Abstract
Pulsed-field gel electrophoresis (PFGE) was used to type 128 Streptococcus infantarius subsp. coli isolates from sea otters and mussels. Six SmaI PFGE groups were detected, with one predominant group representing 57% of the isolates collected over a wide geographic region. Several sea otter and mussel isolates were highly related, suggesting that an environmental infection source is possible.
TEXT
Streptococcus infantarius subsp. coli is a member of the Streptococcus bovis-equinus complex (SBEC), many of whose members are gastrointestinal tract commensals in several mammals and some of which are also associated with bacteremia, septicemia, and endocarditis in humans and animals, including pigeons, mink, and ruminants (3, 8, 10, 15).
S. infantarius subsp. coli is a significant problem in the northern sea otter (Enhydra lutris kenyoni) population along coastal Alaska, where infective endocarditis (IE) and/or septicemia due to this agent was the cause of death in approximately 30% of 613 carcasses collected and necropsied between 2004 and 2010 (V. Gill, personal communication). Further, death due to IE and/or septicemia with isolation of S. infantarius subsp. coli has also occurred in 4 of 281 southern sea otter (Enhydra lutris nereis) carcasses collected and necropsied along the central California coast between 2004 and 2008 (M. Miller, personal communication). S. infantarius subsp. coli in sea otters has not been examined previously.
The aims of the present study were to determine the genetic relatedness of S. infantarius subsp. coli isolates by using pulsed-field gel electrophoresis (PFGE), to evaluate wild mussels as a potential contaminated prey source and the route of transmission of this bacterium to sea otters, and to examine the temporal and spatial distributions of PFGE types. Cultivation and preliminary identification of isolates from sea otter tissues as SBEC members were performed at the Microbiology Laboratory of the University of California, Davis, William R. Pritchard Veterinary Medical Teaching Hospital, and confirmation of S. infantarius subsp. coli isolates was performed at the Centers for Disease Control and Prevention as previously described (2, 4, 5, 7).
Wild blue mussels (Mytilus spp.) that serve as sea otter prey and water quality sentinels were collected from sites within the northern and southern sea otter ranges and screened for S. infantarius subsp. coli in order to identify a potential source of infection (1). Thirty mussels collected at each site at each time point were shucked and collectively homogenized with an equal volume of 0.5× peptone water. Next, 20 100-μl aliquots of homogenate were plated on Edwards modified medium supplemented with colistin sulfate and oxolinic acid (EMCO) to select for Streptococcus spp. Colonies that grew on EMCO were further screened, and the Centers for Disease Control and Prevention definitively identified isolates as S. infantarius subsp. coli by using previously described methods (2, 4, 5, 7). Mussels sampled from Monterey Bay, CA (Laguna Creek, Moss Landing, and Carmel River), in March 2007 produced one S. infantarius subsp. coli isolate from Moss Landing. Mussels collected from Monterey Bay, CA (Scott Creek, Carmel River, and Elkhorn Slough), and Estero Bay, CA (Cayucos, Motel Point, and Morro Bay), in April 2007 yielded three isolates from Scott Creek and two from Cayucos, and the September 2008 sampling yielded no isolates. In Alaska, mussels collected from Ismailof Island, Kasitsna Bay, and Bear Cove in June 2007 and Homer, Ninilchik, Bishop Beach, and Homer Harbor in August 2007 provided no S. infantarius subsp. coli isolates.
PFGE of 128 S. infantarius subsp. coli isolates, 111 from 58 northern sea otters, 11 from 4 southern sea otters, and 6 from mussels, was performed. Agarose plugs were prepared, digested with SmaI restriction enzyme, and subjected to PFGE as previously described (14). Banding pattern analysis was performed using BioNumerics, version 4.6 (Applied Maths, Austin, TX), unweighted-pair group analysis with arithmetic means and Dice's coefficient (1% optimization and 2% position tolerance) (7, 9). Isolates with 80% or greater PFGE pattern similarity were considered closely related and constituted a PFGE group (9). Isolates with 90% or greater PFGE pattern similarity were considered identical (13).
Northern and southern sea otter and mussel S. infantarius subsp. coli isolates generated six major PFGE groups, with 57% of the isolates clustering in group I (Table 1). Unexpectedly, group I contained a mixture of northern and southern sea otter and mussel isolates. The high degree of relatedness among 128 S. infantarius subsp. coli isolates obtained from sea otters and their prey over 5 years was notable. This contrasts with prior studies that examined the PFGE types of SBEC isolates from human cases of IE, which demonstrated considerable diversity. For example, PFGE characterization of seven human S. bovis isolates collected from blood samples of endocarditis patients at a hospital over a 1-year period indicated that all were unrelated (8). The high degree of relatedness among S. infantarius subsp. coli isolates in this study suggests that there may be something unique about these bacteria related to their pathogenicity, their ability to survive in the marine environment, or the susceptibility of sea otters to infection. Alternatively, there could be common infection sources due to human activity or animals in the regions.
TABLE 1.
PFGE groups generated by S. infantarius subsp. coli isolates from northern sea otters, southern sea otters, and mussels
| Group | Total no. of isolates | % Relatednessa | No. of isolates from: |
||
|---|---|---|---|---|---|
| NSOb | SSOc | Mussels | |||
| I | 72 | 84.0 | 66 | 5 | 1 |
| II | 37 | 84.3 | 30 | 6 | 1 |
| III | 6 | 82.5 | 6 | 0 | 0 |
| IV | 4 | 94.1 | 3 | 0 | 1 |
| V | 3 | 100 | 0 | 0 | 3 |
| VI | 2 | 100 | 2 | 0 | 0 |
| None | 4 | 4 | 0 | 0 | |
| Total | 128 | 52.9 | 111 | 11 | 6 |
Percent relatedness was determined by BioNumerics unweighted-pair group analysis with arithmetic means and Dice's coefficient (1% optimization and 2% position tolerance).
NSO, northern sea otters.
SSO, southern sea otters.
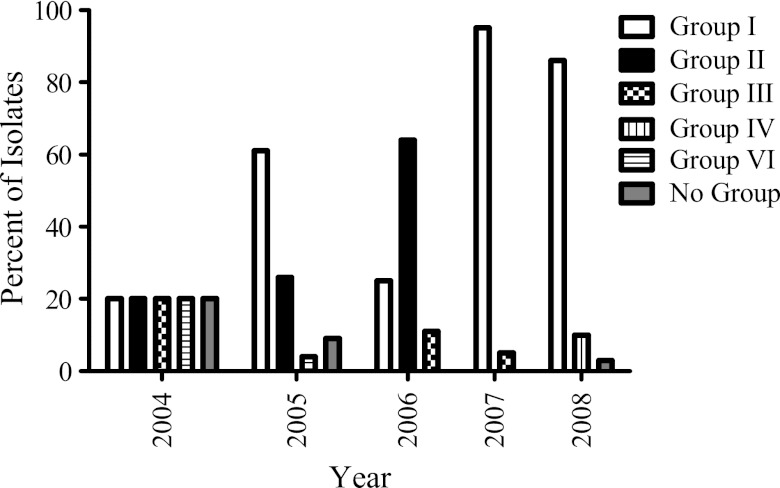
The temporal and spatial distributions of the PFGE groups produced by northern sea otters were investigated. Figure 1 details the yearly prevalence of each group, which fluctuated over time. Group I was generally the predominant strain and was the only group present each year. Additionally, the majority of isolates concentrated around Homer, AK, where there is both a larger human population and a more intensive sampling effort for sea otter carcass recovery. However, isolates were also obtained from more distant geographical regions, such as the Aleutian Islands, Glacier Bay National Park, Resurrection Bay, and Ninilchik. The most notable PFGE type was group IV, which represented isolates from carcasses collected in 2008 near Nordyke Island, on the west side of Cook Inlet. This was the only location where a specific PFGE type was isolated in the absence of all other types. The sources of S. infantarius subsp. coli in that area and whether isolates of this PFGE type are more or less virulent are unknown.
Fig 1.
Temporal distribution of PFGE groups generated by the 111 Alaskan northern sea otter S. infantarius subsp. coli isolates.
Surprisingly, both northern and southern sea otter S. infantarius subsp. coli isolates were grouped together (groups I and II) despite the large geographical separation of the two populations. A previous study describing S. bovis isolated from mink found that highly related strains could be present on different farms not sharing personnel or animals, and an analogous situation seems to be occurring with the two sea otter populations (10). This could indicate a lack of genetic diversity within S. infantarius subsp. coli, that only certain strains infect sea otters, or that a highly conserved strain is widely disseminated. It could also be a reflection of limits in the discriminatory power of PFGE.
The isolation of S. infantarius subsp. coli from mussels in coastal California waters indicates that the bacteria are present in, and can survive in, marine environments. Therefore, the sea otters could acquire the bacteria from contaminated water or prey such as mussels. Many marine organisms accumulate pathogens, but bivalves are especially adept at concentrating bacteria through filter feeding (12). In our study, an S. infantarius subsp. coli mussel isolate from Cayucos, CA, and isolates from a southern sea otter all fell into group I. Another S. infantarius subsp. coli isolate from mussels in Moss Landing, CA, was placed in group II along with isolates from three southern sea otters. These results indicate PFGE types are shared between the environment and southern sea otters and suggest that sea otters may be infected by environmental strains.
Sea otters that died because of S. infantarius subsp. coli IE and/or septicemia were generally septic, and isolates were obtained from multiple tissues. A total of 62 sea otters were sampled, and 30 northern and 2 southern sea otters had S. infantarius subsp. coli isolated from two or more anatomic locations. When isolates from different tissues within the same animal were compared, 69% of the sea otters had all of their isolates fall in the same PFGE group and 59% of the animals had all of their isolates produce identical PFGE patterns. Interestingly, 31% of sea otters were concurrently infected with multiple S. infantarius subsp. coli strains, on the basis of the identification of unrelated PFGE patterns across multiple isolates from the same animal. This is similar to Campylobacter jejuni, an environmentally transmitted bacterium that has been known to cause coinfections with multiple strains. One study found that 5 to 10% of humans were coinfected in sporadic Campylobacter infections, and another study determined that 50% of humans were infected by multiple strains in Campylobacter outbreaks (6, 11).
The S. infantarius subsp. coli isolates evaluated in this study exhibited a high degree of relatedness, with one PFGE type predominating. Notably, several sea otters were infected by multiple strains, and mussel sampling revealed that the bacteria are present in coastal environments. Additional research is needed to determine transmission routes, as well as for identification and remediation of specific sources of S. infantarius subsp. coli that could lead to a reduction of the exposure of sea otters and humans utilizing the coastal environment. Further exploration of the pathogenic properties of S. infantarius subsp. coli will elucidate why certain PFGE types predominate and will clarify complex environmental, host, and pathogen interactions.
ACKNOWLEDGMENTS
This project was supported by the California Department of Fish and Game's Oil Spill Response Trust Fund through the Oiled Wildlife Care Network at the Wildlife Health Center, School of Veterinary Medicine, University of California, Davis. The U.S. Fish and Wildlife Region 7 Marine Mammals program funded the health and disease monitoring program in Alaska that provided the samples. S. infantarius subsp. coli isolates from California sea otters were obtained through collaborations with the California Department of Fish and Game, Office of Spill Prevention and Response, and other members of the Southern Sea Otter Alliance.
Thank you to all of the Alaska and California Marine Mammal Stranding Network members who collected sea otter carcasses for testing and helped with necropsy and sample collection. Thanks also to Dana Jenski for all of her hard work in the necropsy laboratory and for tirelessly shipping samples to the University of California, Davis.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Arkush KD, et al. 2003. Molecular and bioassay-based detection of Toxoplasma gondii oocyst uptake by mussels (Mytilus galloprovincialis). Int. J. Parasitol. 33:1087–1097 [DOI] [PubMed] [Google Scholar]
- 2. Carvalho MDG, et al. 2004. Characterization of three new enterococcal species, Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3, isolated from human clinical specimens. J. Clin. Microbiol. 42:1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Herdt P, et al. 1995. Intracellular survival and multiplication of virulent and less virulent strains of Streptococcus bovis in pigeon macrophages. Vet. Microbiol. 45:157–169 [DOI] [PubMed] [Google Scholar]
- 4. Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Facklam R, Elliot JA. 1995. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8:479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frost JA, Gillespie IA, O'Brien SJ. 2002. Public health implications of Campylobacter outbreaks in England and Wales, 1995-9: epidemiological and microbiological investigations. Epidemiol. Infect. 128:111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jang SS, et al. 2005. Conventional and molecular identification of Streptococcus infantarius ss coli isolated from valvular endocarditis and sepsis in stranded northern and southern sea otters, p 218–219 In 36th Annual Conference of the International Association for Aquatic Animal Medicine, Seward, AK [Google Scholar]
- 8. Mühlemann K, Graf S, Tauber MG. 1999. Streptococcus bovis clone causing two episodes of endocarditis 8 years apart. J. Clin. Microbiol. 37:862–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noguchi N, et al. 2007. Antimicrobial susceptibilities and distribution of resistance genes for β-lactams and macrolides in Streptococcus pneumoniae isolated between 2002 and 2004 in Tokyo. Int. J. Antimicrob. Agents 29:26–33 [DOI] [PubMed] [Google Scholar]
- 10. Pedersen K, Jorgensen JC, Dietz H, Andersen T. 2003. Verrucous endocarditis associated with Streptococcus bovis in mink (Mustela vison). Vet. Rec. 153:264–268 [DOI] [PubMed] [Google Scholar]
- 11. Richardson JF, et al. 2001. Coinfection with Campylobacter species: an epidemiological problem? J. Appl. Microbiol. 91:206–211 [DOI] [PubMed] [Google Scholar]
- 12. Selegean JP, Kusserow R, Patel R, Heidtke TM, Ram JL. 2001. Using zebra mussels to monitor Escherichia coli in environmental waters. J. Environ. Qual. 30:171–179 [DOI] [PubMed] [Google Scholar]
- 13. Shaaly A, Tellevik MG, Langeland N, Hoiby EA, Jureen R. 2005. Comparison of serotyping, pulsed-field gel electrophoresis and amplified fragment length polymorphism for typing of Streptococcus pneumoniae. J. Med. Microbiol. 54:467–472 [DOI] [PubMed] [Google Scholar]
- 14. Vela AI, et al. 2003. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:2498–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waisberg J, Matheus CDO, Pimenta J. 2002. Infectious endocarditis from Streptococcus bovis associated with colonic carcinoma: case report and literature review. Arq. Gastroenterol. 39:177–179 [DOI] [PubMed] [Google Scholar]