point of care (POC) in areas of endemicity or in developing countries with limited laboratory infrastructure.
Isothermal nucleic acid amplification assays provide an alternative to established molecular diagnostics in laboratories with limited resources (reviewed in reference 5). Isothermal helicase-dependent amplification (HDA) technology uses a DNA helicase to untwine the DNA strands and generate single-stranded templates for primer hybridization and subsequent extension (21). This provides an elegant method to perform amplification at a single incubation temperature. In addition, the inclusion of labeled probes in HDA increases assay specificity by enabling nucleic acid detection technologies such as the lateral-flow system (17), with potential applications for “instrument-free” amplicon analysis.
For this study, we developed and evaluated two different YFV molecular diagnosis assays targeting the same conserved region of the YFV which should be suited to different laboratory environments. One is a sensitive and specific TaqMan real-time RT-quantitative PCR (qPCR) protocol. The other is a robust one-step isothermal helicase-dependent amplification (RT-tHDA) assay.
MATERIALS AND METHODS
Except where otherwise noted, molecular biology kits and equipment were used in accordance with the manufacturers' instructions.
Viruses and samples.
Virus stocks were grown in Vero E6 cells that were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (PAA Laboratories, Cölbe, Germany).
The viruses used in this study for testing assay specificity and sensitivity were YFV strains 17D, Brazil, and IvoryC1999 (3); West Nile virus (WNV) strain New York; Japanese encephalitis virus (JEV) strain SA-14-02; St. Louis encephalitis virus (SLEV) strain Parton; tick-borne encephalitis virus (TBEV) strain K063; Chikungunya virus (CHIKV) strain Marseille LR 2006/684-1; and the four dengue virus serotypes DENV-1 VR344 (strain Thai 1958), DENV-2 VR345 (strain TH-36), DENV-3 VR216 (strain H87), and DENV-4 VR217 (strain H241).
The molecular diagnosis external quality assurance (EQA) panel used for the evaluation of the new YFV assays involved 10 YFV-positive samples, as follows: for strain 17D, five sets of 1:10 serial dilutions in the range of 3 × 102 genome equivalents [GE]/sample to 3 × 106 GE/sample; for strain Brazil, two dilutions at 104 GE/sample and 103 GE/sample, respectively; and for strain IvoryC1999, three dilutions in the range of 69 GE/sample to 2 × 104 GE/sample. Two additional plasma samples were used as specificity controls, one containing WNV, JEV, SLEV, and TBEV (strain Absettarov) and the other the four dengue serotypes. Two negative-control plasma samples were also included (8).
The clinical samples studied were sera from suspected cases of YFV infection. Eight sera (samples A to H) were confirmed as representing positive YFV natural infections by molecular assays at the University Medical Center, Göttingen, Germany (22), and the National Institute of Health of Colombia (M. C. Méndez, L. C. Pardo, C. Domingo, G. J. Rey, A. Tenorio, and J. A. Méndez, submitted for publication). Five sera (samples I to M) tested negative by RT-PCR (11) and were further assayed for serology (YFV IgM enzyme-linked immunosorbent assay [ELISA], CDC protocol) at the Pasteur Institute of Bangui, Central African Republic. Three sera (samples I to K) were classified as negative, while 2 sera (samples L and M) were confirmed positive for YFV by the Pasteur Institute of Dakar, Senegal, using a seroneutralization assay. An ELISA for IgM against DENV, WNV, CHIKV, Rift Valley fever virus, and Crimean-Congo hemorrhagic fever virus ruled out infection by these viruses in samples I to M.
Lastly, the study included 17 urine samples from YF-17D healthy vaccinees that had tested positive (n = 9) or negative (n = 8) in a previous study (7) using a real-time RT-qPCR targeted to the NS3 gene (2).
Primers and probes.
Primers for the YFV genome conform to sequences in the GenBank public database, including South American, African, and vaccine strains (accession numbers JN628279, JN628280, JN620362, GQ379163, GQ379162, FJ654700, DQ235229, DQ100292, AY968064, AY968065, DQ118157, AY572535, AY603338, U54798, AY640589, X15062, X03700, AF094612, U89338, U52422, U52419, U52416, U52413, U52410, U52409, U52404, U52403, U52398, U52392, U52395, U21055, U17067, U17066, U21056, D14458, and U52389). The primers and hydrolysis probe described were designed based on multiple sequence alignments generated with Mega 4 software (18). Primer design focused on the 5′-noncoding region (5′-NC) of the YFV genome, as preliminary sequence analysis showed the presence there of well-conserved segments among all YFV strains. To check for specificity and potential mismatches, the sequences of YFV strains from the GenBank database were aligned against the set of candidate primers and probe (see Fig. S1 in the supplemental material).
The primer pair selected permitted efficient amplification of PCR fragments of appropriate size for both the RT-qPCR and RT-tHDA assays. The reverse primer in the RT-tHDA isothermal protocol using a lateral-flow detection system was 5′ end labeled with biotin (YFallR-Biotin). The same 6-carboxyfluorescein (FAM)–6-carboxytetramethylrhodamine (TAMRA) hydrolysis probe, 24-mer, was used for the YF RT-qPCR and the YF RT-tHDA protocols (Table 1). Primers were purchased from Invitrogen (Karlsruhe, Germany), and probes were from TIB Molbiol (Berlin, Germany).
TABLE 1.
Primers and probes for RT-qPCR and RT-tHDA assays
| Primer or probe | Sequencea | Positionb |
|---|---|---|
| Primers | ||
| YFallF | 5′-GCTAATTGAGGTGYATTGGTCTGC-3′ | 15–38 |
| YFallR | 5′-CTGCTAATCGCTCAAMGAACG-3′ | 83–103 |
| YFallR-Biotin | 5′-Biot-CTGCTAATCGCTCAAMGAACG-3′ | 83–103 |
| Probe | ||
| YFallP | 5′-FAM-ATCGAGTTGCTAGGCAATAAACAC-TMR-3′ | 41–64 |
Biot, biotin; TMR, 6-carboxytetramethylrhodamine; FAM, 6-carboxyfluorescein.
Positions are indicated relative to GenBank sequence AY640589.1 for yellow fever virus Asibi strain.
RNA extraction.
Viral RNA was extracted with a QIAamp viral RNA minikit (Qiagen, Hilden, Germany). When 1 ml of sample (i.e., urine samples) was used as the starting material, viral RNA extraction was performed using a QIAamp UltraSens virus kit (Qiagen).
Standard preparations for the YF RT-qPCR assay.
YFV genomic RNA from YF-17D culture supernatants was retrotranscribed using SuperScript II reverse transcriptase and random hexamer primers (Invitrogen) in a Biometra thermoblock (Biometra, Göttingen, Germany). The target YFV cDNA was next amplified using AmpliTaq Gold DNA polymerase (Invitrogen) and primers YFallF and YFallR under the following thermal conditions: 94°C for 2 min; 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; a final extension step at 72°C for 5 min; and a 4°C soak. The PCR product was excised from the gel, extracted using a QIAquick gel extraction kit (Qiagen), and cloned into pDrive cloning vector using a Qiagen PCR cloning kit (Qiagen). After blue-white colony screening using IPTG (isopropyl-β-d-thiogalactopyranoside) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), plasmid DNA (pDrive-5NC) from selected colonies was purified with a plasmid DNA minikit (Qiagen) and the presence of the desired YFV insert confirmed by PCR and sequencing.
RNA copy number standards were synthesized in vitro from the linearized pDrive-5NC template using a Riboprobe T7 in vitro transcription system (Promega, Mannheim, Germany). The RNA product (in vitro-transcribed RNA [ivRNA]) was subjected to DNase digestion twice to remove contaminating plasmid DNA using a Turbo DNA-free kit (Applied Biosystems/Ambion, Foster City, CA). The RNA concentration was determined spectrophotometrically using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Dreieich, Germany) at 260 nm and the copy number estimated with the following formula: RNA copy number = [RNA concentration (g/μl) × 6.02 × 1023]/[345 × RNA length (ribonucleotides)]. Serial dilutions were prepared in molecular biology-grade water to generate a standard curve in the range of 1 RNA copy/assay to 105 RNA copies/assay. ivRNAs were used directly after preparation for evaluation of the RT-qPCR assay and thereafter stored at −80°C.
Standardization of YF real-time quantitative PCR.
The YF RT-qPCR protocol was standardized using a QuantiTect Virus +ROX Vial kit (Qiagen), using 0.4 μM each primer (YFallF and YFallR; Table 1), 0.2 μM fluorescently labeled probe (YFallP), and the following thermal cycling program: 50°C for 20 min, 95°C for 5 min, and then 40 cycles of 95°C for 15 s and 60°C for 45 s.
Two microliters of sample RNA was used in a final volume of 25 μl. Each assay included a negative (nontemplate) control and standards in the range of 1 copy/reaction to 105 copies/reaction for ivRNA or 1 GE/reaction to 106 GE/reaction for the pDrive-5NC vector. The reactions were performed in 96-well plates or 8-tube strips and run on an ABI Prism 7500 real-time PCR system (Applied Biosystems).
One-step helicase-dependent isothermal RT-PCR (RT-tHDA).
The RT-tHDA assay was optimized using a BioHelix IsoAmp II Universal RT-tHDA kit (New England BioLabs, Frankfurt am Main, Germany). Standard reaction temperatures and final concentrations of primers, MgSO4, and NaCl were optimized during preliminary work (data not shown). In the RT-tHDA assay, excess reverse primer is used to preferentially generate biotin-labeled single-stranded amplicon that can be hybridized in the lateral-flow system. The one-step, asymmetric RT-tHDA reaction mixture, at a 25-μl final volume (i.e., one-half of the final volume recommended by the manufacturer of the BioHelix kit), contained 0.08 μM primer YFallF and 0.16 μM primer YFallR-Biotin, 3.5 mM MgSO4, 30 mM NaCl, 0.04 μM YFallP probe, 0.15 μl of ThermoScript reverse transcriptase (Invitrogen) (7.15 U/μl), and 5 μl of sample RNA. The amplification reaction mixture was incubated at 60°C for 2 h in a Biometra thermal block and then subjected immediately to lateral-flow immunochromatography for amplicon detection.
Lateral-flow strip detection.
In this assay, 20 μl of RT-tHDA reaction is applied to the sample pad of a lateral-flow strip (LFS) (RapidSTRIPE detection assay, Analytik Jena AG, Jena, Germany, or HybriDetect MGHD-1, Milenia Biotec, Giessen, Germany), and the LFS is placed into a tube containing 150 μl of running buffer. The carboxyfluorescein- and biotin-labeled amplification-hybridization product migrates by capillary action through the next section of the LFS, where it binds the streptavidin-gold conjugate (detection probe) via the biotin label. As this complex reaches the detection band, it is captured there by anti-fluorescein isothiocyanate (FITC) antibodies (capture probe), resulting in a visible line (13). Noncaptured complexes and excess detection probe are stopped further downstream at the control line, providing a visual check of efficient flow along the stripe. The results are read after 15 min; a positive reading is scored if a test line is observed in addition to the control line, a negative reading if the control line alone is visible.
Determination of analytical performance and intralaboratory repeatability of the YF RT-qPCR.
To accurately determine the limit of detection (LOD) of the assay, seven replicates of serial 1:10 dilutions of YF-17D in the range of 0.01 PFU/ml to 105 PFU/ml were subjected to the RT-qPCR assay, and fractions of positive results for each concentration were subjected to probit regression analysis using SPSS 18 software (IBM, Ehningen, Germany). Likewise, 11 replicates of serial dilutions of in vitro-transcribed RNA in the range of 5 × 102 copies/ml to 5 × 107 copies/ml (i.e., from 105 copies/reaction to 1 copy/reaction) were used to determine the copy number threshold of detection of the RT-qPCR assay. Positive results for each RNA concentration were subjected to probit regression analysis as described above.
To assess the intra-assay repeatability of the YF RT-qPCR, four YF-17D dilutions containing 0.1, 1, 100, and 104 PFU/ml, respectively, were tested 10 times in the same run, and the standard deviation of the quantification cycle values (Cqs) was calculated as a measure of precision.
In assessing specificity, the assay was tested for potential cross-reactivity on RNA from CHIKV and different flaviviruses, namely, WNV, JEV, SLEV, TBEV, and the four DENV serotypes. The analytical sensitivity and specificity of the assay were further verified in the EQA sample panel described above (8).
Analytical performance of the YF RT-tHDA assay.
The LOD of the isothermal RT-tHDA assay was determined on a set of 13 samples containing YF-17D RNA in the range of 1.5 × 103 copies/ml to 3.2 × 109 copies/ml (i.e., from 1.6 × 107 copies/reaction to 7.5 copies/reaction) using six replicates. Fractions of positive results for each concentration were subjected to probit regression analysis using the SPSS 18 software. Detection of different YFV strains was evaluated across the aforementioned EQA panel. Assay specificity was studied as described above for the RT-qPCR.
Interlaboratory reproducibility of the isothermal YF RT-tHDA assay.
A panel of 10 blinded samples were prepared and sent to three expert laboratories involved in YFV diagnostics (Department of Virology of the University Medical Center of Göttingen, Germany; Virology Laboratory of the National Institute of Health of Colombia; and Pasteur Institute de Bangui, Central African Republic).
The panel consisted of 7 positive samples containing 7.3 × 103 to 1.8 × 106 RNA copies/ml (corresponding to 36.4 to 8.8 × 103 RNA copies/reaction) of the YF-17D genome, plus a specificity control containing the four DENV serotypes: 5 × 104 GE/reaction of DENV-1; 1.2 × 104 GE/reaction of DENV-2; 5 × 104 GE/reaction of DENV-3; and 2.5 × 104 GE/reaction of DENV-4. Two negative-control samples were also included. All samples were shipped in RNA extraction buffer (AVL; Qiagen). Each laboratory analyzed the samples blinded and added a negative control for YFV.
Validation of the diagnostic usefulness of the YF RT-tHDA using clinical samples.
A test set of spiked serum (n = 8) and urine (n = 8) samples containing YF-17D RNA and the respective nonpathological, nonspiked controls were processed and tested by the RT-tHDA assay to assess the performance of this method on biological samples. The reliability of the assay in YFV diagnosis was tested using clinical samples, 13 sera from suspected natural YFV infections, and 17 urine samples from YF-17D healthy vaccinees described above under “Viruses and samples.”
RESULTS
Evaluation of the YF TaqMan RT-qPCR assay.
In assessing the analytical performance of the RT-qPCR assay, probit regression analysis determined 95% LOD values of 0.18 PFU/ml (95% confidence interval [CI], 0.067 to 25.9 PFU/ml) and 5.7 × 103 RNA copies/ml (95% CI, 2.8 × 103 to 8.7 × 104 copies/ml). This corresponds to 10.22 RNA copies/reaction (95% CI, 5.48 to 78 copies/reaction). The 50% LOD was 8.16 × 102 RNA copies/ml (95% CI, 2.6 × 102 to 1.35 × 103 copies/ml), corresponding to 1.88 RNA copies/reaction (95% CI, 0.86 to 2.95 RNA copies/reaction). The calculated efficiency of the RT-qPCR was 1.034 for six replicates of serial 1:10 ivRNA dilutions (1 to 105 copies/reaction) were run in parallel. The dispersion of Cqs was greater for the lower viral load samples, and standard deviations ranged overall from 0.73 to 0.11.
Assay sensitivity and specificity were further analyzed across a test set of samples from a YFV molecular diagnostics EQA activity (8), encompassing YFV strains 17D, Brazil, and IvoryC1999, other flaviviral RNAs, and negative controls (see Table S1 in the supplemental material). All the YFV genome-containing samples were correctly detected by the new RT-qPCR regardless of the strain. No cross-reactivity was observed on the samples containing flaviviruses other than YFV. Measured against the assays performed in the EQA exercise (8), the new YF RT-qPCR demonstrated a very good profile of sensitivity and specificity that improved on the highest-scoring EQA methods (see Table S1 in the supplemental material). In parallel, negative results were observed for DENV-1 to -4, WNV, CHIKV, TBEV, SLEV, and JEV tested individually as viral cell supernatants, which confirmed the specificity of this assay for YFV.
Analytical performance evaluation of the YF RT-tHDA isothermal assay.
In the YFV RT-tHDA assay involving lateral-flow detection, probit analysis estimated a 50% LOD of 3.84 × 103 (95% CI, 2.26 × 103 to 6 × 103) copies/ml, corresponding to 45.75 (95% CI, 25.6 to 73) copies/reaction, and a 95% LOD of 1.18 × 104 (95% CI, 7.12 × 103 to 6.95 × 104) copies/ml, corresponding to 180.6 (95% CI, 102.2 to 904.9) copies/reaction for the RT-tHDA.
Tested across the aforementioned EQA panel of YFV strains and other flaviviruses, the RT-tHDA assay correctly detected all YFV samples, except the IvoryC1999 strain sample (sample 6) containing 12 GE/reaction, which falls below the limit of detection of the assay. A faint band distinguishable from the negative controls was observed with the samples containing 66 GE/reaction of YF-17D (sample 14) and 200 GE/reaction of the YFV Ivory Coast strain (sample 1) described above (Fig. 1). Samples containing non-YFV flaviviral RNA were negative in the RT-tHDA assay. In addition to the excellent specificity profile, the performance of the new RT-tHDA was comparable with that of real-time and RT-nested PCR assays, YF specific or panflaviviral, employed in the EQA assay (see Table S1 in the supplemental materials).
Fig 1.
Performance of the isothermal YF RT-tHDA assay. Dilutions of plasma samples representing different strains of YFV (samples labeled 2, 9, 12, 4, 14, 10, 5, 13, 1, and 6 at the top of the image), other related flaviviruses (samples 11 and 3), and negative controls (samples 8 and 7) were subjected to the YFV RT-tHDA assay. The correct performance of the lateral-flow detection method is confirmed by the presence of the control line, C, in all test stripes. Positives samples are identified by the presence of the additional detection line, D.
Interlaboratory reproducibility of the RT-tHDA assay.
Detection consensus using RT-tHDA was observed for those samples containing ≥2.35 × 104 YFV RNA copies/ml (≥280 copies/reaction). Two of the three laboratories correctly detected the YFV RNA in the samples containing 8.4 × 103 copies/ml (100 copies/reaction), and only one laboratory detected as positive the sample containing 3 × 103 copies/ml (36.4 copies/reaction), below the estimated 50% LOD of the assay. The DENV-specific sample and the two negative controls in the set failed to produce amplification or background signals in any of the laboratories, ruling out assay cross-reactivity and sample cross-contamination during this exercise. Together, these results confirmed the expected performance of the assay (Table 2).
TABLE 2.
Study of interlaboratory reproducibility of YFV detection by RT-tHDA
| Test set sample(s) | YFV-17D RNA (no. of copies/reaction) | Test result(s) |
||
|---|---|---|---|---|
| Lab 1 | Lab 2 | Lab 3 | ||
| YFV a | 36.4 | + | – | – |
| YFV b | 40 | + | – | – |
| YFV c | 100 | + | – | + |
| YFV d | 280 | + | + | + |
| YFV e | 480 | + | + | + |
| YFV f | 780 | + | + | + |
| YFV g | 8,800 | + | + | + |
| Blanks a and b | 0 | −/− | −/− | −/− |
| DENV-1 to -4 | 0 | − | − | − |
Application of the assays to clinical samples.
The improved sensitivity and specificity profile led us to apply the RT-qPCR in this work, which targets the highly conserved 5′-NC genomic region of YFV (see Fig. S1 in the supplemental material), to urine samples from 17 healthy YF-17D vaccinees previously studied by a protocol amplifying the NS3 gene (7). The new data corroborated the earlier diagnosis of 9 positive and 8 negative samples. In addition, the new RT-qPCR has become our standard test for suspected YF postvaccinal adverse events during the current WHO mass vaccination campaigns in Africa (G. Breugelmans, R. F. Lewis, E. Agbenu, O. Veit, D. Jackson, C. Domingo, W. Perea, M. Niedrig, B. Gessner, and S. Yactayo, unpublished data).
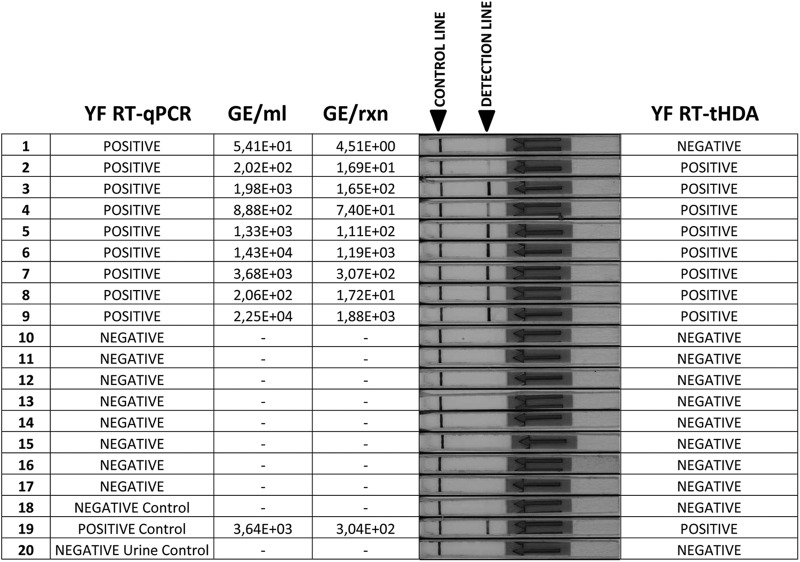
YFV 17D-spiked serum and urine samples were used in a preliminary trial of the RT-tHDA assay on biological fluids. All samples, with YFV RNA copy numbers ranging from 1.3 × 105 to 7.5 × 109 copies/ml, produced a positive signal in the RT-tHDA assay, while the signal from nonspiked serum and urine controls remained negative (data not shown). In addition, the set of 17 urine samples from healthy YF-17D vaccinees (7) described above were retested by the RT-tHDA assay. All the known negative samples (n = 8) tested negative, while 8 of the 9 known positive samples were correctly identified in the new assay. The only false negative was a sample with a very low viral load, 54 RNA copies/ml (4.5 RNA copies/reaction), 1 to 2 orders of magnitude below the 50% LOD determined earlier for this protocol (Fig. 2).
Fig 2.
Parallel detection of the YFV genome in clinical samples by the new RT-qPCR and RT-tHDA assays. The sample set consisted of urine samples from healthy YF vaccinees (samples 1 to 17, YFV strain 17D), a negative (water) control (sample 18), a negative urine control (sample 20), and one positive control (spiked urine, sample 19). In the RT-tHDA assay, a positive detection line was observed on the lateral-flow strip with 8 of the 9 samples that were positive by the more sensitive YF RT-qPCR assay. GE, genome equivalents; rxn, reaction.
In a further assessment involving known wild-type YF patients, all the serum samples that were previously confirmed as YFV positive by RT-PCR yielded a positive result when retested with the RT-tHDA assay (Table 3). Remarkably, one sample from an YFV case confirmed only serologically, i.e., previously considered RT-PCR negative, showed a positive result by RT-tHDA, which further supports the assessment of the sensitivity of the assay in a clinical context. Every sample for which YFV infection had been previously excluded produced a matching negative result in the retrospective RT-tHDA test (Table 3).
TABLE 3.
YFV genome detection by RT-tHDA in clinical samples from confirmed YFV natural infections
| Sample | RT-tHDA result | Previous RT-PCR result in reference laboratora | Reference molecular protocola | Remarks |
|---|---|---|---|---|
| A | + | + | In-house protocol (M. C. Méndez et al., submitted) | |
| B | + | + | ||
| C | + | + | ||
| D | + | + | YFV-specific RT-PCR according to Weidmann et al. (22) | |
| E | + | + | ||
| F | + | + | ||
| G | + | + | ||
| H | + | + | ||
| I | − | − | YFV-specific RT-PCR according to Heraud et al. (11) | YFV infection excluded by molecular and serological methods |
| J | − | − | ||
| K | − | − | ||
| L | − | − | YFV-specific RT-PCR according to Heraud et al. (11) | YFV infection confirmed by serological detection of YFV-specific IgMb |
| M | + | − |
The molecular methods used were different from those described in this article.
YFV IgM ELISA (protocol of CDC, Atlanta, GA) and YF seroneutralization assay.
DISCUSSION
In countries of YFV endemicity, laboratory diagnosis for YFV infection is most often performed using serological assays (16). This methodology is effective after the viremic phase but has definite disadvantages for infection detection. Serology is not diagnostic early after the infection and requires two samples to be taken at least 2 weeks apart to evidence seroconversion or a 4-fold increase in the IgG titer. Most importantly, serological cross-reactions constitute major obstacles in achieving confirmed diagnoses or reliable serosurveys in areas of YFV endemicity where other flaviviruses circulate (e.g., dengue, Zika, St. Louis encephalitis, or West Nile viruses) (12). Seroneutralization is considered the most specific serological technique, but the assays are laborious, time-consuming, and confined to laboratories with appropriate facilities and expertise.
Direct detection of the virus, by cell culture, amplification of genome sequences, or antigen immunodetection, is the procedure of choice for confirmation of YFV infections. In most areas of Africa and South America, however, these methods are not generally operational for first-line diagnosis and surveillance due to the need of specialized equipment and skilled technicians. Thus, once again, testing is limited to a small number of central reference laboratories. All this hampers the confirmation of YF cases, early discovery of outbreaks, and implementation of surveillance programs based on detecting the virus in human and mosquito populations.
In a recent YF EQA exercise, we observed nonspecificity and nonsensitivity in the performance of some of the molecular protocols in routine use by YF diagnosis laboratories. Most worrisome were the inability to detect some strains of the YF virus and cross-reactivity with DENV (8). Strain nondetection has been pointed out also by others with regard to different YFV genome amplification assays (15, 22). Cross-reactivities with DENV can lead to misdiagnosis (1), inasmuch as dengue fever is present in most areas of YF endemicity, especially in Africa, where DENV infections are largely underrecognized.
On this premise, we have developed a new procedure for the sensitive and specific detection of YFV wild-type and vaccine strains in clinical samples using a real-time RT-qPCR approach. This provides a useful YFV molecular diagnosis tool for reference laboratories and fills in the functional gaps left by other protocols. Real-time PCR technology, however, remains outside the reach of the vast majority of laboratories in countries where YFV is endemic, where there is a clear need for dependable YFV molecular diagnostics that are instrument-free and easy to use and interpret. With this aim, we have adapted our YF RT-qPCR into an straightforward isothermal amplification protocol relying on the HDA technology.
Unique among isothermal approaches, HDA is elegantly simple and closely resembles traditional PCR. However, by using a DNA helicase instead of heat to denature DNA and generate single-stranded templates, amplification is accomplished under a simple, noncycling temperature regimen. Different strategies have been explored for amplicon detection downstream of HDA. Probe-based detection allows either real-time monitoring or endpoint analysis in a lateral-flow system. We favored the latter in our YF RT-tHDA assay, as LFS amplicon detection does not require any special instrumentation or complex postamplification analysis. Indeed, amplification products are identified on test strips with the naked eye, and results are easy to interpret. In addition, the RT-tHDA protocol was standardized to a reduced volume of commercially available reagents to decrease the cost per test.
Even at the risk of lower sensitivity, we chose a one-step format in the RT-qPCR and RT-tHDA YFV assays for the sake of workflow simplicity and to minimize the threat of sample cross-contamination. In both cases, however, the sensitivity observed was optimal for diagnostic purposes. The YF real-time RT-qPCR assay demonstrated broad-spectrum detection of YFV strains and an excellent profile of sensitivity and specificity that should make it a valuable tool for reference laboratories or those with real-time amplification capability. In our hands, this assay achieves excellent YFV diagnostic performance on clinical samples and notably facilitates detection of YFV in urine, where the viral load is very low.
The RT-tHDA assay is based on the same primer-probe set and annealing temperature as the RT-qPCR. It showed the same ability to detect different YFV strains, as demonstrated in the analysis of the EQA test set and in clinical samples of South American and African origin. The primers and probe were designed with this aim and contain degenerated positions to enable the detection of all strains with known 5′-NC sequence as of the writing of this article. Like the RT-qPCR, the RT-tHDA was also specific for YFV against other flaviviruses. At 1.18 × 104 RNA copies/ml (180.6 RNA copies/reaction), the detection threshold of the YF RT-tHDA is higher by approximately 1.1 log10 but still compares favorably with those of recent methods with good reported sensitivities in clinical applications (15). The expected level of viremia titers is ≥2 log10 PFU/ml or about 1 × 105 to 1 × 106 GE/ml (4, 10) in cases of yellow fever adverse events after vaccination, and titers far in excess of these are observed in wild-type YFV infections (3, 14, 19). The sensitivity threshold of the RT-tHDA assay is thus suitable for the detection of the YFV genome in serum samples during the viremic phase both in natural infections and in yellow fever adverse events after vaccination.
The YF RT-tHDA assay described was tested on well-characterized samples from YF-17D vaccinees, spiked samples, and infected culture supernatants, with satisfactory results. Likewise, a preliminary evaluation of the RT-tHDA assay was performed using clinical samples from natural YFV infections of different geographical origins (South America and Africa) provided by reference laboratories and the results were consistent with those obtained in parallel by YF RT-PCR diagnostic assays in current use.
Remarkably, the new assay confirmed the initial PCR-negative result for a YF patient confirmed by IgM serology but reversed the initial PCR-negative result for another serologically confirmed infection. In the former case, the high titer (1:640) of neutralizing antibodies pointed to a later stage in the course of the disease congruent with virus clearance. In the latter sample, by contrast, the lower neutralization titer (1:160) appears compatible with the time window when the presence of viremia may overlap that of measurable antibodies. The positive RT-tHDA result probably owes also to improved sensitivity over the earlier molecular diagnostics (11), as demonstrated by our interprotocol comparisons (see Table S1 in the supplemental material).
Moreover, we have been able to detect by this assay the presence of low levels of YF-17D genome in the urine of healthy YF vaccinees, which underscores the sensitivity achieved by the RT-tHDA assay on biological samples. Lastly, good interlaboratory reproducibility of the YF RT-tHDA was observed in the three participating reference laboratories using a validated panel of samples, with the expected detection of positive samples and negative controls in correlation with the YFV genome load and the estimated threshold of detection of the assay. Full corroboration of the diagnostic usefulness of the new assay requires wider testing in patient cohorts screened by reference methods and support by clinical data and point-of-care experience.
When the new RT-qPCR and RT-tHDA assays were compared with the methodologies applied during a previous YFV molecular diagnostics EQA analysis, both tests demonstrated reliable detection of the positive samples in the EQA sample set regardless of the YFV strain and excluded those samples containing other flaviviruses. The YF RT-qPCR proved better than most protocols in the EQA, and the RT-tHDA performed on a par with other real-time and RT-nested PCR initial protocols. Direct method-with-method comparison is not straightforward, however, since the performance achieved by other methods in the course of the EQA depended also on the laboratory carrying out the assay (8).
There is a dearth of YFV diagnostic capability, as even now molecular testing is usually performed only in a few reference laboratories, and some countries depend on international reference centers for case confirmation. Providing robust YF molecular testing nearer the point of care would benefit patients through faster and more reliable diagnosis. Decentralized routine diagnostics would in turn provide physicians with a critical tool for local disease surveillance and response. The ability to activate monitoring programs on detection of initial cases and screen at-risk communities for subclinical yet viremic YF infection would foster early warning and timely control of YF outbreaks (20). In this work, we have developed an isothermal amplification protocol for the detection of the YFV genome, the first one thus far. It is our hope that this reliable, sensitive, and specific assay will provide a useful alternative to current technologies, one that is more readily applicable to YFV detection in areas of endemicity.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Heidenreich and A. Ihlenfeld for technical assistance and J. E. Mejía for critically reviewing the manuscript.
This work was funded by the Federal Ministry of Education and Research (BMBF) under the research program for civil security of the German Federal Government, as part of the high-tech strategy for potential release-oriented biothreat emergency diagnostics (P.R.O.B.E.).
Footnotes
Published ahead of print 10 October 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Amarasinghe A, Kuritsk JN, Letson GW, Margolis HS. 2011. Dengue virus infection in Africa. Emerg. Infect. Dis. 17:1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae HG, Nitsche A, Teichmann A, Biel SS, Niedrig M. 2003. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J. Virol. Methods 110:185–191 [DOI] [PubMed] [Google Scholar]
- 3. Bae HG, et al. 2005. Analysis of two imported cases of yellow fever infection from Ivory Coast and The Gambia to Germany and Belgium. J. Clin. Virol. 33:274–280 [DOI] [PubMed] [Google Scholar]
- 4. Barban V, et al. 2007. High stability of yellow fever 17D-204 vaccine: a 12-year restrospective analysis of large-scale production. Vaccine 25:2941–2950 [DOI] [PubMed] [Google Scholar]
- 5. Craw P, Balachandran W. 2012. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip 12:2469–2486 [DOI] [PubMed] [Google Scholar]
- 6. Domingo C, Patel P, Linke S, Achazi K, Niedrig M. 2011. Molecular diagnosis of flaviviruses. Future Virol. 6:1059–1074 [Google Scholar]
- 7. Domingo C, et al. 2011. Detection of yellow fever 17D genome in urine. J. Clin. Microbiol. 49:760–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Domingo C, et al. 2012. First international external quality assessment study on molecular and serological methods for yellow fever diagnosis. PLoS One 7:e36291 doi:10.1371/journal.pone.0036291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gardner CL, Ryman KD. 2010. Yellow fever: a reemerging threat. Clin. Lab. Med. 30:237–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gershman MD, et al. 2012. Viscerotropic disease: case definition and guidelines for collection, analysis, and presentation of immunization safety data. Vaccine 30:5038–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heraud JM, et al. 1999. First case of yellow fever in French Guiana since 1902. Emerg. Infect. Dis. 5:429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Houghton-Triviño N, Montaña D, Castellanos J. 2008. Dengue-yellow fever sera cross-reactivity; challenges for diagnosis. Rev. Salud Publica (Bogotá) 10:299–307 [DOI] [PubMed] [Google Scholar]
- 13. Kim HJ, et al. 2011. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J. Clin. Virol. 50:26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macnamara FN. 1957. A clinico-pathological study of yellow fever in Nigeria. W. Afr. Med. J. 6:137–146 [PubMed] [Google Scholar]
- 15. Nunes MR, et al. 2011. Evaluation of two molecular methods for the detection of yellow fever virus genome. J. Virol. Methods 174:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan American Health Organization (PAHO) 2005. Control of yellow fever: field guide. PAHO scientific and technical publication no. 603. Pan American Health Organization, Washington, DC: http://www.paho.org/english/ad/fch/im/fieldguide_yellowfever.pdf [Google Scholar]
- 17. Posthuma-Trumpie GA, Korf J, van Amerongen A. 2009. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 393:569–582 [DOI] [PubMed] [Google Scholar]
- 18. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 19. Teichmann D, et al. 1999. A haemorrhagic fever from the Cote d'Ivoire. Lancet 354:1608. [DOI] [PubMed] [Google Scholar]
- 20. Vainio J, Cutts F. 1998. Yellow fever. Division of Emerging and Other Communicable Diseases Surveillance and Control, WHO, Geneva, Switzerland [Google Scholar]
- 21. Vincent M, Xu Y, Kong H. 2004. Helicase-dependent isothermal DNA amplification. EMBO Rep. 5:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weidmann M, et al. 2010. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J. Clin. Virol. 48:187–192 [DOI] [PubMed] [Google Scholar]
- 23. World Health Organization 2010. Yellow fever fact sheet. Wkly. Epidemiol. Rec. 85:33–36 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.