Abstract
Mx1 is a GTPase that is part of the antiviral response induced by type I and type III interferons in the infected host. It inhibits influenza virus infection by blocking viral transcription and replication, but the molecular mechanism is not known. Polymerase basic protein 2 (PB2) and nucleoprotein (NP) were suggested to be the possible target of Mx1, but a direct interaction between Mx1 and any of the viral proteins has not been reported. We investigated the interplay between Mx1, NP, and PB2 to identify the mechanism of Mx1's antiviral activity. We found that Mx1 inhibits the PB2-NP interaction, and the strength of this inhibition correlated with a decrease in viral polymerase activity. Inhibition of the PB2-NP interaction is an active process requiring enzymatically active Mx1. We also demonstrate that Mx1 interacts with the viral proteins NP and PB2, which indicates that Mx1 protein has a direct effect on the viral ribonucleoprotein complex. In a minireplicon system, avian-like NP from swine virus isolates was more sensitive to inhibition by murine Mx1 than NP from human influenza A virus isolates. Likewise, murine Mx1 displaced avian NP from the viral ribonucleoprotein complex more easily than human NP. The stronger resistance of the A/H1N1 pandemic 2009 virus against Mx1 also correlated with reduced inhibition of the PB2-NP interaction. Our findings support a model in which Mx1 interacts with the influenza ribonucleoprotein complex and interferes with its assembly by disturbing the PB2-NP interaction.
INTRODUCTION
Almost 50 years ago, the interferon-induced Mx1 gene was discovered in the mouse as a potent restriction factor of influenza virus. Mice carrying a functional Mx1 locus are resistant to influenza A virus infection. However, most inbred laboratory mouse strains have a deletion of three exons or a nonsense mutation in the Mx1 locus and are susceptible to this virus (20, 30, 31, 46). Mx genes were subsequently identified in other vertebrates, where they are usually represented by one to three isoforms. Many of these Mx proteins, including the human homolog, MxA, have antiviral activity against a wide range of RNA viruses and some DNA viruses (13, 14, 28). This activity seems to depend on the subcellular localization of the protein. Nuclear forms (e.g., mouse Mx1) protect against viruses that replicate in the cell nucleus, such as influenza and Thogoto viruses (7, 12), whereas cytoplasmic forms (e.g., mouse Mx2) inhibit replication of vesicular stomatitis virus (VSV) and some other viruses that replicate in the cytoplasm (23, 54). Remarkably, the human MxA protein is localized in the cytoplasm yet has a broad antiviral spectrum irrespective of the virus's subcellular replication site (38; reviewed in reference 13).
How Mx proteins exert their antiviral activity at the molecular level remains poorly understood. Mx proteins are part of a family of large GTPases that also includes dynamin, and GTP binding is important for the antiviral activity of Mx proteins. This was demonstrated by studying mouse Mx1 and human MxA variants with targeted mutations in one of the three consensus elements that comprise the GTP-binding domain. Mutations leading to loss of GTP binding impaired the antiviral activity against influenza and VSV (33, 40). GTP binding and GTPase activity are closely related. However, because Mx1 or MxA mutants lacking only one of these two functions have not yet been described, it is difficult to determine the relative contributions of GTPase activity and GTP binding to the antiviral activity of Mx1 or MxA.
Influenza A virus has a segmented, negative-stranded RNA genome (vRNA). Each genome segment is complexed with nucleoprotein (NP) molecules and with one RNA-dependent RNA polymerase (RdRp) complex (containing polymerase basic protein 1 [PB1], PB2, and polymerase acidic protein [PA]), forming a viral ribonucleoprotein complex (vRNP). These vRNPs are the minimal functional units for influenza virus replication (vRNA production) and transcription (viral mRNA production). Following virus endocytosis and hemagglutinin-mediated fusion of the viral and host cell membranes, the vRNPs are liberated in the cytoplasm and then transported to the nucleus, where transcription and replication take place (reviewed in references 8 and 27). The incoming vRNPs first produce viral mRNA (primary transcription), which involves cap snatching from cellular mRNAs mediated by PB2 (cap binding) and PA (cap cleavage). The viral mRNA is transported to the cytoplasm and translated. After entry of newly produced proteins into the nucleus, replication of the viral genome can start. Eventually, the progeny vRNAs exit the nucleus in the form of vRNPs that are ready for packaging and budding from the host membrane as new virions (8).
The effect of Mx1 expression on the different steps of influenza virus replication has been studied extensively, sometimes with conflicting results. Mx1 does not appear to affect uncoating of the virus or transport of the vRNPs into the nucleus (3, 19, 34). Rather, inhibition of primary transcription (26) and of viral mRNA translation (34) has been described for Mx1-expressing cells after interferon (IFN) stimulation and influenza virus infection. Pavlovic and colleagues (37) used a stable Mx1-expressing cell line to confirm that Mx1 suppresses primary viral transcription and thereby excluded other IFN-dependent effects. In later studies, reconstituted vRNPs in viral minireplicon systems have also been used to study the effect of Mx1 on viral transcription and replication. These minireplicon systems consist of a virus-like minigenome containing a reporter gene, e.g., firefly luciferase, and the viral proteins PB1, PB2, PA, and NP. Mx1 can inhibit the polymerase activity of such a system, which means that it targets at least one of its components (6, 21, 53). In vivo studies also demonstrated the importance of Mx1 expression for the protection against influenza A virus infection. Mice expressing Mx1 show lower virus titers and pathology in the infected organs. These studies demonstrate a clear protection by Mx1, even against the highly pathogenic human H5N1 and pandemic 1918 virus strains (42, 49).
Overall, there is strong evidence that Mx1 inhibits the activity of the viral polymerase, which is probably present in ribonucleoprotein complexes, even though the detailed molecular mechanism of this inhibition remains unsolved. Huang and coworkers (21) showed that vaccinia-mediated expression of PB2 was necessary and sufficient to outcompete the anti-influenza virus activity of Mx1 in a minireplicon system. In line with this report, Stranden et al. (47), demonstrated that A2G Mx1+/+ cells become sensitive to influenza A virus infection if they overexpress PB2. These results suggest that Mx1 directly or indirectly targets PB2. However, a direct interaction between PB2 and Mx1 has not been reported (21, 47).
More recent reports suggest that NP also modulates the antiviral activity of the Mx1 protein. Influenza A virus strains vary in their sensitivity to the antiviral effect of the Mx1 protein. Human strains (e.g., A/Panama/2007/99) are more resistant, whereas avian strains (e.g., fowl plague virus) are more susceptible to the effect of murine Mx1. This difference in sensitivity was attributed to the identity of the NP (6). The importance of NP for resistance against Mx1 was confirmed by Zimmermann et al., who compared the more resistant A/H1N1 pandemic (pdm) 2009 strain (A/Hamburg/4/09) with a more sensitive highly pathogenic avian H5N1 isolate [A/Thailand/1(KAN1)/04] (53). These studies point to a relationship between resistance against Mx1 activity and the origin (avian or mammalian) of the NP protein.
How Mx1 mechanistically perturbs the biological function of PB2 and NP is not clear. Here, we focused on the molecular interplay between Mx1, NP, and PB2 to identify key events involved in the antiviral activity of Mx1. We demonstrate that Mx1 disturbs the interaction between NP and PB2 by a mechanism that depends on GTP binding and/or GTPase activity. Furthermore, the degree to which Mx1 inhibits this interaction correlates with the susceptibility of NP variants in the minireplicon system. We further demonstrate that the Mx1 protein can interact with the viral NP and PB2 proteins. In contrast to the antiviral activity of Mx1, this interaction is not dependent on the GTPase activity of Mx1. We therefore propose that the antiviral mechanism is a two-step process: Mx1 binds the influenza ribonucleoprotein complexes and subsequently disrupts the PB2-NP interaction by a process requiring the GTPase activity of Mx1.
MATERIALS AND METHODS
Cells and viruses.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.4 mM Na-pyruvate, nonessential amino acids, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. MDCK cells were maintained in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, and 0.1 mg/ml streptomycin.
We used the following virus strains: A/PR/8/34 (H1N1), A/Swine/Ontario/42729A/01 (H3N3) (24), A/New Caledonia/20/99 (H1N1), A/Swine/Iowa/4/1976 (H1N1), mouse-adapted A/Swine/Belgium/1/98 (H1N1), A/Panama/2007/99 (H3N2), and a primary A/H1N1pdm 2009 influenza virus isolate. Influenza viruses were grown on MDCK cells and purified from the culture supernatant by centrifugation at 25,000 × g for 16 h at 4°C. A/Swine/Iowa/4/1976 was grown on 10-day-old embryonated eggs and purified from the allantois fluid by centrifugation at 25,000 × g for 16 h at 4°C. Virions were resuspended in phosphate-buffered saline containing 20% glycerol.
Plasmids.
The mammalian expression plasmids pCAXL-PB1, -PB2, -PA, and -NP were generated by cloning the coding sequences from plasmids pHW191-PB2, pHW192-PB1, pHW193-PA, and pHW195-NP (derived from A/PR/8/34 [H1N1] [18]). The sequences were cloned in pCAXL (derived from pCAGGS, in which we had generated new restriction sites). A V5 tag (GKPIPNPLLGLDST) was added to the C terminus of PB2 to obtain pCAXL-PB2V5. pHW-NSLuc was constructed by placing the firefly luciferase cDNA in a negative-sense orientation between the noncoding regions of the NS segment (3′ 23 nucleotides and 5′ 26 nucleotides) of PHW198-NS (18), followed by removal of the polymerase II promoter sequence using the SnaBI and HpaI restriction sites. The pRL-CMV plasmid (catalog no. E2261; Promega), which contains a Renilla luciferase gene under the control of a cytomegalovirus (CMV) promoter, was used to normalize for transfection efficiency. The mouse Mx1 cDNA (A2G Mx1 sequence, GenBank accession number NP_034976.1) was cloned in pCAXL. Mx1 mutants K49A (40) and T69A (see below) were derived from pCAXL-Mx1 by site-specific mutagenesis using the fusion PCR method.
RNA was purified from virions of the different influenza strains to generate cDNAs, which were used to clone the NP variants into pCAXL. The NP variants were derived from A/Swine/Ontario/42729A/01 (H3N3), A/New Caledonia/20/99 (H1N1), A/Swine/Iowa/4/1976 (H1N1), mouse-adapted A/Swine/Belgium/1/98 (H1N1), A/Panama/2007/99 (H3N2), and a primary A/H1N1pdm 2009 pandemic influenza virus isolate. The cloned NP sequence of the pandemic influenza isolate is the same as that of strain A/Mexico/InDRE4487/2009, except for amino acid positions A6V, K83R, and T379I.
Influenza A virus minireplicon system.
HEK293T cells (seeded at 5 × 104 cells per well in 24-well plates) were transfected in triplicate with the expression plasmids pCAXL-PB1, -PB2, -PA, and -NP (25 ng each), luciferase reporter pHW-NSLuc (100 ng), and pRL-CMV (25 ng) using the calcium phosphate precipitation method. For titration experiments, increasing amounts of the plasmids encoding PB1, PB2, PA, and NP were used (125 ng, 250 ng, or 625 ng). To assess the effect of Mx1 in the minireplicon system, pCAXL, pCAXL-Mx1, pCAXL-Mx1K49A, or pCAXL-Mx1T69A was cotransfected in the amounts mentioned in the figure legends. Cells were lysed 48 h later with luciferase lysis buffer (25 mM Tris-phosphate, 2 mM dithiothreitol [DTT], 2 mM trans-1,2-cyclohexanediaminetetraacetic acid (CDTA), 10% glycerol, and 1% Triton X-100). Luciferase activity was measured with the Dual-Luciferase reporter assay system (catalog no. E-1960; Promega) according to the manufacturer's instructions, using a GloMax 96 microplate luminometer (Promega). The normalized luciferase activity was calculated as the ratio between the activities of firefly and Renilla luciferase (firefly/Renilla luciferase × 1,000).
Chemical reagents and antibodies.
N-Ethylmaleimide was obtained from Sigma (catalog no. E-3876), and protease inhibitor cocktail tables were from Roche (catalog no. 11 873 580 001). A polyclonal antiserum against mouse Mx1 was generated by immunizing New Zealand White rabbits three times subcutaneously with 100 μg of the synthetic, high-performance liquid chromatography-purified peptide CKKFLKRRLLRLDEARQKLAKFSD (C terminus of Mx1) combined with the water-in-oil adjuvant Montanide ISA-720 (SEPPIC SA, Paris, France). The serum IgG fraction was enriched by 50% ammonium sulfate precipitation followed by affinity chromatography with a protein A column (GE Healthcare). Monoclonal anti-V5-horseradish peroxidase (HRP)-conjugated antibody was purchased from Invitrogen (catalog no. R96125). The following reagents were obtained from the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: monoclonal anti-influenza A virus nucleoprotein (NP) antibody, clones A1 and A3 (ascites blend, mouse) and NR-4282, and polyclonal anti-influenza virus RNP antibody, A/Scotland/840/74 (H3N2), (antiserum, goat), NR-3133. The monoclonal anti-influenza A virus PB2 antibody (clone 170-3D5) was kindly provided by J. Yewdell, NIH, Bethesda, MD. Monoclonal mouse antiactin antibody was purchased from MP Biomedicals Europe N.V. (catalog no. 691002). Fluorescently labeled Alexa Fluor 488 donkey anti-rabbit IgG (catalog no. A-21206), Alexa Fluor 488 donkey anti-mouse IgG (catalog no. A-21202), and Alexa Fluor 555 donkey anti-goat IgG (catalog no. A21432) were purchased from Life Technologies Europe B.V.
Coimmunoprecipitation.
HEK293T cells (seeded at 1.2 × 106 cells per 9-cm dish) were transfected by the calcium phosphate method with equal amounts of the expression plasmids pCAXL-PB1, -PB2V5, -PA, -NP, and pHW-NSLuc: 1 μg of each of these plasmids was transfected to assess a dose response of increasing amounts of cotransfected pCAXL-Mx1 (0 μg, 0.5 μg, 1 μg, 2 μg, or 5 μg). To compare the effects of different Mx1 mutants, HEK293T cells were transfected with 0.5 μg each of pCAXL-PB1, -PB2V5, -PA, -NP, and pHW-NSLuc together with an empty control plasmid or expression vectors of the different Mx1 mutants (2 μg of pCAXL, 1 μg of pCAXL-Mx1WT [expressing wild-type Mx1], 2 μg of pCAXL-Mx1K49A, or 1 μg of pCAXL-Mx1T69A). Total lysates were prepared 24 h after transfection in low-salt lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, and a protease inhibitor cocktail without [to analyze PB2-NP association] or with [for Mx1 association with PB2 or NP] 25 mM N-ethylmaleimide). The cells were lysed for 20 min on ice and centrifuged for 3 min at 16,000 × g to remove insoluble proteins. PB2V5 and NP were immunoprecipitated from the cleared lysates with anti-V5-HRP and monoclonal anti-NP antibodies, respectively, for 3 h at 4°C. Immune complexes were collected by incubation for 1 h at 4°C in the presence of protein G Sepharose beads (catalog no. 17-0618-01; GE Healthcare) followed by centrifugation. Immunoprecipitates were washed 4 times with high-salt lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM EDTA, and 1% NP-40). Proteins were eluted from the beads by boiling at 99°C for 13 min in 2× Laemmli buffer. The samples were separated by SDS-PAGE (8%), and the relevant bands were visualized by Western blotting with antibodies directed against the V5 tag, NP (polyclonal goat anti-RNP antibody), or Mx1.
Coimmunoprecipitation from mixed lysates.
HEK293T cells were seeded at 1.2 × 106 cells per 9-cm dish and transfected with 2.5 μg pCAXL-Mx1 (Mx1 lysates), 2.5 μg pCAXL (lysates with no vRNPs), or 1 μg each of pCAXL-PB1, -PB2V5, -PA, and -NP and 1 μg of pHW-NSLuc (lysates with transfected vRNPs). Lysates from transfected cells were prepared 30 h after transfection. To obtain lysates containing vRNPs from infected cells, HEK293T cells were infected with A/PR/8/34 (multiplicity of infection [MOI] of 10). The virus was allowed to attach to the cells for 1 h at 4°C, and the cells with the inoculum virus in place were subsequently incubated at 37°C for 4 h. Transfected and infected cells were lysed for 20 min on ice in low-salt lysis buffer containing NEM (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 25 mM N-ethylmaleimide, and a protease inhibitor cocktail). The lysates were centrifuged for 3 min at 16,000 × g to remove insoluble proteins. vRNPs were isolated from A/PR/8/34 virions (6.25 × 108 PFU/ml lysis buffer) in low-salt lysis buffer containing N-ethylmaleimide to release the vRNPs. The lysates were centrifuged for 3 min at 16,000 × g to remove insoluble proteins. Lysates were then mixed (Mx1 plus vRNPs) and immediately used for immunoprecipitation with monoclonal anti-NP antibody for 17 h at 4°C. Immune complexes were collected by incubation for 1 h at 4°C in the presence of protein G Sepharose beads (catalog no. 17-0618-01; GE Healthcare) followed by centrifugation. Immunoprecipitates were isolated and washed as described above and visualized by Western blotting using antibodies directed against NP (polyclonal goat anti-RNP antibody) or Mx1.
FLIM.
HEK293T cells (seeded at 2 × 104 cells per well of a Lab-Tek chambered coverglass with 8 wells) were transfected with 25 ng each of pCAXL-PB1, -PB2V5, -PA, -NP, and pHW-NSLuc using the calcium phosphate method. In addition, 125 ng of pCAXL or pCAXL-Mx1 was cotransfected. After 24 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and stained to reveal PB2V5 (with anti-V5-HRP antibody diluted 1/1,500 and Alexa Fluor 488-labeled donkey anti-mouse IgG diluted 1/600 as donor) and NP (with polyclonal anti-RNP antibody diluted 1/4,000 and Alexa Fluor 555-labeled donkey anti-goat IgG diluted 1/600 as acceptor). In the control setting without acceptor, the Alexa Fluor 555-labeled secondary antibody was left out of the staining. Fluorescence lifetime-imaging microscopy (FLIM) measurements were performed with a confocal microscope (Fluoview 1000 Olympus) equipped with a FLIM module (Picoquant, Berlin, Germany). This module consists of a pulsed diode laser, PDL-800-B 440 nm, a single-photon avalanche diode with a time resolution of 10 ps, and a time-correlated single-photon counting unit, PicoHarp 200. The software that controls the FLIM module and synchronizes it with the confocal microscope is Symphotime (Picoquant). To detect the fluorescence of Alexa Fluor 488, a 520/35-25 bandpass filter was positioned in front of the FLIM detector. The cells were visualized with a 60×/1.2 UplanSapo water immersion objective. For each setting, 8 to 12 fields of 64 by 64 pixels were measured, covering approximately 30 cells per experimental setup. The mean fluorescent lifetime for each field was calculated, and the averages of different conditions were compared. Statistical analysis was done by performing a Kruskal-Wallis test.
Mx1 immunofluorescence.
HEK293T cells (5 × 104 cells per well in 24-well plates seeded on glass coverslips) were transfected using the calcium phosphate method with 50 ng of pCAXL-Mx1WT, -Mx1K49A, or -Mx1T69A. After 24 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and stained for Mx1 with anti-Mx1 antibody (diluted 1/1,000) and Alexa Fluor 488-labeled donkey anti-rabbit IgG (diluted 1/600). Cell nuclei were visualized with Hoechst stain (diluted 1/1,000, catalog no. H21492; Invitrogen). Images were recorded with a confocal microscope (Leica Sp5 AOBS confocal system) using a 63× HCX PL Apo 1.4 oil immersion objective. A 510-to-555-nm bandpass filter was positioned before the detector to measure the fluorescence of Alexa 488 after excitation with an Argon 488-nm laser.
RESULTS
PB2 and NP counteract Mx1 activity in an influenza minireplicon system.
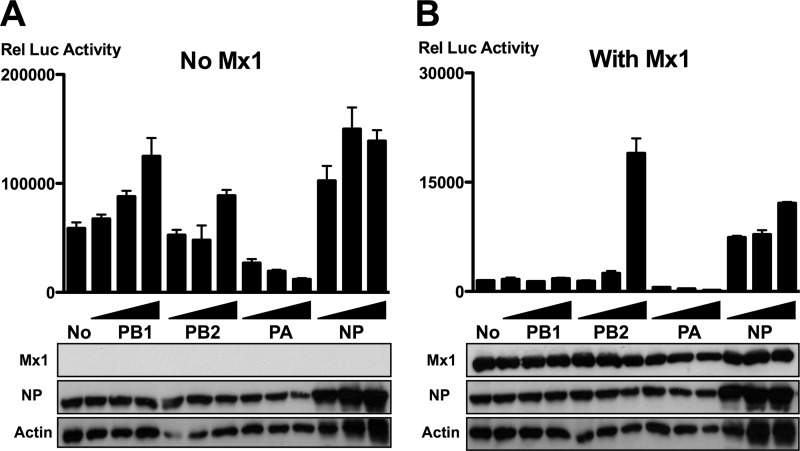
To quantify the inhibition of the influenza A virus polymerase complex by mouse Mx1 protein, we used a minireplicon system based on reconstituted vRNPs from A/PR/8/34. HEK293T cells were transfected with expression plasmids encoding the components of the vRNP complex (PB1, PB2, PA, and NP, as well as a firefly luciferase vRNA-like reporter) in combination with either an Mx1 expression plasmid or a control plasmid. As expected, coexpressed Mx1 strongly reduced the luciferase reporter activity in this assay (Fig. 1A and B; compare lanes 1). To investigate which components of the vRNP complex are involved in the Mx1-mediated repression of vRNP activity, increasing amounts of each polymerase subunit or NP were introduced into the minireplicon system in the absence (Fig. 1A) or presence (Fig. 1B) of Mx1. In the absence of Mx1, increasing amounts of PB1, PB2, or NP increased the polymerase activity, whereas increasing amounts of PA lowered the polymerase activity (Fig. 1A). This inhibitory effect of PA has been described before and is probably due to the expression of the PA-X protein (16, 22, 43, 44). Coexpression of increasing amounts of PB2 and, to some extent, of NP counteracted the inhibition of viral polymerase activity by Mx1 (Fig. 1B). This observation is in line with previous results and suggests that PB2 and NP are directly involved in Mx1-dependent inhibition of influenza A virus replication (6, 21, 47, 53).
Fig 1.
PB2 and NP outcompete Mx1 activity. HEK293T cells were transfected in triplicate with PB1, PB2, PA, and NP expression plasmids (25 ng each), together with pHW-NSLuc (100 ng) and pRL-CMV (25 ng). In addition, 125 ng of pCAXL (A) or pCAXL-Mx1 (B) was cotransfected. Increasing amounts of PB1, PB2, PA, or NP were introduced by increasing the amount of the corresponding expression vectors (125 ng, 250 ng, or 625 ng). The normalized luciferase activity in the lysates was determined 48 h after transfection. Bars represent the average of the triplicates, and the error bars depict one standard deviation. This graph is representative of three independent experiments. Mx1, NP, and actin expression were determined in the lysates by Western blotting.
Mx1 inhibits the interaction between PB2 and NP.
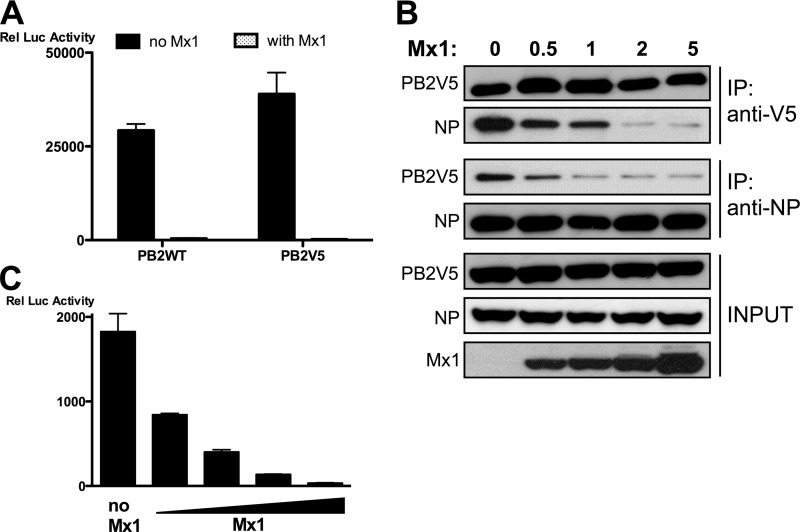
Interaction between PB2 and NP is essential for the formation of an enzymatically active influenza RNA-dependent RNA polymerase (RdRp) in ribonucleoprotein complexes (1). Therefore, we analyzed the PB2-NP interaction in the presence or absence of Mx1 by coimmunoprecipitation after transfection of all the components of the minireplicon system in HEK293T cells. We were unable to pull down unmodified PB2 from the complex using a monoclonal anti-PB2 antibody (monoclonal antibody 170-3D5; kindly provided by J. Yewdell, NIH, Bethesda, MD), presumably because the N-terminal 170-3D5 epitope in PB2 was shielded by interaction with PB1 or PA (17, 39, 48). Therefore, we used V5-tagged PB2 (PB2V5) and unmodified NP to assess whether they coexist in one complex. The V5 tag of PB2V5 did not affect its polymerase activity in the minireplicon system or its sensitivity to Mx1-mediated inhibition (Fig. 2A). We performed pulldown experiments in both directions, i.e., (i) pull down of PB2V5 followed by analysis of coimmunoprecipitated NP and (ii) pull down of NP followed by analysis of coimmunoprecipitated PB2V5. Analysis of these immunoprecipitates revealed that PB2V5 and NP reside in the same complex, most likely the functional ribonucleoprotein complex (Fig. 2B, lane 1). We found that this interaction was inhibited by Mx1 in a dose-dependent manner (Fig. 2B, lanes 2 to 5). The degree of inhibition of the PB2-NP interaction by Mx1 correlated with the antiviral activity of Mx1, as determined by minireplicon-driven reporter gene activity (Fig. 2C). Taken together, these data suggest that Mx1 can reduce influenza A virus polymerase activity by inhibiting the PB2-NP interaction.
Fig 2.
Mx1 inhibits the interaction between PB2 and NP. (A) HEK293T cells were transfected in triplicate with PB1, PA, and NP expression plasmids (25 ng each), together with pHW-NSLuc (100 ng) and pRL-CMV (25 ng). In addition, 25 ng pCAXL-PB2 or pCAXL-PB2V5 and 125 ng of pCAXL or pCAXL-Mx1 were cotransfected. The relative luciferase activity in the lysates was determined 48 h after transfection. Bars represent the average of the triplicates, and the error bars depict one standard deviation. This graph is representative of two independent experiments. (B) HEK293T cells were transfected with plasmids for expression of PB1, PB2V5, PA, and NP (1 μg each) and with pHW-NSLuc (1 μg). In addition, increasing amounts of pCAXL-Mx1 were cotransfected (0 μg, 0.5 μg, 1 μg, 2 μg, or 5 μg). Total lysates were made 24 h after transfection, and PB2V5 and NP were immunoprecipitated (IP) with anti-V5 and anti-NP antibodies, respectively. Proteins were visualized by Western blotting with antibodies recognizing the V5 tag, NP (anti-RNP antibody), and Mx1. (C) HEK293T cells were transfected in triplicate with plasmids for expression of PB1, PB2, PA, and NP (25 ng each), together with pHW-NSLuc (100 ng) and pRL-CMV (25 ng). Increasing amounts of pCAXL-Mx1 were cotransfected (0 ng, 12.5 ng, 25 ng, 50 ng, or 125 ng). The normalized luciferase activity in the lysates was determined 48 h after transfection. Bars represent the average of triplicates, and the error bars depict one standard deviation.
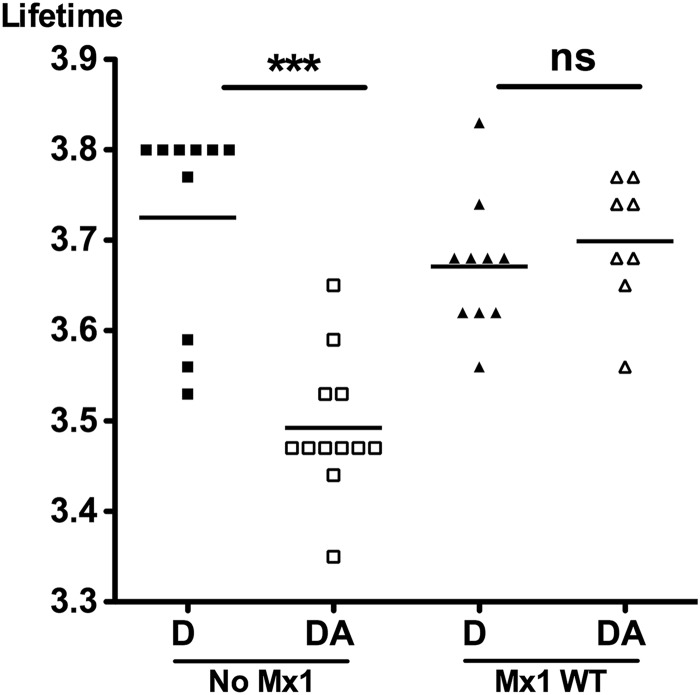
We also studied the PB2-NP interaction by an alternative method, called fluorescence lifetime-imaging microscopy (FLIM). This technique is based on the energy transfer (fluorescence resonance energy transfer [FRET]) between two fluorophores that are close together (<10 nm). PB2V5 and NP were labeled in situ with antibodies, each carrying a different fluorophore but with overlapping spectra. If both antibodies are sufficiently close together, excitation of the donor fluorophore (coupled to PB2) emits fluorescence that can excite the acceptor fluorophore (coupled to NP). This energy transfer results in a shortening of the lifetime of the donor fluorophore, the characteristic that is measured in FLIM (15). The advantage of FLIM over conventional FRET analysis is that the fluorescence lifetime does not depend on the concentration or brightness of the fluorophores. We transfected cells with expression vectors for PB1, PB2V5, PA, and NP and the vRNA-like reporter to reconstitute the viral ribonucleoprotein complexes, either together with or without pCAXL-Mx1. We labeled the proteins PB2V5 and NP with the fluorophores Alexa Fluor 488 (donor) and Alexa Fluor 555 (acceptor), respectively. The lifetime of the donor was measured in the absence and in the presence of the Alexa Fluor 555 acceptor. In the absence of Mx1, the fluorescence lifetime was reduced significantly, from 3.725 ns to 3.493 ns, by the presence of the acceptor, demonstrating an interaction between PB2V5 and NP. However, in the presence of Mx1, the fluorescence lifetime of the donor fluorophore was restored (lifetime from 3.671 ns to 3.699 ns), indicating inhibition of the interaction between PB2V5 and NP (Fig. 3). Confocal imaging of the donor and acceptor showed that both proteins are expressed mainly in the nucleus (data not shown). This indicates that the interaction between PB2 and NP and disruption of this interaction by Mx1, as determined by FLIM, occur mainly in the nucleus.
Fig 3.
Mx1 inhibits the interaction between PB2 and NP as determined by FLIM. HEK293T cells were transfected with 25 ng each of plasmids for expression of PB1, PB2V5, PA, and NP and pHW-NSLuc. In addition, 125 ng of pCAXL or pCAXL-Mx1 was cotransfected. After 24 h, cells were fixed and stained for PB2V5 (anti-V5 antibody with Alexa Fluor 488 as FLIM donor) and NP (anti-NP antibody with Alexa Fluor 555 as FLIM acceptor). For each setting, FLIM measurements were performed on 8 to 12 fields (altogether about 30 cells) and the mean fluorescence lifetime per field was determined. For each setting, the average of the means was calculated and statistical analysis was performed with a Kruskal-Wallis test (***, P < 0.001). D, donor only; DA, donor and acceptor.
An intact GTPase domain of Mx1 is required for inhibiting the PB2-NP interaction.
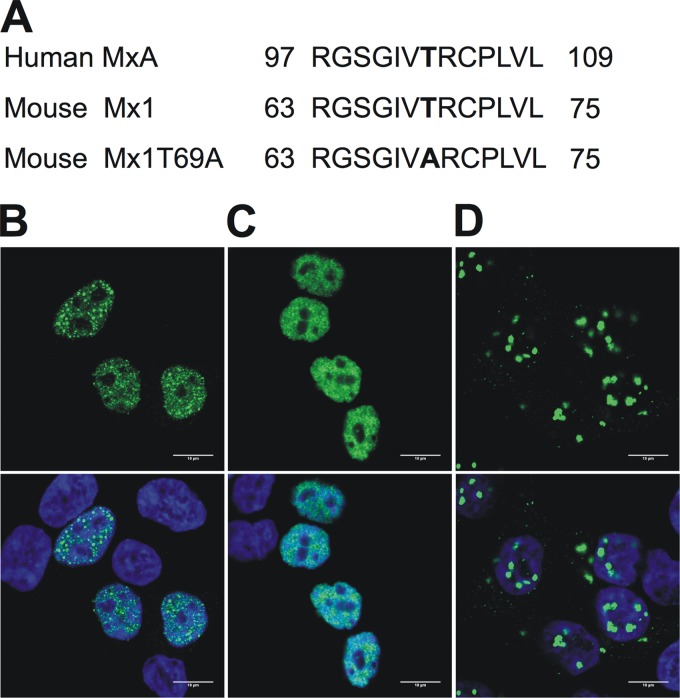
The GTPase activity of Mx1, or at least its GTP-binding activity, is important for its antiviral function (40). Therefore, we tested whether this activity is also important for inhibition of the PB2-NP interaction. We used two Mx1 mutants to investigate this question. The first, Mx1K49A, has a mutation in one of the three elements forming the GTP-binding domain and is therefore defective in GTP binding (40). The second, Mx1T69A, was generated on the basis of the previously described human MxAT103A mutant (Fig. 4A), which lacks both GTPase activity and GTP-binding capacity (41). It is unclear whether the mouse Mx1T69A mutant can still bind GTP, as this function is lost in the corresponding MxAT103A mutant (41) but is still present in the corresponding MxBT151A mutant (25). Since nuclear localization of mouse Mx1 is important for its antiviral activity (55), we first determined the subcellular localization of the different Mx1 mutants by immunofluorescence confocal microscopy (Fig. 4B to D). In transfected HEK293T cells, wild-type Mx1 (Mx1WT) shows a punctuate nuclear staining, in accordance with previous findings (7). We noted that transfected cells with higher Mx1 expression levels also displayed cytoplasmic Mx1 (not shown). The Mx1K49A mutant showed exclusively nuclear staining, which was more diffuse than that of Mx1WT. Mx1T69A appeared as relatively large aggregates both in the nucleus and in the cytoplasm.
Fig 4.
GTPase-deficient Mx1 mutants. (A) Alignment of the amino acid sequence of the human MxAT103A mutant, wild-type mouse Mx1, and the mouse Mx1T69A mutant. The mutation is localized between the first and second GTP-binding consensus motifs. Numbers denote the position in the primary amino acid sequence. (B to D) Subcellular localization of Mx1 (B), Mx1K49A (C), and Mx1T69A (D). HEK293T cells were transfected with pCAXL-Mx1WT, -Mx1K49A, or -Mx1T69A (50 ng). After 24 h, cells were fixed and stained with Hoechst stain (DNA, blue, not shown) and anti-Mx1 antibody (green, top). An overlay was made of the two images (bottom). Scale bar, 10 μm.
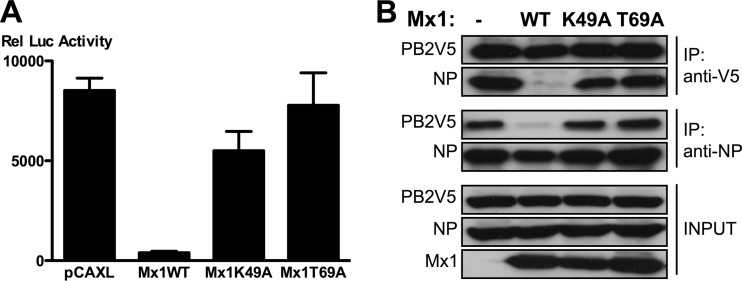
In contrast to Mx1WT, both Mx1K49A and -T69A largely failed to inhibit reporter gene expression in the minireplicon system (Fig. 5A). As we noted that Mx1K49A expression levels were consistently lower than those of Mx1 and Mx1T69A, we doubled the amount of transfected Mx1K49A plasmid to obtain similar Mx1 protein levels in our minireplicon and immunoprecipitation assays. We next analyzed the effect of WT and mutant Mx1 proteins on the PB2-NP interaction in a coimmunoprecipitation experiment. Whereas wild-type Mx1 protein significantly blocked the interaction between PB2 and NP, neither Mx1K49A nor Mx1T69A interfered with this interaction (Fig. 5B). These results indicate that inhibition of the PB2-NP interaction by Mx1 is dependent on its GTP binding and/or GTPase activity.
Fig 5.
An intact GTPase domain of Mx1 is required to inhibit the PB2-NP interaction. (A) HEK293T cells were transfected in triplicate with PB1, PB2, PA, and NP expression plasmids (25 ng each), pHW-NSLuc (100 ng), and pRL-CMV (25 ng). Empty control and expression vectors for the different Mx1 mutants were cotransfected (50 ng of pCAXL, 50 ng of pCAXL-Mx1WT, 100 ng of pCAXL-Mx1K49A, or 50 ng of pCAXL-Mx1T69A). After 48 h, the relative luciferase activity in the lysates was determined. Bars represent the average of triplicates, and the error bars depict one standard deviation. This graph is representative of at least three independent experiments. (B) HEK293T cells were transfected with PB1, PB2V5, PA, and NP expression plasmids (0.5 μg each) and with pHW-NSLuc (0.5 μg). Empty control and expression vectors for the different Mx1 mutants were cotransfected (2 μg of pCAXL, 1 μg of pCAXL-Mx1WT, 2 μg of pCAXL-Mx1K49A, or 1 μg of pCAXL-Mx1T69A). Total lysates were made 24 h after transfection, and PB2V5 and NP were immunoprecipitated with anti-V5 and anti-NP antibodies, respectively. Proteins were visualized by Western blotting with antibodies directed against the V5 tag, NP (anti-RNP antibody), or Mx1.
Mx1 interacts with PB2 and NP.
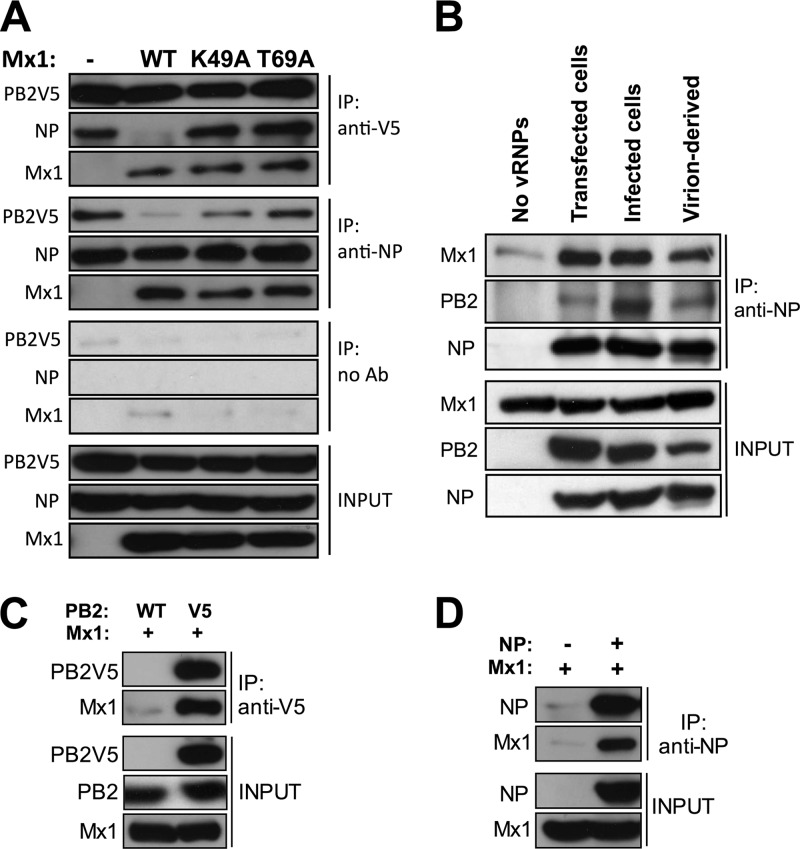
Mx1 might have inhibited the PB2-NP interaction by forming a competing complex with the heterotrimeric influenza RdRp or with the fraction of NP that is associated with the polymerase. However, we could not demonstrate the presence of coexpressed Mx1 in the NP or PB2 immunoprecipitates when we used the lysis and coimmunoprecipitation conditions under which we demonstrated the PB2-NP interaction and its absence in the presence of Mx1. To optimize the lysis and immunoprecipitation conditions, we added N-ethylmaleimide (NEM) to the lysis buffer because NEM can inhibit the GTPase activity of the related dynamin protein (45) and, probably, the GTPase activity of Mx1 as well. Using these conditions, we found that Mx1 coimmunoprecipitated with PB2V5 and NP in lysates prepared from cells that had been transfected with the minireplicon components. This result indicates that Mx1 interacts with PB2 and with NP. Remarkably, Mx1K49A and Mx1T69A coimmunoprecipitated as efficiently as wild-type Mx1 coimmunoprecipitated with PB2V5 and NP. Nevertheless, neither of the Mx1 mutants disturbed the PB2-NP interaction (Fig. 6A). Taken together, these results suggest that although Mx1 can interact with PB2 and NP independently of its GTPase activity, it requires this activity to inhibit the PB2-NP interaction (Fig. 6A).
Fig 6.
Mx1 interacts with PB2 and NP. (A) HEK293T cells were transfected with PB1, PB2V5, PA, and NP expression plasmids (0.5 μg each) and with pHW-NSLuc (0.5 μg). Empty control and expression vectors for the different Mx1 mutants were cotransfected (2 μg of pCAXL, 1 μg of pCAXL-Mx1WT, 2 μg of pCAXL-Mx1K49A, or 1 μg of pCAXL-Mx1T69A). After 24 h, total lysates were made in the presence of 25 mM N-ethylmaleimide, and PB2V5 and NP were immunoprecipitated with anti-V5 and anti-NP antibodies, respectively. We also included a control immunoprecipitation in the absence of monoclonal antibody (no Ab). Proteins were visualized by Western blotting with antibodies recognizing the V5 tag, NP (anti-RNP antibody), and Mx1. The results shown are representative of two independent experiments. (B) HEK293T cells were transfected with 2.5 μg pCAXL-Mx1 (Mx1 lysates), with 2.5 μg pCAXL (lysates without vRNPs), or with 1 μg of pCAXL-PB1, -PB2V5, -PA, and -NP and 1 μg pHW-NSLuc (lysates with transfected vRNPs). To obtain lysates containing vRNPs from infected cells, HEK293T cells were infected for 4 h with A/PR/8/34 (MOI of 10). vRNPs isolated from virions were generated by lysing influenza A virions (A/PR/8/34, 6.25 × 108 PFU/ml lysis buffer). Lysates from all conditions were made in the presence of 25 mM N-ethylmaleimide. The mixed lysates (Mx1 plus vRNPs) were immunoprecipitated with monoclonal anti-NP antibody. Proteins were visualized by Western blotting with antibodies recognizing NP (anti-RNP antibody), PB2, or Mx1. The results shown are representative of three independent experiments. (C) HEK293T cells were transfected with 2 μg pCAXL-Mx1 and 1 μg pCAXL-PB2V5 or -PB2WT as control. After 24 h, total lysates were made in the presence of 25 mM N-ethylmaleimide, and PB2V5 was immunoprecipitated with anti-V5 antibody. Proteins were visualized by Western blotting with antibodies recognizing the V5 tag, PB2, and Mx1. The results shown are representative of two independent experiments. (D) HEK293T cells were transfected with 2 μg pCAXL-Mx1 and 1 μg pCAXL-NP or pCAXL as control. After 24 h, total lysates were made in the presence of 25 mM N-ethylmaleimide, and NP was immunoprecipitated with anti-NP antibody. Proteins were visualized by Western blotting with antibodies recognizing NP (anti-RNP antibody) and Mx1. The results shown are representative of two independent experiments.
Next, we investigated whether Mx1 can also interact with vRNPs isolated from infected cells or from detergent-disrupted virions. We also used vRNPs isolated from cells that had been transfected with the minireplicon components or with an empty control plasmid (no vRNPs). To examine the interaction between Mx1 and vRNPs, we combined cell lysates containing Mx1 with lysates containing vRNPs (both lysates contained NEM) and used these mixtures in a coimmunoprecipitation experiment. This experiment revealed that Mx1 coimmunoprecipitated with NP from all three vRNP sources tested (Fig. 6B). These results suggest that Mx1 can interact with the viral NP protein in infected cells and that it might target incoming virion-derived vRNPs, leading to suppression of viral transcription and replication.
The aforementioned experiments clearly demonstrate an interaction between Mx1 and PB2 or NP when all components of the minireplicon system are present. We wondered if Mx1 could interact with PB2 or NP in the absence of other viral proteins. To address this question, we expressed Mx1 together with PB2V5 or NP in the absence of the other minireplicon components and immunoprecipitated PB2V5 (Fig. 6C) or NP (Fig. 6D) with anti-V5 or anti-NP antibodies, respectively. In both cases, Mx1 could be coimmunoprecipitated, demonstrating that Mx1 can interact with PB2V5 and NP in the absence of other viral proteins.
Natural NP variants display different sensitivities to Mx1.
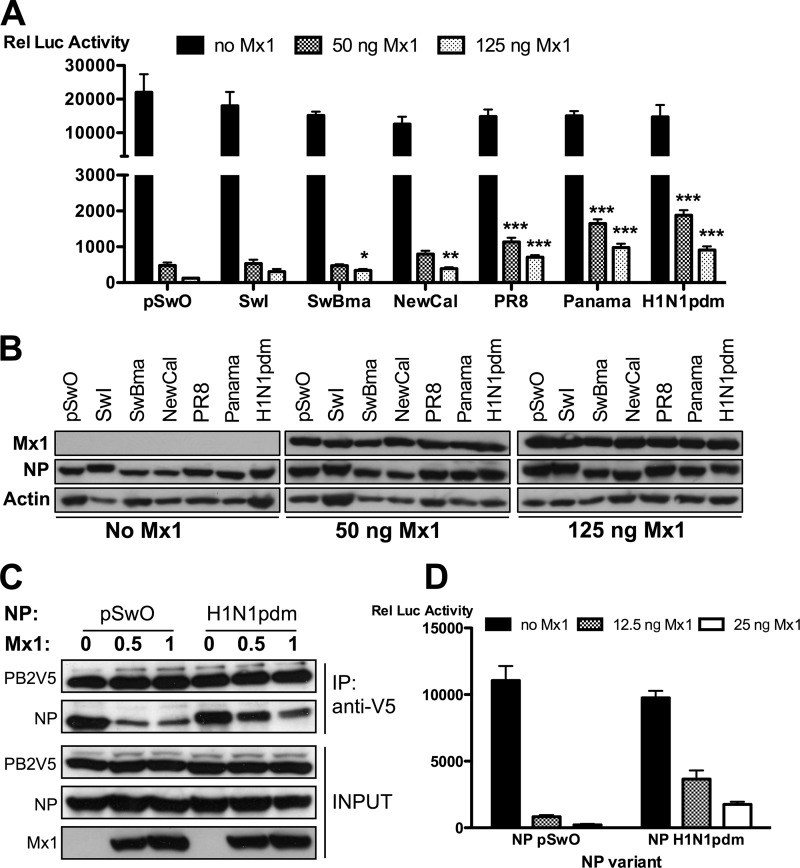
It has been reported that the NPs of various influenza A virus strains vary in their sensitivity to the activity of the Mx1 protein in a minireplicon system (6, 53). In general, NP expressed by human influenza A viruses is more resistant to Mx1 than NP from avian viruses. Since we demonstrated a correlation between Mx1 activity and disruption of the PB2-NP interaction, we next investigated whether the reported differential sensitivity of NP variants is also reflected in our coimmunoprecipitation assay. We compared NP proteins from seven different influenza strains for their Mx1 sensitivity in the minireplicon system. In the absence of Mx1, the relative luciferase activities were comparable, indicating that the PR8 virus (A/Puerto Rico/8/34)-derived polymerase components used in our minireplicon assay are equally functional in combination with the different NP variants that we tested (Fig. 7A). However, we observed a significant differential sensitivity of the NP variants for the inhibitory activity of Mx1 (Fig. 7A). The outcome of the minireplicon assay made it possible to arrange the different NP variants based on their Mx1 sensitivity. Starting with the most sensitive NP variant, the order is as follows: NP-pSwO > NP-pSwI, NP-SwBma > NP-NewCal > NP-PR8 > NP-panama, NP-H1N1pdm (pSwO, A/Swine/Ontario/42729A/01; pSwI, A/Swine/Iowa/4/1976; SwBma, mouse-adapted A/Swine/Belgium/1/98; NewCal, A/New Caledonia/20/99; Panama, A/Panama/2007/99; H1N1pdm, A/Mexico/InDRE4487/2009). This comparison demonstrates that NP derived from an avian influenza virus isolated from a pig (NP-pSwO) (24) is the most sensitive and that NP derived from the 2009 pandemic H1N1 virus (NP-H1N1pdm) is the most resistant to Mx1 inhibition. To further validate the association between inhibition of the polymerase activity by Mx1 and disruption of the PB2-NP interaction, the effect of Mx1 on the PB2–NP-pSwO interaction was compared with the effect of Mx1 on the PB2–NP-H1N1pdm interaction. In the absence of Mx1, the two NP variants coimmunoprecipitated with PB2V5 with equal efficiency (Fig. 7B). However, in agreement with the differential sensitivity to Mx1 in the minireplicon system, the interaction between PB2V5 and pandemic H1N1pdm-derived NP was less affected by coexpressed Mx1 than the interaction of PB2V5 with NP of pSwO (Fig. 7B). This result agrees with the relative resistance of these two NP variants in the minireplicon system when increasing amounts of Mx1 were introduced (Fig. 7C). We conclude that the ability of Mx1 to inhibit the PB2-NP interaction correlates with its antiviral potency.
Fig 7.
Natural NP variants display differential sensitivity to Mx1. (A and B) HEK293T cells were transfected in triplicate with expression plasmids for PB1, PB2, and PA (25 ng each), pHW-NSLuc (100 ng), and pRL-CMV (25 ng). In addition, 25 ng of expression plasmids for different NP variants were cotransfected (pSwO, A/Swine/Ontario/42729A/01; SwI, A/Swine/Iowa/4/1976; SwBma, mouse-adapted A/Swine/Belgium/1/98; NewCal, A/New Caledonia/20/99; PR8, A/Puerto Rico/8/34; Panama, A/Panama/2007/99; H1N1pdm, A/Mexico/InDRE4487/2009). Increasing amounts of pCAXL-Mx1 were cotransfected (0 ng, 50 ng, and 125 ng). (A) The normalized luciferase activity in the lysates was determined 48 h after transfection. Bars represent the average of triplicates, and the error bars depict one standard deviation. This graph is representative of three independent experiments. In the absence of Mx1 (black bars), no significant difference between the NP variants was observed (P = 0.0528). In the presence of Mx1, the NP variants differed significantly in their sensitivity to the inhibitory activity of Mx1. Results for all groups were compared with results for the most sensitive variant, NP-pSwO. NP variants whose results differ significantly are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Statistical analysis was performed with a nonparametric one-way analysis of variance followed by Bonferroni's multiple comparison test. (B) Mx1, NP (polyclonal anti-RNP antibody), and actin expression were determined in the lysates by Western blotting. (C) HEK293T cells were transfected with 1 μg each of expression plasmids for PB1, PB2V5, PA, and NP (from pSwO or H1N1pdm) and with 1 μg pHW-NSLuc. Increasing amounts of pCAXL-Mx1 were cotransfected (0 μg, 0.5 μg, and 1 μg). Total lysates were made 24 h later, and PB2V5 was immunoprecipitated by an anti-V5-HRP antibody. Proteins were visualized by Western blotting with antibodies specific for the V5 tag, NP (anti-RNP antibody), and Mx1. (D) HEK293T cells were transfected in triplicate with expression plasmids for PB1, PB2, and PA (25 ng each), together with pHW-NSLuc (100 ng) and pRL-CMV (25 ng). In addition, 25 ng of pCAXL-NP-pSwO or 25 ng of pCAXL-NP-H1N1pdm was cotransfected. Increasing amounts of pCAXL-Mx1 were cotransfected (0 ng, 12.5 ng, and 25 ng). The normalized luciferase activity in the lysates was determined 48 h after transfection. Bars represent the average of triplicates, and the error bars depict one standard deviation.
DISCUSSION
Mx1 inhibits influenza polymerase activity, but the viral targets of this inhibition and the molecular mechanisms involved are still largely unknown. In previous studies, two components of the viral ribonucleoprotein complex have been proposed as the Mx1 target: PB2 and NP (6, 21, 47, 53). We confirmed the importance of these two proteins by noting that overexpression of PB2 or NP decreases Mx1 activity in the minireplicon system (Fig. 1). This indicates that Mx1 may interact with at least one of these proteins or that they compete for the same cellular factor required for proper viral nucleoprotein complex assembly and activity. We focused on the molecular interplay between Mx1, NP, and PB2 to identify the mechanism of the antiviral activity of Mx1. To this end, we investigated the interaction of Mx1 with components of the ribonucleoprotein complex and the interactions within this complex. We demonstrate that the interaction between PB2 and NP is inhibited by the presence of Mx1 (Fig. 2B). The degree of inhibition depended on the Mx1 dose and correlated with the inhibitory effect of Mx1 on the polymerase activity in the minireplicon system (Fig. 2C). This correlation strongly suggests that the loss of this PB2-NP interaction might be crucial for the mechanism of Mx1 activity against influenza A viruses.
As the GTPase activity of Mx1 is required for its antiviral function (40), we investigated whether Mx1 also requires its GTPase activity to inhibit the PB2-NP interaction. Mutations that abrogate the GTPase and antiviral activities of Mx1 did not interfere with the PB2-NP interaction (Fig. 5). This observation indicates that the disruption is an active process that requires enzymatically active Mx1 protein. Alternatively, the GTPase activity or the GTP-binding activity could be necessary to keep the Mx1 protein in the conformation required for interference with the PB2-NP interaction. As the interaction between PB2 and PB1 is not influenced by the presence of the Mx1 protein, Mx1 inhibits the interaction between PB2 and NP but probably leaves the ternary RdRp complex intact (data not shown). This finding is in line with the results of Stranden et al. (47), who also did not observe an effect of Mx1 on the integrity of the RdRp. This suggests that Mx1 inhibits the interaction between the RdRp and the NP protein in ribonucleoprotein complexes (vRNPs). If this hypothesis is correct, it is quite possible that Mx1 also inhibits the interaction between PB1 and NP (1), but this remains to be investigated.
We cannot exclude the possibility that the observed inhibition of the PB2-NP interaction by Mx1 is (partly) attributable to a reduced production of de novo-synthesized vRNPs as a result of the inhibitory activity of Mx1 on the viral RNA polymerase.
In an attempt to stabilize the surmised interaction between Mx1 and PB2 or NP, we added GTPγS, a nonhydrolyzable GTP analog, during immunoprecipitation. However, by using this approach, we could not document an association between Mx1 and RdRp or NP. As an alternative, we tried to stabilize the active conformation of Mx1 by adding NEM to the lysis buffer. NEM can inhibit the GTPase activity of the related dynamin protein (45) and, probably, the GTPase activity of Mx1 as well. Inhibition of Mx1 GTPase activity could block Mx1 in a GTP-bound state, allowing detection of its interaction with vRNP components. We demonstrate that in the presence of NEM, Mx1 interacts with both PB2 and NP (Fig. 6A). This suggests that Mx1 has a direct effect on the polymerase complex, which contains PB2 and NP. The observed interaction between Mx1 and both PB2 and NP also explains why both proteins can titrate out the Mx1 activity when overexpressed in the minireplicon system. Including NEM in the lysis buffer also allowed us to demonstrate an interaction between Mx1 and the viral NP protein present in vRNPs isolated from infected cells or from virions (Fig. 6B). The strongest interaction was seen with vRNPs isolated from infected cells when compared to the interaction with vRNPs from virions. This result might indicate the need for an additional cellular factor for the interaction between Mx1 and vRNPs or the involvement of a modification of the vRNPs inside the cell, e.g., phosphorylation, that facilitates the interaction with Mx1. Other viral proteins might also interfere with the interaction of Mx1 with the vRNPs isolated from virions, e.g., the viral M1 protein, which is released in the endosomes during viral entry. The seemingly more efficient coimmunoprecipitation of Mx1 with vRNPs isolated from infected cells could also reflect an additional interaction between Mx1 and free NP protein that is produced during infection. We could not examine the interaction between Mx1 and the viral PB2 protein because of the lack of a suitable antibody to pull down untagged PB2.
It is not clear why the interactions between Mx1 on the one hand and PB2 and NP on the other cannot be detected in the absence of NEM. NEM might preserve the conformation of viral proteins or a cellular factor that mediates the interaction between Mx1 and PB2 or NP. It is also possible that the interactions are too transient or too weak, and inhibiting the GTPase activity of Mx1 by using NEM could stabilize these interactions. However, as the presence of NEM is also required to demonstrate the interaction of the two GTPase-deficient Mx1 variants with PB2 and NP, a specific effect of NEM on Mx1 GTP binding or GTPase activity is unlikely. NEM mainly modifies the sulfhydryl group of cysteine residues, thereby preventing various −SH-dependent functions, such as cysteine protease activity and the formation of disulfide bridges. At a pH of 8.0, NEM can also modify amine groups. However, at pH 7.2, we could still demonstrate an interaction between Mx1 and PB2 or NP (data not shown). This suggests that modification of free sulfhydryl groups by NEM is a requirement, whereas modification of amino groups is not needed. Identifying the sites where such modifications occur could help to elucidate the interaction interface of Mx1 with PB2 and/or NP.
The interaction of Mx1 with PB2 and NP could be direct, but it could also require the help of another cellular protein. This cellular protein could function as a bridge between Mx1 and the viral ribonucleoprotein complex. Multiple cellular proteins interact with PB2 and/or NP and might fulfill this function. One of these proteins is the RNA helicase, UAP56, which interacts with NP and with Mx1 (50, 51). We investigated the involvement of this helicase in the effect of Mx1 on the PB2-NP interaction. By using coimmunoprecipitation, we demonstrated an interaction between UAP56 and NP but not with the Mx1 protein (data not shown). Coexpression of UAP56 also did not influence the inhibition of the PB2-NP interaction by Mx1 (data not shown), suggesting that UAP56 is not directly involved in the activity of the Mx1 protein. Other cellular factors that might be involved include nucleophosmin (32), one of the importin-alpha isoforms (2, 10), CDK9 (52), USP11 (29), and Hsp90 (4, 35, 36), all of which have been shown to interact with PB2 and/or NP. Only a few proteins have been reported to interact with the Mx1 protein, and these interacting proteins are potential candidates as bridging factors (9).
The NP proteins of various influenza A virus strains have different sensitivities to the activity of the Mx1 protein in the minireplicon system (6, 53). These differences were also reflected in the interaction between PB2 and the different NP variants, as determined for the most resistant and most sensitive NP variant. The interaction between PB2 and NP-H1N1pdm was less affected by coexpressed Mx1 than the interaction between PB2 and NP-pSwO (Fig. 7C).
Comparing the different NP variants in the minireplicon system revealed a higher Mx1 sensitivity for NPs isolated from swine virus strains than for NPs isolated from human strains. This higher Mx1 sensitivity is not unexpected, as these swine NP variants are more closely related to avian NP sequences than to human NP sequences. The most sensitive NP (NP-pSwO) is actually from an avian influenza strain isolated from a pig (24). The higher Mx1 resistance of the NP derived from A/H1N1pdm is remarkable, as its sequence is more closely related to the more sensitive swine viruses than to the more resistant human NP sequences. We tried to identify the residues important for this increased resistance of the NP-H1N1pdm variant by mutating candidate positions in the NP-pSwO variant to the corresponding amino acids in NP-H1N1pdm, guided by the alignment of the different NP variants. However, these mutations in NP did not alter the sensitivity for the antiviral activity of the Mx1 protein in the minireplicon system (data not shown). More research will be needed to fully characterize the Mx1 sensitivity determined by the NP protein.
We provide evidence that Mx1 blocks the interaction between PB2 and NP in reconstituted viral ribonucleoprotein complexes. But is the same true during viral infection, especially in the early phase of infection when parental vRNPs enter the nucleus? We investigated this possibility extensively, particularly by focusing on the integrity of the incoming vRNPs after infection. Presumably, Mx1 targets these incoming vRNPs in the nucleus, disrupting the interaction between PB2 and NP and leading to a block in viral transcription. But our attempts to demonstrate the PB2-NP interaction early in infection were unsuccessful for technical reasons: there are too few PB2 molecules per infected cell before new viral proteins are produced to allow analysis by coimmunoprecipitation. The presence of Mx1 also interferes strongly with the production of viral proteins and thereby with the analysis of the interaction between newly produced PB2 and NP proteins. The interaction (after combining lysates) between Mx1 and vRNPs isolated from infected cells or from isolated virions indicates that Mx1 can also interact with vRNPs generated during infection and, probably, also with the incoming vRNPs in the nucleus.
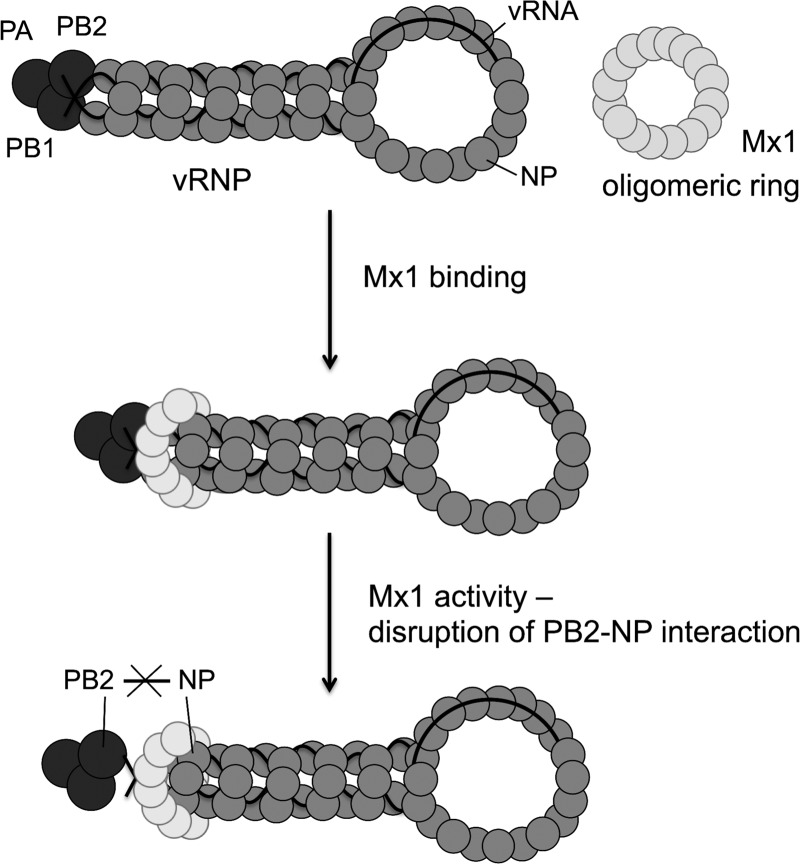
Two main conclusions can be drawn from this study. First, Mx1 interacts with the viral proteins PB2 and NP independently of its GTP-binding activity. Second, inhibition of the PB2-NP interaction by Mx1 is dependent on GTP binding. The relative ease with which this interaction is inhibited correlates with the Mx1 sensitivity of the different NP variants. These conclusions are in line with but do not prove the following mechanistic model for the antiviral activity of the Mx1 protein (Fig. 8). First, Mx1 interacts with the viral PB2 and NP proteins, presumably those present in the incoming vRNPs. This interaction might be mediated by an Mx1 oligomeric ring, as was proposed for MxA (5, 11). After this initial binding, Mx1 actively disrupts the interaction between PB2 and NP in a GTPase-dependent way, leading to a block of viral transcription and replication. Elucidation of the PB2-Mx1 and NP-Mx1 interaction domains will be required to corroborate or modify this model and might help predict potential future escape mechanisms of influenza A viruses against this remarkable IFN-induced host restriction factor.
Fig 8.
Proposed mechanism of Mx1's antiviral activity against influenza A virus.
ACKNOWLEDGMENTS
We thank Anouk Smet and Tine Ysenbaert for excellent technical support, Amin Bredan for critically reading the manuscript, and Nadia Naffakh (Institut Pasteur, Paris, France) for helpful discussions. We are grateful to Erich Hoffmann for providing the A/PR/8/34-based eight-plasmid system for generating recombinant influenza A viruses. Alan Hay (MRC National Institute for Medical Research, Mill Hill, London, United Kingdom), Suzan Carman (University of Guelph, Guelph, Ontario, Canada), D. Desmecht (University of Liège, Liege, Belgium), K. Van Reeth (Ghent University, Merelbeke, Belgium), and Isabel Thomas (Scientific Institute of Public Health, Brussels, Belgium) provided us valuable influenza virus strains. We thank J. Yewdell (NIH, Bethesda, MD) for providing us with the monoclonal anti-PB2 antibody (clone 170-3D5). We are grateful to the company SEPPIC SA (Paris, France) for the generous gift of the adjuvant Montanide ISA-720.
J.V. is a research fellow of the FWO-Vlaanderen. This work was supported by FWO-Vlaanderen and IOF project IOF10/StarTT/027.
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Biswas SK, Boutz PL, Nayak DP. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boivin S, Hart DJ. 2011. Interaction of the influenza A virus polymerase PB2 C-terminal region with importin alpha isoforms provides insights into host adaptation and polymerase assembly. J. Biol. Chem. 286:10439–10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broni B, et al. 1990. Parental influenza virion nucleocapsids are efficiently transported into the nuclei of murine cells expressing the nuclear interferon-induced Mx protein. J. Virol. 64:6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chase G, et al. 2008. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 377:431–439 [DOI] [PubMed] [Google Scholar]
- 5. Daumke O, Gao S, von der Malsburg A, Haller O, Kochs G. 2010. Structure of the MxA stalk elucidates the assembly of ring-like units of an antiviral module. Small Gtpases 1:62–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dittmann J, et al. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of the Mx-GTPase. J. Virol. 82:3624–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dreiding P, Staeheli P, Haller O. 1985. Interferon-induced protein Mx accumulates in nuclei of mouse cells expressing resistance to influenza viruses. Virology 140:192–196 [DOI] [PubMed] [Google Scholar]
- 8. Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329–345 [DOI] [PubMed] [Google Scholar]
- 9. Engelhardt OG, Ullrich E, Kochs G, Haller O. 2001. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp. Cell Res. 271:286–295 [DOI] [PubMed] [Google Scholar]
- 10. Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11 doi:10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao S, et al. 2011. Structure of myxovirus resistance protein A reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525 [DOI] [PubMed] [Google Scholar]
- 12. Haller O, Frese M, Rost D, Nuttall PA, Kochs G. 1995. Tick-borne thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J. Virol. 69:2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haller O, Kochs G. 2011. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31:79–87 [DOI] [PubMed] [Google Scholar]
- 14. Haller O, Stertz S, Kochs G. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9:1636–1643 [DOI] [PubMed] [Google Scholar]
- 15. Hallworth R, Currall B, Nichols MG, Wu X, Zuo J. 2006. Studying inner ear protein-protein interactions using FRET and FLIM. Brain Res. 1091:122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hara K, et al. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87–97 [DOI] [PubMed] [Google Scholar]
- 17. Hemerka JN, et al. 2009. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay. J. Virol. 83:3944–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170 [DOI] [PubMed] [Google Scholar]
- 19. Horisberger MA, Haller O, Arnheiter H. 1980. Interferon-dependent genetic resistance to influenza virus in mice: virus replication in macrophages is inhibited at an early step. J. Gen. Virol. 50:205–210 [DOI] [PubMed] [Google Scholar]
- 20. Horisberger MA, Staeheli P, Haller O. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. U. S. A. 80:1910–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang T, Pavlovic J, Staeheli P, Krystal M. 1992. Overexpression of the influenza virus polymerase can titrate out inhibition by the murine Mx1 protein. J. Virol. 66:4154–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jagger BW, et al. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin HK, Takada A, Kon Y, Haller O, Watanabe T. 1999. Identification of the murine Mx2 gene: interferon-induced expression of the Mx2 protein from the feral mouse gene confers resistance to vesicular stomatitis virus. J. Virol. 73:4925–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karasin AI, West K, Carman S, Olsen CW. 2004. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J. Clin. Microbiol. 42:4349–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. King MC, Raposo G, Lemmon MA. 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U. S. A. 101:8957–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krug RM, Shaw M, Broni B, Shapiro G, Haller O. 1985. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J. Virol. 56:201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lakadamyali M, Rust MJ, Zhuang X. 2004. Endocytosis of influenza viruses. Microbes Infect. 6:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee SH, Vidal SM. 2002. Functional diversity of Mx proteins: variations on a theme of host resistance to infection. Genome Res. 12:527–530 [DOI] [PubMed] [Google Scholar]
- 29. Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. 2010. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 29:3879–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindenmann J. 1962. Resistance of mice to mouse-adapted influenza A virus. Virology 16:203–204 [DOI] [PubMed] [Google Scholar]
- 31. Lindenmann J, Lane CA, Hobson D. 1963. The resistance of A2G mice to myxoviruses. J. Immunol. 90:942–951 [PubMed] [Google Scholar]
- 32. Mayer D, et al. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melen K, Julkunen I. 1994. Mutational analysis of murine Mx1 protein: GTP binding core domain is essential for anti-influenza A activity. Virology 205:269–279 [DOI] [PubMed] [Google Scholar]
- 34. Meyer T, Horisberger MA. 1984. Combined action of mouse alpha and beta interferons in influenza virus-infected macrophages carrying the resistance gene Mx. J. Virol. 49:709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Momose F, et al. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306–45314 [DOI] [PubMed] [Google Scholar]
- 36. Naito T, Momose F, Kawaguchi A, Nagata K. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pavlovic J, Haller O, Staeheli P. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 66:2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pavlovic J, Zurcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64:3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perales B, de la Luna S, Palacios I, Ortin J. 1996. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70:1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pitossi F, et al. 1993. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67:6726–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ponten A, Sick C, Weeber M, Haller O, Kochs G. 1997. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71:2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salomon R, et al. 2007. Mx1 gene protects mice against the highly lethal human H5N1 influenza virus. Cell Cycle 6:2417–2421 [DOI] [PubMed] [Google Scholar]
- 43. Sanz-Ezquerro JJ, de la Luna S, Ortin J, Nieto A. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanz-Ezquerro JJ, Zurcher T, de la Luna S, Ortin J, Nieto A. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J. Virol. 70:1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shpetner HS, Vallee RB. 1992. Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature 355:733–735 [DOI] [PubMed] [Google Scholar]
- 46. Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stranden AM, Staeheli P, Pavlovic J. 1993. Function of the mouse Mx1 protein is inhibited by overexpression of the PB2 protein of influenza virus. Virology 197:642–651 [DOI] [PubMed] [Google Scholar]
- 48. Sugiyama K, et al. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tumpey TM, et al. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81:10818–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wisskirchen C, Ludersdorfer TH, Muller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 85:8646–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wisskirchen C, Ludersdorfer TH, Muller DA, Moritz E, Pavlovic J. 2011. Interferon-induced antiviral protein MxA interacts with the cellular RNA helicases UAP56 and URH49. J. Biol. Chem. 286:34743–34751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J, Li G, Ye X. 2010. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J. Virol. 84:12619–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 85:8133–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zurcher T, Pavlovic J, Staeheli P. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187:796–800 [DOI] [PubMed] [Google Scholar]
- 55. Zurcher T, Pavlovic J, Staeheli P. 1992. Nuclear localization of mouse Mx1 protein is necessary for inhibition of influenza virus. J. Virol. 66:5059–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]