Abstract
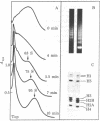
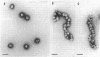
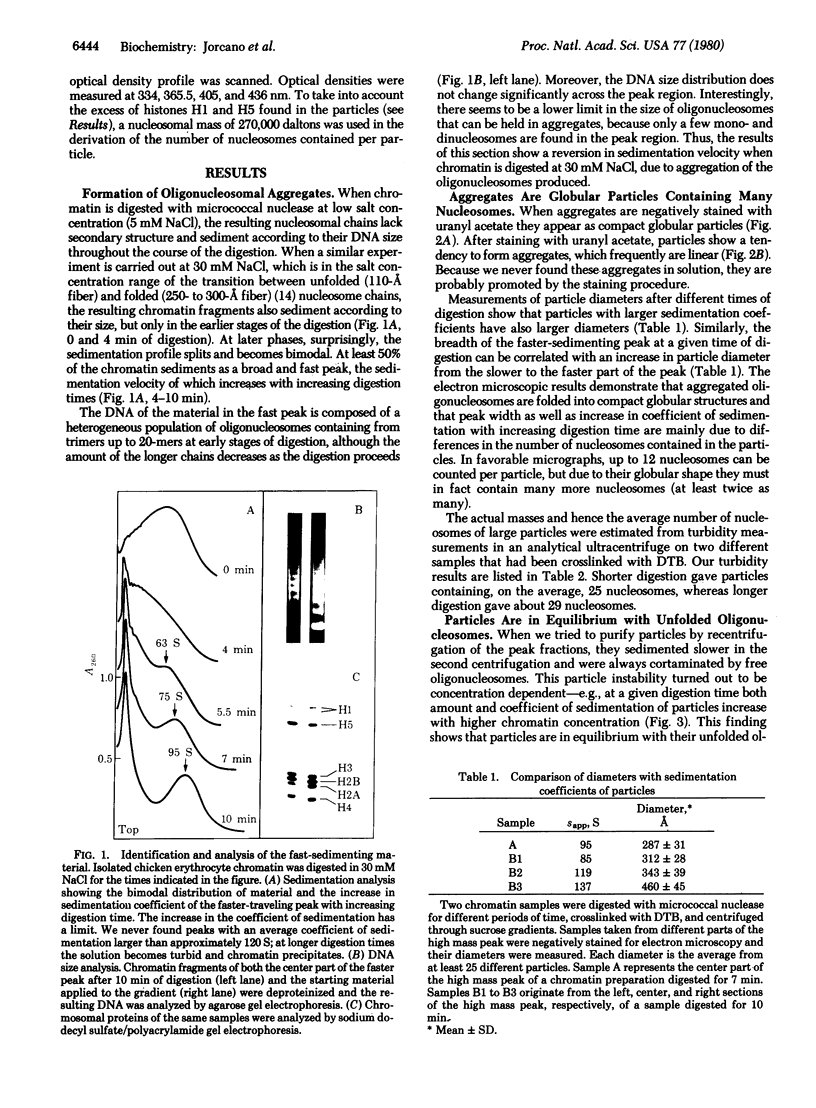
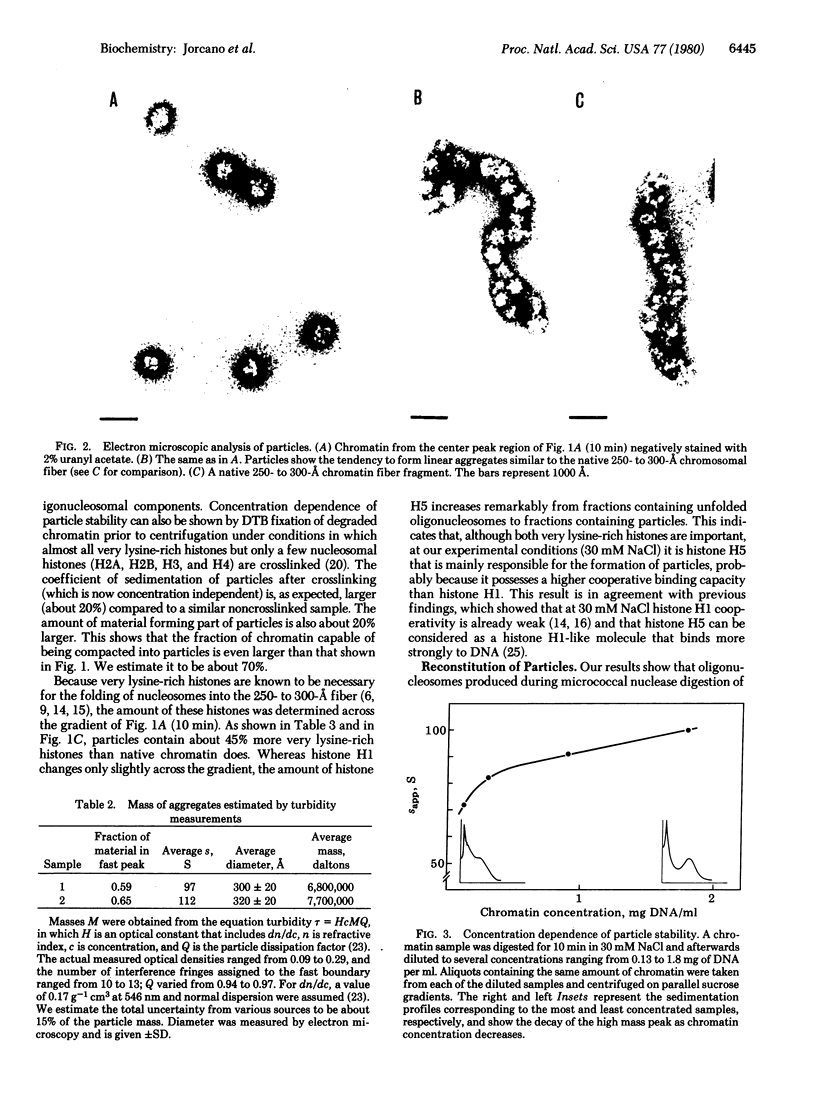
Chicken erythrocyte oligonucleosomes (trimers to about 20-mers) are able to interact with each other through the very lysine-rich histones (H1 and H5) and form heterogeneous globular particles with a mean diameter of about 300 A. These particles assemble spontaneously during micrococcal nuclease digestion of chromatin in the presence of 30 mM NaCl and contain approximately 25 nucleosomes. They are sensitive to ionic strength and unfold at lower salt concentrations but can be reconstituted by restoring the initial salt concentration. Even at 30 mM NaCl, the particles remain dynamic structures, being in equilibrium with their oligonucleosomal components as revealed by the fact that particle stability depends on the concentration of oligonucleosomes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkowitz S. A., Day L. A. Turbidity measurements in an analytical ultracentrifuge. Determinations of mass per length for filamentous viruses fd, Xf, and Pf3. Biochemistry. 1980 Jun 10;19(12):2696–2702. doi: 10.1021/bi00553a025. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I., Pardon J. F. Light scattering measurements supporting helical structures for chromatin in solution. Nucleic Acids Res. 1978 May;5(5):1571–1580. doi: 10.1093/nar/5.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G. Chromosome fibers studied by a spreading technique. Chromosoma. 1966;20(2):221–233. doi: 10.1007/BF00335209. [DOI] [PubMed] [Google Scholar]
- Hozier J., Renz M., Nehls P. The chromosome fiber: evidence for an ordered superstructure of nucleosomes. Chromosoma. 1977 Jul 18;62(4):301–317. doi: 10.1007/BF00327030. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Jorcano J. L., Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979 Mar 6;18(5):768–774. doi: 10.1021/bi00572a005. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Manamshjan T. A., Polyakov V. Y., Fais D., Chentsov J. S. Levels of granular organization of chromatin fibres. FEBS Lett. 1976 Sep 1;67(3):323–327. doi: 10.1016/0014-5793(76)80557-1. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeStourgeon W. M., Beyer A. L., Christensen M. E., Walker B. W., Poupore S. M., Daniels L. P. The packaging proteins of core hnRNP particles and the maintenance of proliferative cell states. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):885–898. doi: 10.1101/sqb.1978.042.01.090. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G. F., Renz M. Native and reconstituted chromosome fiber fragments. Chromosoma. 1979 Nov;75(2):177–184. doi: 10.1007/BF00292206. [DOI] [PubMed] [Google Scholar]
- Muyldermans S., Lasters I., Wyns L. Histone H1 can be removed selectively from chicken erythrocyte chromatin at near physiological conditions. Nucleic Acids Res. 1980 Feb 25;8(4):731–739. [PMC free article] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Renz M., Day L. A. Transition from noncooperative to cooperative and selective binding of histone H1 to DNA. Biochemistry. 1976 Jul 27;15(15):3220–3228. doi: 10.1021/bi00660a010. [DOI] [PubMed] [Google Scholar]
- Renz M. Heterogeneity of the chromosome fiber. Nucleic Acids Res. 1979 Jun 25;6(8):2761–2767. doi: 10.1093/nar/6.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau P., Bradbury E. M., Baldwin J. P. Higher-order structures of chromatin in solution. Eur J Biochem. 1979 Jul;97(2):593–602. doi: 10.1111/j.1432-1033.1979.tb13148.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Nedospasov S. A., Schmatchenko V. V., Bakayev V. V., Chumackov P. M., Georgiev G. P. Compact form of SV40 viral minichromosome is resistant to nuclease: possible implications for chromatin structure. Nucleic Acids Res. 1977 Oct;4(10):3303–3325. doi: 10.1093/nar/4.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978 Apr;5(4):1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Zentgraf H., Müller U., Franke W. W. Supranucleosomal organization of sea urchin sperm chromatin in regularly arranged 40 to 50 nm large granular subunits. Eur J Cell Biol. 1980 Feb;20(3):254–264. [PubMed] [Google Scholar]