Abstract
HIV-1 buds as an immature, noninfectious virion. Proteolysis of its main structural component, Gag, is required for morphological maturation and infectivity and leads to release of four functional domains and the spacer peptides SP1 and SP2. The N-terminal cleavages of Gag and the separation of SP1 from CA are all essential for viral infectivity, while the roles of the two C-terminal cleavages and the role of SP2, separating the NC and p6 domains, are less well defined. We have analyzed HIV-1 variants with defective cleavage at either or both sites flanking SP2, or largely lacking SP2, regarding virus production, infectivity, and structural maturation. Neither the presence nor the proteolytic processing of SP2 was required for particle release. Viral infectivity was almost abolished when both cleavage sites were defective and severely reduced when the fast cleavage site between SP2 and p6 was defective. This correlated with an increased proportion of irregular core structures observed by cryo-electron tomography, although processing of CA was unaffected. Mutation of the slow cleavage site between NC and SP2 or deletion of most of SP2 had only a minor effect on infectivity and did not induce major alterations in mature core morphology. We speculate that not only separation of NC and p6 but also the processing kinetics in this region are essential for successful maturation, while SP2 itself is dispensable.
INTRODUCTION
The main structural components of human immunodeficiency virus type 1 (HIV-1) and other retroviruses are derived from the viral Gag polyprotein. All retroviral Gag proteins contain three conserved functional domains: (i) matrix (MA), which harbors the membrane binding domain; (ii) capsid (CA), a protein composed of two domains, which forms the protective capsid encasing the genome; and (iii) nucleocapsid (NC), which is responsible for packaging and condensing the viral RNA genome (45). In addition, the 55-kDa HIV-1 Gag polyprotein includes the C-terminal p6 protein, which acts as an adapter to recruit the cellular ESCRT machinery responsible for virus budding (47), and two short spacer peptides, SP1 and SP2, which separate CA and NC and NC and p6, respectively. SP1 plays an important role in immature particle assembly and in regulating the formation of the mature HIV capsid essential for virus infectivity (1, 14, 24, 39). In contrast, the function of SP2 in virus morphogenesis and infectivity is currently unclear.
HIV morphogenesis involves the assembly of Gag molecules at the cytoplasmic face of the plasma membrane of the virus-producing cell (44). Approximately 2,500 copies of Gag form a truncated spherical shell within an approximately spherical virus particle. The particle buds from the cell in an immature, noninfectious form. Cryo-electron microscopy (cEM) and tomography (cET) analyses of immature HIV-1 revealed a radial organization of Gag molecules, with the MA domain adjacent to the membrane (19). Toward the interior of the particle, CA domains are organized in a hexameric lattice with a unit cell size of ∼8 nm (6, 7, 49). Rod-like structures presumably formed by SP1 connect CA to the NC-RNA-p6 layer, which is not hexagonally ordered and not well resolved in these structural analyses (6, 15, 16, 49).
Concomitant with or shortly after HIV budding, the viral protease (PR) cleaves Gag in five positions (Fig. 1A). This process, termed maturation, is accompanied by dramatic structural and morphological changes within the virion, resulting in the formation of mature, infectious HIV (5, 21). In the mature virus, MA remains associated with the viral membrane, while CA is detached from MA and forms a fullerene cone with a hexagonal lattice spacing of ∼10 nm (8, 20, 30). The wider spacing of the hexagonal lattice, together with the fact that only a subset of CA molecules are required to form the mature core (26), indicates that maturation involves disassembly of the immature lattice followed by assembly of a new lattice within the confined space of the virion. The ribonucleoprotein (RNP), including NC and viral replication proteins, forms a condensed mass within the cone-shaped CA core.
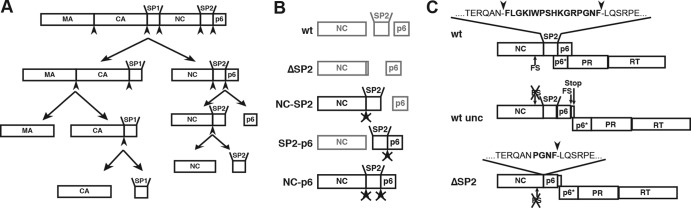
Fig 1.
Schematic overview of the constructs analyzed in this study. (A) Outline of the proteolytic cleavages within Gag that occur during the maturation of HIV-1. Arrowheads indicate the position of PR recognition sites. The domain sizes are not drawn to scale. The displayed order of cleavage events is based on cleavage rates determined for peptides in vitro as well as in vitro studies of Gag proteolysis (35–38) (B) Schematic representation of Gag processing products after completion of proteolytic maturation for each variant analyzed. The uncleaved products remaining due to mutation of the protease recognition site(s) are highlighted. Crossed arrowheads indicate that the cleavage site cannot be processed due to mutation. (C) Schematic representations of the uncoupled gag and pol open reading frames in pNL4-3unc and the deletion introduced in the ΔSP2 variant.
Morphological maturation is apparently controlled by sequential proteolysis of Gag. The five PR recognition sites within Gag are cleaved at different rates in vitro, with an ∼400-fold difference in rates between the fastest cleavage event (separating SP1 and NC) and the slowest one (separating CA and SP1) (18, 37, 38). These different cleavage rates appear to translate into an ordered intravirion cleavage process, which is required for morphological maturation and infectivity (21, 48) (see Fig. 1A for a scheme of sequential proteolysis of HIV-1 Gag). Distinct morphological intermediates have not been detected for wild-type (wt) HIV-1, indicating that maturation is a rapid process. The function of individual cleavage sites has therefore been mainly investigated by introducing mutations that block processing at specific sites or by using the drug bevirimat (31), which selectively inhibits processing between CA and SP1. These studies showed that cleavage sites separating MA and CA, CA and SP1, and SP1 and NC are all essential for viral infectivity (1, 24, 39, 48). Initial cleavage between SP1 and NC (Fig. 1A) is required for condensation of the RNP in the center of the virus particle but is not sufficient to disrupt the immature CA lattice (16). Cleavage between MA and CA is necessary to detach CA from the membrane-associated MA, but by itself is also not sufficient for immature CA lattice disassembly (16). Cleavage at both sides of the CA-SP1 module must occur for dissociation of the immature lattice, while removal of SP1 is further needed for assembly of the regular mature CA lattice (16, 23, 48).
The functional role of the two C-terminal Gag cleavages removing SP2 from p6 and from NC (Fig. 1A) and the importance of the SP2 region are much less clear. The lack of clarity is largely due to the problem that the 3′ portion of the gag open reading frame (ORF) comprising the SP2-p6 coding sequences overlaps a frameshift signal required for the synthesis of the Gag-Pol fusion protein and the 5′ portion of the pol ORF. The interpretation of viral phenotypes resulting from point mutations and deletions in this region of Gag is thus confounded by effects on the overlapping frameshift signal and pol sequence. Accordingly, the effect of deletion of the SP2 region on viral maturation and infectivity has not been studied so far.
Blocking of both cleavages between NC and p6 did not affect processing at upstream cleavage sites, but severely reduced viral infectivity to close to background levels (10, 22, 35). The defective phenotype was mainly caused by a block in proviral integration, while earlier stages of infection, including reverse transcription, were only mildly affected (10). Mutation of the faster cleavage site between SP2 and p6 also led to a severe reduction in infectivity, while discrepant phenotypes were observed when the slower cleavage site between NC and SP2 was blocked. Coren et al. (10) reported only minor differences in infectivity, while Kafaie et al. (22) observed a complete loss of infectivity for a similar mutation. This difference may be due to the specific change introduced and to the viral background, but the results of Coren et al. (10) clearly indicate that separation of SP2 from NC is not essential for HIV-1 infection in tissue culture. In vitro studies have shown that both NC and NC-SP2 function as nucleic acid chaperones, while uncleaved NC-p6 is poorly active in this regard (11, 12, 27, 32, 33, 43). Nucleic acid chaperone activity contributes to genomic RNA dimerization and condensation, to tRNA primer placement, and to efficient reverse transcription and thus plays an important role in HIV replication (13, 29, 42).
Here we have studied the effect of prevention of Gag processing between NC and p6 on structural maturation using cET. In addition, we have deleted SP2 without affecting the frameshift signal and pol reading frame and have analyzed the effects on HIV-1 morphogenesis, structure, and infectivity. Our results indicate that correct processing between NC and p6 is relevant for regular assembly of the mature capsid core, while SP2 itself is dispensable for structural maturation and viral infectivity.
MATERIALS AND METHODS
Cell lines and plasmids.
293T cells were grown in Dulbecco's modified Eagle's medium supplemented with glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (FCS). C8166 cells were grown in RPMI 1640 medium, supplemented with penicillin-streptomycin, 20 mM HEPES (pH 7.4), and 10% FCS. Proviral plasmids used for virus production were constructed based on plasmid pNLC4-3 (3) or pNL4-3uncoupled (pNL4-3unc). pNL4-3unc was constructed by insertion of a synthetic DNA fragment (Geneart) whose sequence was based on plasmid pNL4-3AL (28) between the ApaI and MluI restriction sites of pNL4-3 (2). Plasmids pNLC4-3.NC-SP2 and pNLC4-3.NC-p6 have been described previously (35). Plasmid pNLC4-3.SP2-p6 was generated by introduction of mutation F16S (10) into pNLC4-3 by overlap PCR using oligonucleotides 5′-GGGAAGGCCAGGGAATTCTCTTCAGAGCAGACCAGA-3′ and 5′-TATGGTCTGCTCTGAAGAGAATTCCCTGGCCTTCCC-3′. pNL4-3uncΔSP2 was also cloned by overlap PCR, using oligonucleotides 5′-AGATTGTACTGAGAGACAGGCTAACCCGGGCGCCGC-3′ and 5′-GCGGCGCCCGGGTTAGCCTGTCTCTCAGTACAATCT-3′ to generate a mutated ApaI-AgeI fragment.
Virus preparation.
293T cells were transfected with the respective proviral plasmids using a standard calcium phosphate transfection procedure. At 44 h posttransfection, tissue culture supernatant was harvested and passed through a 0.45-μm-pore filter. Virions were purified from the cleared supernatant by pelleting through a 20% (wt/wt) sucrose cushion and subsequent ultracentrifugation through an iodixanol gradient as described previously (17). Particles were resuspended in phosphate-buffered saline (PBS) and stored at −20°C. For cET analyses, virions were subjected to inactivation by mild fixation (1% paraformaldehyde, 1 h on ice). Purity was assessed by SDS-PAGE and silver staining. Immunoblot analyses were performed to confirm the processing pattern of the respective variants. Samples were separated by SDS-PAGE and transferred by semidry blotting to nitrocellulose (for MA or CA) or Immobilon (for NC) membranes, respectively. Membranes were incubated with polyclonal antisera raised against recombinant HIV-1 CA (sheep), MA (rabbit), NC (rabbit), or RT (rabbit), respectively. Bound antibodies were detected by quantitative immunoblotting using a LiCor Odyssey system and labeled secondary antibodies (LiCor) according to the manufacturer's instructions.
RT activity assay.
Supernatants from 293T cells transfected with the indicated proviral plasmids were harvested at 44 h posttransfection. Samples were treated with lysis buffer (0.125% Triton X-100, 25 mM KCl, 50 mM Tris-HCl [pH 7.4], 20% glycerol, supplemented with 1 μl of RNase inhibitor/100 μl) to disrupt virus particles. Reverse transcriptase (RT) activity was determined by a SYBR green-based product-enhanced RT assay using a CXR 96 real-time PCR detection system (Bio-Rad) as described by Pizzato et al. (40), using particles purified from the supernatant of 293T cells transfected with a wt proviral plasmid as a standard.
Infectivity assay.
Tissue culture supernatants from 293T cells transfected with the respective proviral plasmids were harvested and passed through a 0.45-μm-pore filter. Infectivity was determined by endpoint titration. For this titration, C8166 T cells were seeded in 96-well plates (2.5 × 104 cells/well) and infected in quadruplicate using serial 10-fold dilutions of virus stocks. Seven days postinfection, wells harboring a spreading virus infection were scored based on syncytium formation, and the 50% tissue culture infective dose (TCID50) was calculated (Spearman-Kärber). Virus titers were normalized to the virus concentration as determined by quantitative immunoblotting using purified recombinant CA (kindly provided by L. Castillo) as a standard.
Cryo-electron tomography.
Virions were mixed with 10-nm-diameter gold beads, deposited on C-flat holey carbon grids, blotted, and vitrified by plunge freezing in liquid ethane. Tilt series were collected on a FEI Tecnai F30 “Polara” transmission electron microscope (EM) with a Gatan GIF 2002 post column energy filter and 2kx2k Multiscan charge-coupled device camera (CCD) camera. The data collection was performed at 300 kV using the SerialEM software package, and tomograms were reconstructed using the IMOD software package (25). Tilt series were typically collected between −60 and +60° with a tilt increment of 3° and a total electron dose of approximately 100 e/Å2. The defocus range was between 6.0 and 8.0 μm, with a magnification of 27,500× resulting in a pixel size at the specimen level of 4.7 Å.
Segmentation and volume measurement.
To measure the volume of the virion cores, cores were manually segmented using Amira (Visage) from cryo-electron tomograms low-pass filtered to 80 Å. To refine the position of the CA layer, subtomograms centered on the surface of the segmented volume were extracted with an even spacing of 6 pixels, preoriented to the normal of the surface of the segmented object. The subtomograms underwent iterative translational alignment along the normal vectors. A closed surface in three dimensions (3D) based on the final coordinate of the subtomograms was spline fitted to define the refined surface. From this, the surface area and the volume of the cores were calculated. To determine the virion volume, for each virion, three orthogonal radii were measured and the volume at the inner side of the envelope was calculated. All calculations and image processing were done using Matlab (Mathworks).
RESULTS
Production and functional characterization of HIV-1 variants.
To elucidate the relative importance of the individual processing sites between NC and p6 for structural maturation of HIV-1 and to determine the relevance of SP2 for virion morphogenesis, maturation, and infectivity, we generated and analyzed a panel of mutant HIV-1 derivatives. Variants NC-SP2, SP2-p6 and NC-p6 have been described previously; they carry point mutations disabling the NC-SP2 and/or the SP2-p6 cleavage site (Fig. 1B) (10, 35). Mutational analysis of the SP2 region in the viral context is hindered by the fact that SP2 overlaps the gag-pol frameshift signal and the p6* region of the pol ORF. Accordingly, deletion of SP2 affects both Pol protein expression (41) and sequence. This problem was overcome by constructing the proviral derivative pNL4-3uncoupled (pNL4-3unc), in which gag and pol ORFs are uncoupled due to mutational inactivation of the original frameshift signal and insertion of a copy of the frameshift region just upstream of the gag stop codon with subsequent duplication of the overlap region (Fig. 1C, wt unc) as previously described (28). In this case, translational frameshifting should occur just after the last codon of the authentic gag reading frame, eliminating the sequence overlap of gag with the subsequent p6* region of pol. Accordingly, mutations in the gag ORF do not affect pol genes or Gag-Pol frameshifting in this construct. A ΔSP2 variant, which lacks the first 10 codons of SP2 but retains the fast cleavage site upstream of p6 to allow its separation from NC, was constructed in the uncoupled virus context (Fig. 1C). To confirm that the sequence duplications introduced in the case of the pNL4-3unc and ΔSP2 variants were retained over multiple rounds of virus replication in tissue culture, the presence of the modified region was verified by RT-PCR of viral RNA 8 days postinfection (see Fig. S1 and the methods described in the supplemental material).
Virus particles were produced by transfection of proviral plasmids into 293T cells and analyzed for release efficiency, polyprotein processing, and infectivity. All variants displayed release efficiencies similar to that of wt HIV-1, as determined by quantitative immunoblotting of cell lysates and tissue culture supernatants (Fig. 2A). These quantitative analyses demonstrated that SP2 is not required for particle release, in agreement with the observations of Popova et al. (41). Immunoblot analysis of particles using antisera against MA and CA revealed similar processing patterns for all cleavage site mutants as well as for the uncoupled variant and its ΔSP2 derivative (Fig. 2B, upper and middle panels). This result showed that the presence or cleavage of SP2 is not required for upstream polyprotein processing. It further indicated that sufficient amounts of the pol gene product PR were produced to yield complete proteolysis in all cases despite the introduced alterations in the frameshift region. The presence of wt amounts of pol products was confirmed by analyzing the ratio of reverse transcriptase (RT) to CA in viral particles by immunoblotting and by determining particle-associated RT activity (Fig. 2C).
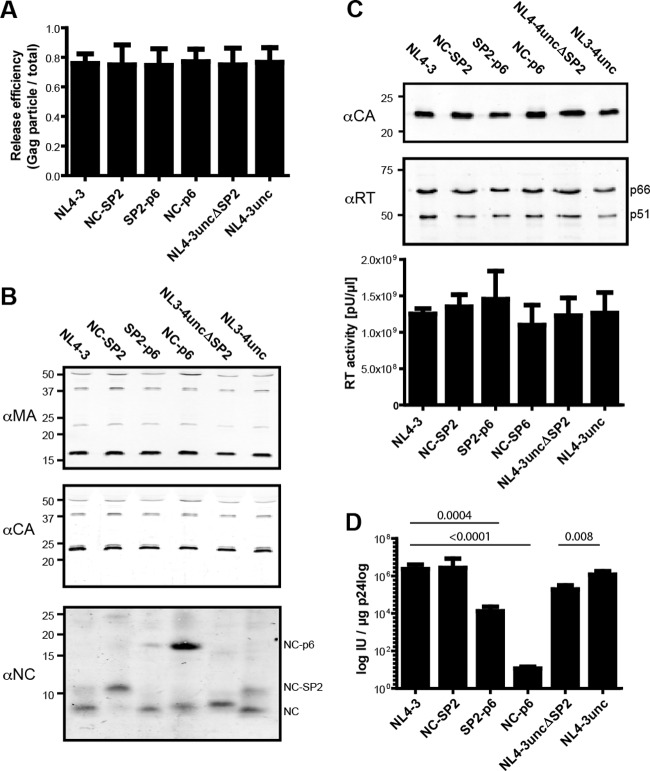
Fig 2.
Characterization of the mutated HIV-1 derivatives. (A) Relative release efficiency of the mutated variants. 293T cells were transfected with the indicated proviral plasmids, and cell lysates and tissue culture supernatants were harvested at 44 h posttransfection. Samples were separated by SDS-PAGE, and the amount of proteins comprising the CA domain in the samples was determined by quantitative immunoblotting (LiCor) using polyclonal antiserum raised against HIV-1 CA. These data were used to calculate the amounts of CA released relative to total Gag expressed. The graph shows mean values and standard deviations (SD) from triplicate transfections. (B) Immunoblot analysis of virus particles. Virions were prepared from the tissue culture supernatants of 293T cells transfected with the respective proviral plasmids. Samples were separated by SDS-PAGE, and Gag-derived proteins were detected by quantitative immunoblotting using polyclonal antisera raised against the indicated proteins (αMA, αCA, and αNC). The positions of molecular mass standards are indicated to the left. (C) RT content of virions. (Top) Immunoblot analysis. Virions prepared as described above were subjected to SDS-PAGE followed by quantitative immunoblot analysis using the indicated antisera (αCA and αRT). (Bottom) Virion-associated RT activity. The bar graph displays RT enzymatic activity in supernatants from transfected 293T cells determined by a PCR-enhanced RT assay as described in Materials and Methods. Mean values and SD from triplicate transfections are shown. (D) Infectivities of the variants used in this study. Relative infectivities were determined by endpoint titration of tissue culture supernatants from transfected 293T cells on C8166 T cells. The values shown were normalized for the amount of CA detected in the samples by quantitative immunoblotting. Bars represent mean values and standard deviations from triplicate transfections. P values were calculated by an unpaired Student's t test on the logs of titers.
Detection of NC-reactive proteins in viral particles (Fig. 2B, lower panel) revealed fully processed NC and some remaining NC-SP2 in wt HIV-1 and confirmed that NC-SP2 and NC-p6 were predominantly found in the respective variants. The stronger signal for NC-p6 is caused by preferential detection of NC precursor proteins by this antiserum compared to NC and NC-SP2; thus, the relative band intensities do not provide a direct measure of relative protein amounts. Variant SP2-p6, which harbors a mutation at the cleavage site between SP2 and p6, displayed mostly cleaved NC, but also contained small amounts of partially processed NC-p6, which was not observed in wt HIV-1 (compare lanes 1 and 3). NC processing in the case of the uncoupled variant was similar to that of wt HIV-1 (compare lanes 1 and 6). NC was also fully processed in the case of the SP2 deletion (lane 5); the resulting NC protein displayed a slightly lower electrophoretic mobility than wt NC due to the presence of four additional amino acids at its C terminus derived from the fast cleavage site retained in the respective construct.
The infectivity of all variants was determined by endpoint titration on C8166 T cells (Fig. 2D). Consistent with published results (10, 35), infectivity in C8166 cells was not affected by blocking cleavage at the slow processing site between NC and SP2, while blocking the cleavage at the faster site between SP2 and p6 caused a 2-log reduction in infectious titer (Fig. 2C). Viral infectivity was almost abolished when cleavage at both sides of SP2 was blocked, as described previously (10, 35). Uncoupling of the gag and pol ORFs had no significant effect on viral infectivity, as previously described for a similar construct (28). Unexpectedly, deletion of most of SP2, retaining only the fast cleavage site to allow rapid processing between NC and p6, caused only a minor reduction in HIV infectivity. This effect was reproducible and statistically significant (P = 0.008, determined by an unpaired Student's t test), but amounted to only an ∼10-fold reduction in titer compared to wt HIV-1. Similar results were observed when the single-round infectivity of the respective variants was analyzed in TZM-bl indicator cells. Blocking cleavage between SP2 and p6 or blocking both cleavages flanking SP2 led to a severe decrease in infectivity, while the other mutations had only a minor effect (see Fig. S2 in the supplemental material).
Structural analysis of HIV-1 variants.
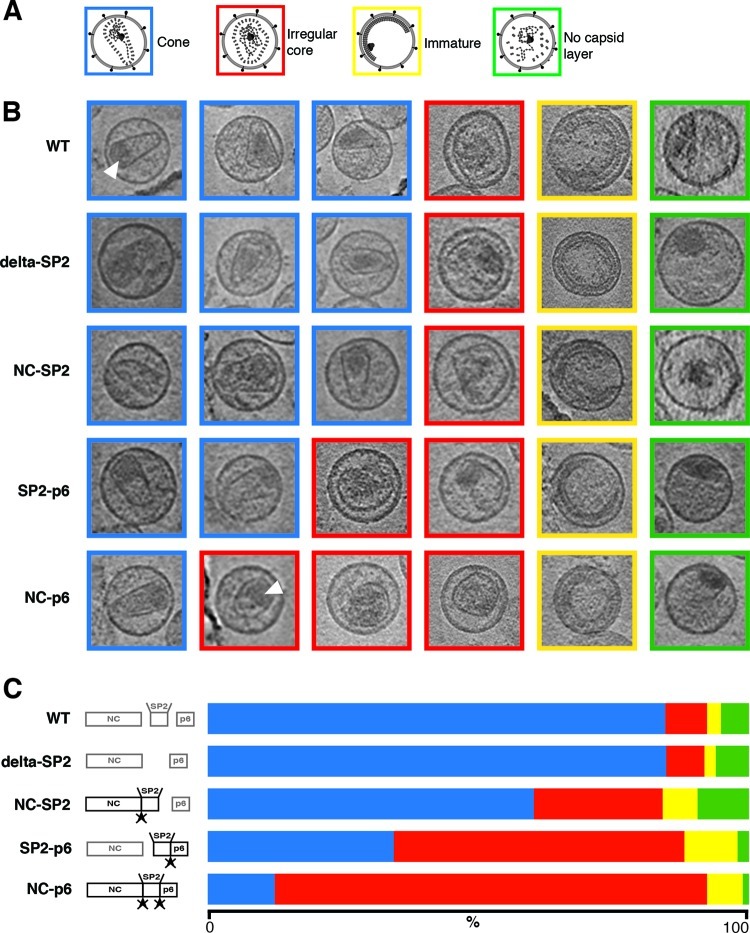
Purified wt and variant HIV-1 particles were inactivated and subjected to cET. The purity of particle preparations was confirmed by SDS-PAGE followed by silver staining. Since HIV particle preparations display morphological heterogeneity (8), analyses were performed for two independent virus preparations of all variants, and the results were compared to those of wt controls. More than 50 particles were analyzed for each variant and particle preparation. Individual particles were manually classified into one of four structural groups (Fig. 3A): (i) mature, conical core (blue); (ii) irregular, sometimes open, core structure (red); (iii) striated, spherical immature Gag shell (yellow); or (iv) no discernible Gag layer, but containing a condensed RNP (green). Computational sections exemplifying the different classes and their relative distribution among the variants analyzed are shown in Fig. 3B, while Fig. 3C provides the results of quantification. Data from both independent preparations were pooled for analysis since there were no substantial differences (Table 1).
Fig 3.
Morphological analysis of the HIV-1 variants. Virus particles were assigned to one of four categories, based upon the core structure: conical (blue), irregular (red), immature (yellow), and no lattice detected (green). (A) Schematic diagrams of the four categories. (B) Overview of the different morphological phenotypes and their relative abundance. The images represent 18-Å-thick computational slices through cryo-electron tomograms. The boxes have a size of 150 by 150 nm. White arrowheads indicate examples of electron-dense structures presumed to represent the condensed RNP. (C) Bar graph showing the relative abundance of the phenotypes determined for the different virus variants. Crossed arrowheads indicate that the cleavage site cannot be processed due to mutation. The color code is as in panel A.
TABLE 1.
Morphological analysis of the variantsa
| Construct | Mean virion diam (nm) | Preparation no. | % of particles: |
No. of virions measured | |||
|---|---|---|---|---|---|---|---|
| With conical core | With irregular core | Immature | Without capsid layer | ||||
| pNL4-3 | 129 ± 23 | 1 | 84 | 10 | 4 | 1 | 68 |
| 2 | 87 | 6 | 1 | 6 | 67 | ||
| ΔSP2 | 135 ± 24 | 1 | 97 | 0 | 2 | 1 | 93 |
| 2 | 70 | 15 | 2 | 13 | 86 | ||
| NC-SP2 | 132 ± 22 | 1 | 68 | 13 | 6 | 13 | 63 |
| 2 | 58 | 31 | 5 | 7 | 98 | ||
| SP2-p6 | 127 ± 17 | 1 | 39 | 46 | 11 | 3 | 61 |
| 2 | 30 | 61 | 6 | 2 | 87 | ||
| NC-p6 | 136 ± 28 | 1 | 12 | 75 | 12 | 1 | 69 |
| 2 | 12 | 84 | 2 | 0 | 89 | ||
| pNL4-3uncoupled | 137 ± 22 | 1 | 86 | 5 | 8 | 2 | 65 |
| 2 | 78 | 13 | 2 | 7 | 54 | ||
The table summarizes the mean diameters of the virus particles and the percentage of virus particles assigned to each of the morphological classes for each variant analyzed. Data obtained from two independent preparations per variant are shown.
The wt HIV-1 preparations displayed a range of morphologies, comparable with previously published data (8). The vast majority (85%) showed a conical core, with some particles containing more than one core. A small proportion (3%) displayed the striated Gag density characteristic of immature virions. These may represent immature particles prior to maturation or may correspond to virions defective in maturation. Irregularly shaped, sometimes open cores of various sizes were observed in 8% of all wt particles, while 4% contained no discernible capsid layer but did contain a condensed electron-dense mass resembling the RNP detected inside mature cores (Fig. 3). The uncoupling of gag and pol had no significant effect on the distribution of viral morphologies (Table 1; compare NL4-3unc and NL4-3), as expected from its wt Gag processing pattern and the similar infectious titer. Deletion of the SP2 region also had no apparent influence on the morphology of HIV-1 particles (Fig. 3), consistent with its modest effect on viral infectivity. An increase in the proportion of irregular cores was observed when SP2 was present but could not be removed from NC (NC-SP2) (Fig. 3), although this variant also displayed wt infectivity. The fraction of irregular cores was higher in the case of the SP2-p6 variant (54%), with only 35% of particles displaying the regular mature core. Irregular cores were found in 80% of all particles in the case of the NC-p6 variant, where all cleavages between NC, SP2, and p6 were prevented, and only 12% of particles exhibited a conical morphology in this case. The proportions of immature particles and of particles lacking any capsid layer did not vary systematically between virus variants (Fig. 3 and Table 1).
One important functional role of mature NC is condensation of the viral genome into a dense RNP (13, 34). This condensation is thought to constitute a prerequisite for subsequent capsid assembly since noncondensed viral RNA may not fit into the confined space of the regular mature capsid. To determine whether the described alterations affected RNP condensation, we subjected particles to visual inspection regarding differences in the sizes of electron-dense structures presumed to represent the RNP (Fig. 3). No consistent differences were detected, but variations in densities within these structures and their irregular shapes did not allow a sharp definition of structure borders and thus prevented a precise measurement of RNP volumes. We therefore determined whether the volume enclosed by irregular HIV cores differed from that encapsidated by regular conical cores. Core volumes were plotted against virion volumes for both conical and irregular cores observed in wt and NC-p6 variant particles (Fig. 4). The volume of conical cores was approximately proportional to the diameter of the virion in both cases, consistent with the relationship between core size and virion size previously observed for wt HIV-1 (4). Irregular cores comprised a larger volume in relation to virion size than conical cores and appeared to be more variable in size; this correlation was observed for both wt and NC-p6 variant particles (Fig. 4).
Fig 4.
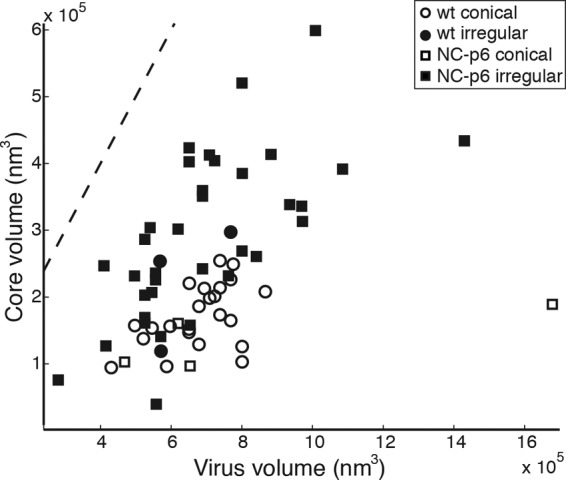
Relationship between core volume and virus volume. The scatterplot shows the relationship between the volume of each virion and the volume of the core it contains. Circles represent data from wt HIV-1, and squares represent data from NC-p6. Open data points represent conical cores, and solid data points represent irregular cores. The dotted line indicates the total internal volume of the virion.
DISCUSSION
Here, we have explored the role of SP2 and its separation from the upstream NC domain and the downstream p6 domain of HIV-1 Gag for viral morphogenesis, maturation, and infectivity without confounding effects on the pol reading frame. While both the SP1 peptide and its separation from CA are essential for HIV-1 maturation and infectivity (1, 24, 48), the presence of SP2 and its separation from the flanking NC region do not appear to be equally important. Deletion of SP2 had no apparent effect on viral morphogenesis and structure and led only to a minor reduction in viral infectivity in tissue culture. Furthermore, cleavage of SP2 from NC was also dispensable for HIV-1 maturation and infectivity, as previously described (10), while lack of any cleavage downstream of NC abolished infectivity. Strikingly, inhibition of cleavage between NC and p6 (and to a lesser extent between SP2 and p6) severely affected formation of the regular cone-shaped capsid, although CA processing itself appeared unimpaired.
The SP2 peptide and the flanking cleavage sites are highly conserved in HIV-1 (www.hiv.lanl.gov), which can only in part be explained by the necessity for conservation of the overlapping translational frameshift signal. A spacer peptide separating NC and p6 is also present in other primate lentiviruses (www.hiv.lanl.gov). It was therefore surprising that SP2 deletion caused only a minor reduction in infectivity, at least in the T-cell line tested, with no apparent effect on viral morphogenesis. Conceivably, this deletion may have a more pronounced phenotype under other conditions, and this will be the subject of further investigations. It cannot be excluded that the minor defect of the ΔSP2 variant may be due to the presence of four additional amino acids (Pro-Gly-Asn-Phe) at the C terminus of NC, which were retained to allow rapid processing from p6. However, the presence of the complete 14-amino-acid spacer peptide at the C terminus of NC did not affect virus morphogenesis and infectivity under the conditions tested. We therefore consider it more likely that the small defect is due to the lack of amino acids 1 to 10 of SP2.
Although SP2 itself is clearly not needed for morphological maturation and is dispensable for infectivity, processing of the NC-p6 region appears to be essential. Blocking both cleavages between NC and p6 almost abolished viral infectivity, with severely altered core morphology in at least 80% of all particles. This defect in formation of the regular, cone-shaped core occurred despite complete processing of CA, suggesting that it may not be due to a direct effect on CA. Separation of the NC-p6 segment of Gag from the MA-CA-SP1 region is required for condensation of the RNP (16), which in turn is thought to be essential for fitting the viral genome and replication proteins into the confined space of the mature capsid. A simple explanation would thus be that cleavage between NC and p6 is also essential for condensation of the RNP. This explanation does not seem to be correct, however, since NC-p6 particles contained a condensed RNP as well, and we saw no clear difference in the dimensions of the RNP between wt and variant particles. Since, however, NC-p6 particles had a much higher proportion of irregular cores than wt particles and the irregular cores occupied a larger fraction of the inner virion volume in both variants, a kinetic delay rather than a failure of RNP condensation may be the cause of irregular capsid formation. NC and NC-SP2 have been shown to be significantly more active nucleic acid chaperones than NC-p6 (11, 12, 27, 32, 33, 36, 43, 50), and this chaperone activity is needed to condense the RNA genome into the most thermodynamically stable form. The lower chaperone activity of NC-p6 may translate into delayed condensation of the RNP upon maturation and thereby indirectly affect capsid condensation. While the RNP is well condensed in virions harvested at late time points after infection, it may not yet be sufficiently condensed during the rapid process of maturation, thereby hindering the assembly of the mature capsid shell. It should be noted, however, that the observed drop in infectivity dramatically exceeds the increased proportion of irregular capsid shells (>5-log reduction in infectivity versus a 1-log difference in the proportion of irregular cores), indicating additional defects besides structural maturation of the core. The apparent presence of additional effects is consistent with the observation that the NC-p6 variant has a mild defect in reverse transcription (which could be caused by an increased proportion of irregular cores that may be defective for proper uncoating), but a much more severe defect in provirus integration (10), which is unlikely to be caused by a structural effect on the viral capsid shell alone.
Analysis of the relative contributions of the two cleavage sites flanking SP2 indicated that blocking removal of SP2 from NC had no detectable effect on viral infectivity despite a significant and reproducible increase in irregular capsid shells. Since HIV-1 preparations have been shown to contain a proportion of defective, noninfectious particles, it appears most likely that the observed structural alteration mainly affected defective particles, and thus it was not scored in infectivity assays.
In contrast to mutation of the NC-SP2 cleavage site, altering the SP2-p6 cleavage site had a significant effect on viral infectivity and also led to a higher proportion of irregular cores. This differential effect on infectivity of mutation of the two cleavage sites had been reported previously (10). Based on this observation, it has been suggested that NC-SP2 may play an important role in early HIV-1 infection, which cannot be performed in the case of the SP2-p6 mutant due to the lack of the NC-SP2 processing intermediate (10). This hypothesis is, however, not consistent with our results because the ΔSP2 variant, which cannot generate the NC-SP2 intermediate, had only a minor defect in infectivity and no alteration in structural maturation.
Instead, we propose several, mutually nonexclusive, explanations for the observed differential effects of cleavage site mutations upstream and downstream of SP2. (i) The data are confounded by contributions of the introduced amino acid changes, which go beyond prevention of cleavage, such as introducing structural changes in Gag. (ii) Free p6, but not free NC is important for replication capacity in tissue culture, and free p6 cannot be formed if the SP2/p6 site is blocked. (iii) SP2-p6 has a trans-dominant negative effect on early HIV replication. (iv) The observed effect on viral infectivity is caused by residual NC-p6, which is completely processed in NC-SP2 and in wt HIV-1, but not in SP2-p6 or NC-p6 (Fig. 2B). (v) The kinetics of cleavage separating the NC and p6 domains, but not the actual cleavage products, matter most, and infectivity requires at least one rapid cleavage in this region. Currently, none of these explanations can be excluded. However, we favor the final hypothesis (no. v), since the infectious phenotype of our variants directly correlated with the presence or absence of a fast cleavage site in this region. HIV-1 wt, the NC-SP2 variant, and HIV-1uncΔSP2 all retained this fast cleavage site and showed no decrease or a minor decrease in infectivity. In contrast, the NC-p6 and SP2-p6 variants that either lost all cleavages or only retained a slow cleavage site in this region exhibited a much more severe phenotype. One would therefore hypothesize that the SP2-p6 variant could be rescued by improving cleavage at the upstream NC/SP2 site, and this is under investigation.
The hypothesis that cleavage kinetics in the NC-SP2-p6 region affect mature particle formation would also be consistent with the observed selection of improved cleavage sites in the NC-p6 region in PR-resistant HIV-1 variants compensating for decreased proteolytic activity (9). Furthermore, studies on the replication capacity of such variants in the presence and absence of protease inhibitors also indicate that temporal control of NC-SP2 cleavage is important for efficient HIV-1 replication (46). In such a scenario, it may not be critical whether either the upstream or the downstream site is improved as long as faster processing of this region can be achieved at all. Of course, both cleavage sites and the presence of an intervening spacer peptide are likely to contribute to optimal HIV-1 replication in vivo as they are highly conserved, but additional defects of the respective mutations may be minor and thus may not easily be scored in the experimental systems described.
In summary, SP2 appears to be dispensable for HIV-1 morphogenesis, maturation, and infectivity in tissue culture, while not only separation of NC and p6 but also the kinetics of separation may be important. The importance of processing of SP2 for virion morphology could be due to a need for rapid condensation of the viral genome once proteolytic maturation has been initiated in order to achieve an appropriately condensed RNP before CA is fully processed inside the virion and mature core formation commences. NC-p6 has a significantly lower nucleic acid chaperone activity than processed NC, and a fast processing site downstream of NC may thus be essential for regular maturation and viral infectivity.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the gifts of plasmid pNL4-3unc from Ivonne Morales and purified HIV-1 CA from Luis Castillo. We thank Sonja Welsch for helpful comments and discussions and Benjamin Radestock for establishing the PCR analysis of NL4-3unc.
This work was supported by the Molecular Medicine Partnership Unit of the EMBL and the Medical Faculty of the University of Heidelberg and by grants from the Deutsche Forschungsgemeinschaft within SPP 1175 to H.-G.K. and to J.A.G.B., from European Union FP6 grant LSHP-CT-2007-036793 (HIV PI resistance) to H.-G.K. and to J.A.G.B. and European Union FP7 grant HEALTH-F3-2008-201095 (HIV-ACE) to H.-G.K. and B.M.
A.d.M., H.-G.K., B.M., and J.A.G.B. conceived and designed the experiments, A.d.M., A.M.H., and B.G. performed the experiments, A.d.M., H.-G.K., B.M., and J.A.G.B. analyzed the data, and A.d.M., H.-G.K., B.M., and J.A.G.B. wrote the manuscript.
Footnotes
Published ahead of print 10 October 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Accola MA, Hoglund S, Gottlinger HG. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adachi A, et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bohne J, Krausslich HG. 2004. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 563:113–118 [DOI] [PubMed] [Google Scholar]
- 4. Briggs JA, et al. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20 [DOI] [PubMed] [Google Scholar]
- 5. Briggs JA, Krausslich HG. 2011. The molecular architecture of HIV. J. Mol. Biol. 410:491–500 [DOI] [PubMed] [Google Scholar]
- 6. Briggs JA, et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briggs JA, et al. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672–675 [DOI] [PubMed] [Google Scholar]
- 8. Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clavel F, Mammano F. 2010. Role of Gag in HIV resistance to protease inhibitors. Viruses 2:1411–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coren LV, et al. 2007. Mutational analysis of the C-terminal gag cleavage sites in human immunodeficiency virus type 1. J. Virol. 81:10047–10054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. 2006. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 363:867–877 [DOI] [PubMed] [Google Scholar]
- 12. Cruceanu M, et al. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34:593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darlix JL, et al. 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 410:565–581 [DOI] [PubMed] [Google Scholar]
- 14. Datta SA, et al. 2011. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J. Virol. 85:4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Marco A, et al. 2010. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84:11729–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Marco A, et al. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 6:e1001215 doi:10.1371/journal.ppat.1001215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dettenhofer M, Yu XF. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erickson-Viitanen S, et al. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retroviruses 5:577–591 [DOI] [PubMed] [Google Scholar]
- 19. Fuller SD, Wilk T, Gowen BE, Krausslich HG, Vogt VM. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729–738 [DOI] [PubMed] [Google Scholar]
- 20. Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83 [DOI] [PubMed] [Google Scholar]
- 21. Ganser-Pornillos BK, Yeager M, Pornillos O. 2012. Assembly and architecture of HIV. Adv. Exp. Med. Biol. 726:441–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kafaie J, et al. 2009. Role of capsid sequence and immature nucleocapsid proteins p9 and p15 in human immunodeficiency virus type 1 genomic RNA dimerization. Virology 385:233–244 [DOI] [PubMed] [Google Scholar]
- 23. Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J. Virol. 85:1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krausslich HG, Facke M, Heuser AM, Konvalinka J, Zentgraf H. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 26. Lanman J, et al. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11:676–677 [DOI] [PubMed] [Google Scholar]
- 27. Le Cam E, et al. 1998. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers 45:217–229 [DOI] [PubMed] [Google Scholar]
- 28. Leiherer A, Ludwig C, Wagner R. 2009. Uncoupling human immunodeficiency virus type 1 Gag and Pol reading frames: role of the transframe protein p6* in viral replication. J. Virol. 83:7210–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286 [DOI] [PubMed] [Google Scholar]
- 30. Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413 [DOI] [PubMed] [Google Scholar]
- 31. Martin DE, Salzwedel K, Allaway GP. 2008. Bevirimat: a novel maturation inhibitor for the treatment of HIV-1 infection. Antivir. Chem. Chemother. 19:107–113 [DOI] [PubMed] [Google Scholar]
- 32. Mirambeau G, et al. 2007. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One 2:e669 doi:10.1371/journal.pone.0000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mirambeau G, et al. 2006. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 364:496–511 [DOI] [PubMed] [Google Scholar]
- 34. Mirambeau G, Lyonnais S, Gorelick RJ. 2011. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 7:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muller B, et al. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J. Biol. Chem. 284:29692–29703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohishi M, et al. 2011. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 39:3404–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. 2005. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 2:66 doi:10.1186/1742-4690-2-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettit SC, et al. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pizzato M, et al. 2009. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods 156:1–7 [DOI] [PubMed] [Google Scholar]
- 41. Popova E, Popov S, Gottlinger HG. 2010. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J. Virol. 84:6590–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rein A. 2010. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol. 7:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stoylov SP, et al. 1997. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers 41:301–312 [DOI] [PubMed] [Google Scholar]
- 44. Sundquist W. I., Krausslich. H. G. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2:a006924 doi:10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swanstrom R, Wills JW. 1997. Synthesis, assembly, and processing of viral proteins, p 263–334 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 46. van Maarseveen NM, et al. 2012. Modulation of HIV-1 Gag NC/p1 cleavage efficiency affects protease inhibitor resistance and viral replicative capacity. Retrovirology 9:29 doi:10.1186/1742-4690-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weiss ER, Gottlinger H. 2011. The role of cellular factors in promoting HIV budding. J. Mol. Biol. 410:525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wiegers K, et al. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wright ER, et al. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu T, et al. 2010. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology 405:556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.