Abstract
Dried blood spots (DBS) are useful for molecular assays but are prone to false positives from cross-contamination. In our malaria DBS assay, cross-contamination was encountered despite cleaning techniques suitable for HIV-1. We therefore developed a contact-free laser cutting system that effectively eliminated cross-contamination during DBS processing.
TEXT
Dried blood spots (DBS) are easily collected, stored, and transported and are suitable for many diagnostic tests (2, 3, 5, 7, 8, 12). However, cross-contamination of molecular DBS assays is a potential problem since the sample-containing region is usually physically touched by manual or automated punching devices. Contaminating DNA and/or RNA carried over from a positive sample can result in subsequent false-positive results.
The risk of cross-contamination is related to the concentration of the nucleic acid template and has been most well studied for HIV-1 DBS. False positives are infrequent at low viral loads but increase (2.5 to 3.0%) at higher levels (5 × 103 pol copies/punch) (10). Cleaning between samples by repeatedly punching negative-control paper can eliminate most false positives, whereas bleach cleaning corrodes the cutting edge and increases the false-positive rate (10). False positives are also more common for manual versus automated disc punching (4). Collectively, the literature suggests that disc punching with cleaning precautions is suitable for most HIV-1 DBS.
Assays for pathogens that reach higher template concentrations than HIV-1 may be at greater risk for significant nucleic acid carryover. For instance, false-positive results for beak and feather disease virus (BFDV) were propagated into 13 samples processed after a BFDV-positive sample (1). BVDF contamination was removed by bleach, water, and ethanol washes, followed by air drying, but such steps are laborious and poorly suited for high-throughput labs. Although a negative-control sample is usually included in each test run to control for gross contamination of common reagents, negative controls are not interspersed between every specimen. Without definitive ways to eliminate or detect cross-contamination in the routine workflow of clinical laboratories, samples with high pathogen loads processed using contact-dependent methods remain at risk for false positives from cross-contamination.
Here, we adapted a Plasmodium falciparum reverse transcription (RT)-PCR assay (11) to utilize TaqMan probe chemistry and a noncompetitive internal control (IC) RNA on liquid blood and DBS. Cultured parasites were diluted into whole blood as in reference 11 at high, medium, and low parasitemias (4 × 105, 8 × 103, and 80 parasites/ml, respectively). Liquid blood samples (50 μl) were stored in NucliSens lysis buffer (bioMérieux, Marcy l'Etoile, France) (11). Total nucleic acids were extracted on an Abbott m2000sp (Abbott Molecular, Niles, IL). The noncompetitive IC RNA was synthesized in vitro using T7 RNA polymerase from an AlwNI-digested pCRII plasmid (Invitrogen, Carlsbad, CA) containing a 94-bp fragment of the Drosophila white gene (5′-CAAGCAGCCATGCAAATGTTAGCTAGTGCATCCAGTGCATGCAGGGCCGTCCTACCAACTACAATCGAGAGAACCAAGGGGAAGTGACATAGCA-3′) and was added upon extraction. Eluates were amplified by multiplex RT-PCR using the AgPath one-step RT-PCR kit (Ambion, Grand Island, NY), P. falciparum 18S rRNA primers (9, 11), a BHQplus-labeled P. falciparum probe [5′-(6-carboxyfluorescein {FAM})-ATTTATTCAGTAATCAAATTAGGAT-3′-(black hole quencher 1) (courtesy of Abbott Molecular and Biosearch Technologies, Novato, CA)], IC primers (forward, CAAGCAGCCATGCAAATGTT; reverse, TGCTATGTCACTTCCCCTTGGTTCTCT), and an IC probe [5′-(VIC)ATTGTAGTTGGTAGGAC-(minor groove binder/nonfluorescent quencher) (Applied Biosystems, Carlsbad, CA)]. Assay performance was comparable to that of the first-generation assay (11), with the following values: analytical sensitivity, 20 parasites/ml; target recovery, 107% (95% confidence interval [CI], 60 to 154%); linear correlation (r2 = 0.9911, slope = 1.03); minimal bias of +0.143 log10 RNA copies/ml (95% CI, −0.379 to +0.367 log10 RNA copies/ml); and no false-positive or -negative results. RNA copies were converted to parasites/ml using an m2000-specific conversion factor (3.56 mean log10 18S rRNA copies/parasite; 95% CI, 3.51 to 3.61 log10) determined by testing P. falciparum samples and RNA standards as in reference 11. Testing of paired samples on the first- and second-generation assays indicated that platform-specific conversion factors were appropriate for both assays (data not shown). Notably, there was no cross-contamination of negative liquid blood samples. To date, the second-generation assay for whole blood was used to support a clinical malaria trial with >1,000 RT-PCR samples (our unpublished data).
We next adapted the assay to DBS. Fifty-microliter aliquots were spotted onto Whatman 903 cards (GE/Whatman, Kent, United Kingdom) that were dried for 4 h and stored at −80°C with desiccant (Fisher, Pittsburgh, PA). DBS samples were processed by punching the entire spot into 2 ml lysis buffer using a manual puncher; the puncher was cleaned as described previously (10). Lysis buffer containing the excised DBS was agitated for 2 h at room temperature and then tested as for liquid samples. With this method, we immediately observed numerous false positives in malaria-negative DBS processed after high-parasitemia samples.
We hypothesized that preanalytical contamination can be prevented by eliminating contact with the blood-containing surfaces of the DBS. Therefore, we developed a touchless DBS laser cutting system. DBS cards were positioned above a collection tube without making contact with blood-containing portions of the card. A 10W laser cutter (Universal Laser Systems, Scottsdale, AZ) excised an 8.89-mm disc (corresponding to 27.4 μl of blood), which fell into the tube below; this was the largest disc that would enter the 4-ml tubes (Simport, Beloeil, Canada) without manual manipulation.
Cross-contamination was evaluated by repeatedly testing a malaria-positive sample followed by a malaria-negative sample. Using DBS cards containing five spots of the same parasitemia, a high-, medium-, or low-parasitemia sample was processed, followed by a negative sample from a separate card. Positive/negative paired samples are shown in Table 1. Carryover was not detected in laser-cut DBS but was detected following 7/8 high- and 2/6 medium-parasitemia samples processed by punching (Table 1 and Fig. 1). There were no false positives following low-parasitemia samples processed by either method. Only 2/9 false positives generated values ≥20 parasites/ml (limit of quantification for this assay). However, almost all false positives were due to carryover of >102 contaminating template copies/sample, a concerning level of template easily detected by most molecular assays. We had not previously encountered such carryover during several years of liquid whole-blood testing in our laboratory (our unpublished data). However, during the posttreatment periods of infection, we occasionally detect 18S rRNA in the low-positive range (<20 parasites/ml) (11). Subjects with low-positive results posttreatment eventually drop to undetectable levels. Thus, while not quantitatively reportable, low-positive results can sometimes be useful for monitoring the rise and fall of parasitemia in exposed persons. The frequent presence of false positives from carryover would invalidate such evaluations.
TABLE 1.
P. falciparum cross-contamination evaluationa
| Sample | Values for each method |
|||||
|---|---|---|---|---|---|---|
| Conventional punch |
Laser cut |
|||||
| CT | Log10 copies (12.0-mm spot) | Parasites/ml | CT | Log10 copies (8.9-mm spot) | Parasites/ml | |
| +++ | 22.49 | 6.76 | 65,173 | 21.85 | 6.95 | 184,330 |
| − | 32.34 | 3.83 | 77 | ND | ND | ND |
| +++ | 21.79 | 6.96 | 105,249 | 21.75 | 6.98 | 197,394 |
| − | 37.81 | 2.20 | 2 | ND | ND | ND |
| +++ | 21.39 | 7.08 | 138,409 | 21.47 | 7.06 | 239,108 |
| − | 37.09 | 2.41 | 3 | ND | ND | ND |
| +++ | 19.98 | 6.96 | 103,338 | 20.10 | 6.92 | 173,978 |
| − | 41.99 | 0.48 | <1 | ND | ND | ND |
| +++ | 19.30 | 7.15 | 163,121 | 20.79 | 6.72 | 109,478 |
| − | 34.55 | 2.71 | 6 | ND | ND | ND |
| +++ | 19.59 | 7.07 | 134,265 | 20.82 | 6.71 | 107,296 |
| − | ND | ND | ND | ND | ND | ND |
| +++ | 19.33 | 7.15 | 159,869 | 20.73 | 6.74 | 113,978 |
| − | 32.73 | 3.24 | 20 | ND | ND | ND |
| +++ | 19.57 | 7.08 | 136,080 | 20.03 | 6.94 | 182,349 |
| − | 36.02 | 2.28 | 2 | ND | ND | ND |
| ++ | 27.18 | 5.36 | 2,627 | 27.5 | 5.27 | 3,850 |
| − | 35.45 | 2.90 | 9 | ND | ND | ND |
| ++ | 26.84 | 5.46 | 3,315 | 27.69 | 5.21 | 3,381 |
| − | 43.66 | ND | <1 | ND | ND | ND |
| ++ | 27.00 | 5.41 | 2,971 | 26.95 | 5.43 | 5,611 |
| − | ND | ND | 0 | ND | ND | ND |
| ++ | 25.36 | 5.39 | 2,791 | 25.89 | 5.23 | 3,568 |
| − | ND | ND | 0 | ND | ND | ND |
| ++ | 24.64 | 5.60 | 4,525 | 26.53 | 5.05 | 2,322 |
| − | ND | ND | 0 | ND | ND | ND |
| ++ | 24.77 | 5.56 | 4,147 | 26.82 | 4.96 | 1,911 |
| − | ND | ND | 0 | ND | ND | ND |
Shading denotes cross-contaminated samples based on detection of any 18S rRNA template in the sample. Data were from two experiments using different lots of reagent, a switch which accounts for the shift in threshold cycle (CT) values; assays were recalibrated for each reagent lot. ND, not detected; +++, 4 × 107 parasites/ml; ++, 8 × 103 parasites/ml; −, negative controls.
Fig 1.
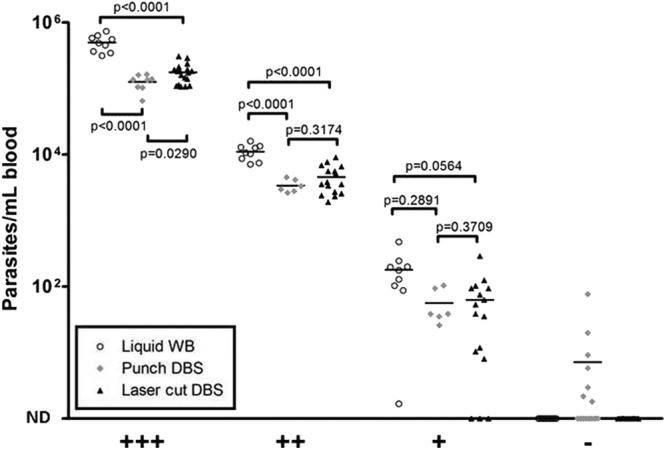
Comparison of liquid samples and punched and laser-cut DBS samples. +++, 4 × 107 parasites/ml; ++, 8 × 103 parasites/ml; +, 80 parasites/ml; −, negative controls.
Judging by expected template concentrations for malaria and HIV-1, it is apparent that the risk of cross-contamination differs markedly for these pathogens. Whereas HIV-1 viral loads usually peak at around 1 × 106 RNA copies/ml during acute infection and remain lower thereafter (13), blood from P. falciparum-infected individuals can exceed 1% parasitemia (∼5 × 107 parasites/ml), corresponding to ∼1 × 108 A-type 18S rRNA genomic (DNA) copies/ml and ∼1.75 × 1011 actual 18S rRNA copies/ml (based on 5 × 106 red blood cells [RBC]/μl). For example, an 8.89-mm disc from a 1% parasitemia card contains 2.75 × 106 18S rRNA genomic (DNA) copies and 4.75 × 109 18S rRNA copies/punch, values that are 550-fold higher for DNA and 950,000-fold higher for RNA than for the most concentrated HIV-1 samples reported in reference 10. Even at the limit of detection for thick blood smears (e.g., ∼5 × 103 parasites/ml), the template concentration (∼4.8 × 105 18S rRNA copies/disc) exceeds the limits tested for HIV-1 (10). Thus, malaria DBS require more-stringent approaches to avoid cross-contamination.
Beyond carryover, we assessed template recovery across methods and parasitemias. Recovery did not differ between punched or laser-cut DBS but was moderately reduced (∼0.5 log10 copies/ml) for all DBS compared to that for liquid whole-blood samples (Fig. 1). The mean differences between the liquid blood and punched and laser-cut DBS were −0.60 and −0.45 log10 parasites/ml for high positives and −0.51 and −0.38 log10 parasites/ml for medium positives, respectively. There were no significant differences between low-positive liquid blood and DBS; P values were calculated using unpaired t tests. Similar losses were reported for HIV-1 (6) and may reflect degradation of the template or an inability to elute template from the DBS. Several laser-cut low-positive samples were not detected by RT-PCR, whereas all punched samples were detected (Fig. 1). When two low-positive laser-cut discs were processed in a single tube (equivalent to 54.9 μl whole blood), 13/13 samples were positive (mean parasite density, 117 parasites/ml). Since detection of low positives was restored by testing two discs/sample, the false-negative findings for low-positive samples were likely due to Poisson statistics and not laser cutting.
We briefly evaluated laser-cut HIV-1 DBS. HIV-1-containing plasma was diluted into whole blood, spotted onto DBS cards (50 μl/spot), processed by manual punching or laser cutting, and tested using the Abbott RealTime HIV-1 plasma assay. Quantitative HIV-1 results were equivalent for punched and laser-cut samples (data not shown). Cross-contamination was not studied for HIV-1 DBS.
In summary, we developed a second-generation RT-PCR assay for P. falciparum that was suitable for liquid blood and DBS. Laser cutting of DBS eliminated carryover during processing. This method also reduces repetitive hand movements required during manual punching. Contamination-free laser cutting may expand the use of DBS in molecular diagnostics.
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health (1K08AI097238 to S.C.M.), an AIDS Clinical Trials Group Virology Specialty Laboratory grant (AI-38858), and the University of Washington Center for AIDS Research (CFAR), which is an NIH-funded program (P30 AI027757) supported by the following NIH Institutes and Centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NIA.
Abbott Molecular, Inc., provided the research instrument and the BHQplus-labeled probe for this assay.
Production of the BHQplus probe was with the permission of Biosearch Technologies.
Abbott provided travel for S.C.M. for a project review meeting.
S.C.M. filed a provisional patent on the use of laser cutting for processing DBS.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Bonne N, Clark P, Shearer P, Raidal S. 2008. Elimination of false-positive polymerase chain reaction results resulting from hole punch carryover contamination. J. Vet. Diagn. Invest. 20:60–63 [DOI] [PubMed] [Google Scholar]
- 2. Cassol S, et al. 1997. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J. Clin. Microbiol. 35:2795–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desbois D, Roque-Afonso AM, Lebraud P, Dussaix E. 2009. Use of dried serum spots for serological and molecular detection of hepatitis A virus. J. Clin. Microbiol. 47:1536–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Driver GA, Patton JC, Moloi J, Stevens WS, Sherman GG. 2007. Low risk of contamination with automated and manual excision of dried blood spots for HIV DNA PCR testing in the routine laboratory. J. Virol. Methods 146:397–400 [DOI] [PubMed] [Google Scholar]
- 5. Elbin CS, et al. 2011. The effect of preparation, storage and shipping of dried blood spots on the activity of five lysosomal enzymes. Clin. Chim. Acta 412:1207–1212 [DOI] [PubMed] [Google Scholar]
- 6. Garrido C, et al. 2009. Correlation between human immunodeficiency virus type 1 (HIV-1) RNA measurements obtained with dried blood spots and those obtained with plasma by use of Nuclisens EasyQ HIV-1 and Abbott RealTime HIV load tests. J. Clin. Microbiol. 47:1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jorgez CJ, Bischoff FZ. 2008. Stability of placental RNA using dried maternal blood spots. Reprod. Biomed. Online 17:716–721 [DOI] [PubMed] [Google Scholar]
- 8. Leelawiwat W, et al. 2009. Dried blood spots for the diagnosis and quantitation of HIV-1: stability studies and evaluation of sensitivity and specificity for the diagnosis of infant HIV-1 infection in Thailand. J. Virol. Methods 155:109–117 [DOI] [PubMed] [Google Scholar]
- 9. Mangold KA, et al. 2005. Real-time PCR for detection and identification of Plasmodium spp. J. Clin. Microbiol. 43:2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitchell C, Kraft K, Peterson D, Frenkel L. 2010. Cross-contamination during processing of dried blood spots used for rapid diagnosis of HIV-1 infection of infants is rare and avoidable. J. Virol. Methods 163:489–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy SC, et al. 2012. Real-time quantitative reverse transcription PCR for monitoring of blood-stage Plasmodium falciparum infections in malaria human challenge trials. Am. J. Trop. Med. Hyg. 86:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pritsch M, et al. 2012. Stability of gametocyte-specific Pfs25-mRNA in dried blood spots on filter paper subjected to different storage conditions. Malar. J. 11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson BA, et al. 2003. Comparison of human immunodeficiency virus type 1 viral loads in Kenyan women, men, and infants during primary and early infection. J. Virol. 77:7120–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]


