Abstract
Infection with severe acute respiratory syndrome coronavirus (SARS-CoV) causes acute lung injury (ALI) that often leads to severe lung disease. A mouse model of acute SARS-CoV infection has been helpful in understanding the host response to infection; however, there are still unanswered questions concerning SARS-CoV pathogenesis. We have shown that STAT1 plays an important role in the severity of SARS-CoV pathogenesis and that it is independent of the role of STAT1 in interferon signaling. Mice lacking STAT1 have greater weight loss, severe lung pathology with pre-pulmonary-fibrosis-like lesions, and an altered immune response following infection with SARS-CoV. We hypothesized that STAT1 plays a role in the polarization of the immune response, specifically in macrophages, resulting in a worsened outcome. To test this, we created bone marrow chimeras and cell-type-specific knockouts of STAT1 to identify which cell type(s) is critical to protection from severe lung disease after SARS-CoV infection. Bone marrow chimera experiments demonstrated that hematopoietic cells are responsible for the pathogenesis in STAT1−/− mice, and because of an induction of alternatively activated (AA) macrophages after infection, we hypothesized that the AA macrophages were critical for disease severity. Mice with STAT1 in either monocytes and macrophages (LysM/STAT1) or ciliated lung epithelial cells (FoxJ1/STAT1) deleted were created. Following infection, LysM/STAT1 mice display severe lung pathology, while FoxJ1/STAT1 mice display normal lung pathology. We hypothesized that AA macrophages were responsible for this STAT1-dependent pathology and therefore created STAT1/STAT6−/− double-knockout mice. STAT6 is essential for the development of AA macrophages. Infection of the double-knockout mice displayed a lack of lung disease and prefibrotic lesions, suggesting that AA macrophage production may be the cause of STAT1-dependent lung disease. We propose that the control of AA macrophages by STAT1 is critical to regulating immune pathologies and for protection from long-term progression to fibrotic lung disease in a mouse model of SARS-CoV infection.
INTRODUCTION
Infection with a variety of respiratory viruses, including influenza virus H1N1 (25, 37, 50) and severe acute respiratory syndrome coronavirus (SARS-CoV) (9, 13, 18, 49), is known to cause acute lung injury (ALI) and acute respiratory distress syndrome (ARDS), leading to high rates of severe morbidity and mortality (1, 26). In the case of the 2002 SARS-CoV pandemic, there were nearly 8,000 cases and 800 deaths worldwide, with many of these deaths occurring due to development of ARDS in elderly patients (28, 29, 32). It is hypothesized that ALI and ARDS develop from both intrinsic viral infection and dysregulation of the host immune response to infection, although the exact mechanism is not fully understood.
In acute infections with SARS-CoV, lung damage rapidly progresses to diffuse alveolar damage (DAD), resulting clinically in ARDS, which is considered to be the most severe form of ALI (13, 18, 28, 49). ARDS is known to develop from a variety of etiologies that cause severe lung damage and presents a major challenge in critical care medicine, with nearly 1 million deaths attributed to ARDS each year (1). The predominant pathological feature of SARS-CoV infection is diffuse alveolar damage, including hyaline membrane formation, which together clinically manifest as ARDS (9, 49). Even after resolution of SARS, DAD can still be observed by a computed-tomography (CT) scan, and a high proportion of these patients progress to develop pulmonary fibrosis (18).
The host response to viral infection is critical for proper resolution of disease. When that response is altered by the pathogen, the incorrect response can lead to significant tissue damage and further disease. The innate immune response has been shown to protect mice from SARS-CoV replication both in vitro and in vivo (14, 15, 19, 27, 45, 46, 65, 72, 78). Specifically, infection of STAT1−/− mice on the 129/Sv background with a human isolate of SARS-CoV demonstrated that STAT1 (signal transducers and activators of transcription 1) was necessary to control viral replication and spread (15, 79).
We have identified STAT1 as a key mediator controlling the proper host response to SARS-CoV infection (15, 79). STAT1 is a member of the signal transducers and activators of transcription family of transcription factors and plays an important role in both innate and adaptive immune function, as well as cell cycle regulation (33, 35). STAT1's function in cell cycle control makes it of interest in potentially contributing to pulmonary fibrosis development following acute lung injury. In instances where epidermal growth factor (EGF) signaling mediates cell cycle arrest rather than growth, it has been shown that STAT1 is an important signaling component that negatively regulates the EGF pathway (2, 47). It has also been demonstrated that STAT1−/− mice are prone to the development of spontaneous tumors and are more sensitive to varying in vivo models of cancer, including esophageal cancer and melanoma (71, 73). STAT1-deficient mice have also been shown to be sensitive to radiation- and bleomycin-induced fibrosis, again illustrating STAT1's role in control of the regulation of cell cycle progression and proliferation (70).
The involvement of STAT1 in viral infections is associated with its activity in the interferon signaling pathway and the innate immune response (11, 40, 62). In the case of SARS-CoV infection, we have shown that in 129/Sv mice, STAT1 is important in pathways other than the interferon signaling pathway (15). In this model, we found that SARS-CoV infection of mice lacking the alpha/beta interferon receptor (IFNAR1), gamma interferon (IFN-γ) receptor (IFNGR), or interferon lambda receptor (IL28Ra) and IFNAR/IFNGR double mutants display no enhanced pathogenesis, while STAT1−/− mice are highly susceptible and display severe lung disease and pathology (15). STAT1−/− mice in this genetic background have an inability to clear virus by 9 days postinfection and proceed to develop a severe lung disease phenotype that closely replicates the pathology seen in human patients during the SARS-CoV outbreak (31, 49). This includes extensive mixed inflammatory infiltrates, predominantly neutrophils, lymphocytes, and macrophages, as well as pathological features of DAD. The data suggest that a different STAT1-dependent pathway is critical for host protection rather than STAT1's role in IFN signaling. We have shown that STAT1−/− mice do show an altered immune response to SARS-CoV infection, including an enhanced TH2 profile with a significant increase in genes related to the alternatively activated subset of macrophages (15, 79).
Alternatively activated (AA) macrophages have been implicated in models of lung disease, including asthma and pulmonary fibrosis (16, 51, 75). In contrast to classical macrophages, which become activated and differentiate in response to IFN-γ, AA macrophages differentiate in response to interleukin 4 (IL-4) and IL-13 (66). Induction of the AA macrophage phenotype is dependent upon IL-4Rα signaling, as well as the presence and activation of STAT6 (7, 44, 67). Their functions also differ from that of classically activated macrophages, as they are associated with wound repair and healing rather than direct pathogen killing (16). In mice, AA macrophages secrete the effector proteins YM1, FIZZ1, and arginase 1 (16). These effector protein functions are not yet fully understood, but their presence has been associated with liver fibrosis (36, 51, 56), as well as allergic airway inflammation (3, 42). In this paper, we utilize a mouse model of SARS-CoV infection to elucidate the role of STAT1 in ALI as it relates to the induction of AA macrophage subsets in the lung. We demonstrate that the lack of STAT1 in the macrophage and monocyte lineage can shift the immune response and produce significant lung injury, as seen in total STAT1−/− mice. We also show that inhibition of AA macrophage development in STAT1−/− mice, by knocking out STAT6 in concert, protects them from severe lung disease and prefibrotic lesions during SARS-CoV infection. These data demonstrate the role of STAT1 in SARS-CoV pathogenesis, as well as the role that macrophage subtypes play in repair of acute lung injury and development of pulmonary-fibrosis-like diseases.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All mice in this study were treated following IACUC guidelines, and procedures were approved by the University of Maryland School of Medicine IACUC. For infection, the mice were pretreated with ketamine and xylazine as an anesthetic. Mice were euthanized if their weight dropped below approved levels or if clinical symptoms warranted it according to our IACUC protocol. Animal housing and care and experimental protocols were in accordance with all Animal Care and Use Committee guidelines.
Viruses and cells.
SARS-CoV rMA15 was constructed as previously described (57). All virus stocks were stored at −80°C until they were ready to use. VeroE6 cells were purchased from ATCC (catalog number CRL-1586; Manassas, VA) and were used for growing rMA15 virus, as well as plaque assays to determine the viral load in lung tissue. Cells were grown in minimal essential medium (MEM) (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Gemini Bioproducts, West Sacramento, CA).
Mouse breeding.
C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). STAT1−/− mice were a gift from David Levy (New York University [NYU]) and were bred and housed in the animal facilities at the University of Maryland, Baltimore, MD (UMB).
For Cre/LoxP crosses, C57BL/6 LysM/Cre mice (catalog number 004781) were purchased from Jackson Laboratory (Bar Harbor, ME), C57BL/6 FoxJ/Cre mice were a gift from Michael Holtzman (Washington University in St. Louis), and C57BL/6 STAT1/LoxP mice were a gift from Lothar Hennighausen (NIDDK, NIH) (24). C57BL/6 STAT1/LoxP female mice were crossed with male LysM/Cre and FoxJ/Cre mice. Initial F1 pup tails were genotyped for the presence of Cre with primers 5′-GCATTACCGGTCGATGCAACGAGTGATGAG-3′ and 5′-GAGTGAACGAACCTGGTCGAAATCAGTGCG-3′ and for the STAT1/LoxP allele with primers 5′-GCATGACATGATCAGCATTGC-3′ and 5′-ACTGACGTCAACCAAGCCTG-3′. F1-positive female pups were crossed back to STAT1/LoxP males, and F2 pups were generated. These pups were screened for the presence of Cre and 2 copies of the STAT1/LoxP allele. Dual-positive pups were bred, and the lines were screened by immunohistochemistry (IHC) and PCR for the presence of STAT1 in the affected tissue. The methods of this screen are described below.
For STAT1−/− × STAT6−/− double-knockout production, STAT6−/− mice were purchased from Jackson Laboratory (Bar Harbor, ME). STAT6−/− mice were crossed with STAT1−/− mice, and F1 pups were generated. The F1 pups were crossed, and F2 double-positive pups were produced. These pups were screened for deletion of both genes by PCR, and selected pups were bred for use in the infection experiments.
Histological analysis.
Lung sections were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for a minimum of 48 h, after which they were sent to the Histology Core at the University of Maryland, Baltimore, for paraffin embedding and sectioning. Five-micrometer sections were prepared and used for hematoxylin and eosin (H&E) staining by the Histology Core Services (University of Maryland). Stained sections were blinded and analyzed by Lindsay Goicochea at the University of Maryland Medical Center Department of Pathology. Images of histological sections were modified across all sections using Adobe Photoshop CS5 for only minimal modification of images.
Histology scoring.
Slides were prepared as 5-μm sections and stained with hematoxylin and eosin. A pathologist (L.G.) was blinded to mouse genetic background, as well as infection status. Fields were examined by light microscopy and analyzed by Lindsay Goicochea (University of Maryland at Baltimore). The degrees of interstitial, peribronchiolar, and perivascular inflammation was scored from 0 to 3 (Table 1 provides a description of the scoring methodology). For each experimental group, Goicochea blindly scored 3 mice, and those scores were tabulated to compare strains, time points, and infections. Other histologic features, such as the presence of reactive bronchiolar epithelial and pleural changes and the extent of peribronchovascular neutrophilia, were also noted for each group in the text. A combined score, based on the described pathological descriptors (Table 1), was generated for each mouse, and then, 3 mice per group were averaged and the standard deviation for the scoring was computed. These scores are presented in the histological scoring figures.
TABLE 1.
Histological scoring criteria
| Inflammation | Criteria for score of: |
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Interstitial | <2 inflammatory cells/alveolar septal diam | 2–5 inflammatory cells/alveolar septal diam | 6–25 inflammatory cells/alveolar septal diam involving <5% of the parenchyma | 6–25 inflammatory cells/alveolar septal diam involving >5% of the parenchyma |
| Peribronchiolar | Rare inflammatory cells | Mild; 250–1,000 inflammatory cells/mm2 involving <10% of bronchiolar circumference | Moderate; >1,000 inflammatory cells/mm2 involving <25% of bronchiolar circumference | Severe; >1,000 inflammatory cells/mm2 involving >25% of bronchiolar circumference |
| Perivascular | Rare inflammatory cells | Mild; 250–1,000 inflammatory cells/mm2 involving <10% of arteriolar circumference | Moderate; >1,000 inflammatory cells/mm2 involving <25% of arteriolar circumference | Severe; >1,000 inflammatory cells/mm2 involving >25% of arteriolar circumference |
Bone marrow chimeras.
Bone marrow (BM) chimeras of C57/B6 wild-type (WT) and C57/B6 STAT1−/− mice were created using standard chimera techniques. We utilized the congenitally marked CD45.2 C57/B6 WT mice and CD45.1-marked C57/B6 STAT1−/− mice to be able to track the bone marrow reconstitution. We harvested bone marrow from the hind leg bones of both STAT1−/− CD45.1 mice and WT CD45.2 mice. This was done by removing and cleaning the leg bones and then flushing the bones with Dulbecco's modified Eagle's medium (DMEM) to remove the bone marrow. We then washed the bone marrow cells and resuspended them in 10% bovine serum albumin (BSA) in PBS. Both STAT1−/− CD45.1 and WT CD45.2 mice were then irradiated with 1,080 rads to achieve complete bone marrow ablation. Immediately following the irradiation, we injected 4 × 106 STAT1−/− BM cells per mouse into CD45.2 WT mice, and 4 × 106 WT BM cells per mouse into CD45.1 STAT1−/− mice, via intravenous injection into the tail vein. The mice were then given enrofloxacin (Baytril) in their drinking water to ward off infection and allowed to recover and reconstitute for 6 weeks. After 6 weeks, the mice were screened by fluorescence-activated cell sorter (FACS) on the CD45 marker (Santa Cruz Biotechnology, Santa Cruz, CA) to check for successful reconstitution. Mice that were successfully reconstituted (>95% reconstitution) were moved to an animal biosafety level 3 (ABSL3) facility and allowed to rest for 48 h prior to infection.
We then intranasally infected the mice with 1 × 105 PFU SARS-CoV rMA15 or mock infected them with a PBS control and monitored the disease course to identify differences in pathogenesis. We infected and treated age-matched, unaltered wild-type and STAT1−/− mice that were treated similarly with antibiotics prior to their infection as control groups. We monitored the weight of the mice daily and sacrificed them at day 9 postinfection for analysis. We harvested the lung tissue for pathology, immunohistochemistry, RNA, protein, and viral-titer assays.
In vivo mouse infections.
All infections were performed in an animal biosafety level 3 facility at the University of Maryland, Baltimore, MD, using appropriate practices, including a HEPA-filtered bCON caging system, HEPA-filtered powered air-purifying respirators (PAPRs), and Tyvek suiting. All animals were grown to 10 weeks of age prior to use in experiments. The animals were anesthetized using a mixture of xylazine (0.38 mg/mouse) and ketamine (1.3 mg/mouse) in a 50-μl total volume by intraperitoneal injection. The mice were inoculated intranasally with 50 μl of either PBS or 1 × 105 PFU of rMA15 SARS-CoV, after which all animals were monitored daily for weight loss. Mice were euthanized at days 2, 5, or 9 postinfection, and lung tissue was harvested for further analysis. All animals were housed and used in accordance with the University of Maryland, Baltimore, Institutional Animal Care and Use Committee guidelines.
YM1 and Fizz1 immunohistochemistry.
Five-micrometer paraffin sections of fixed lung tissue were cut by the Histology Core at the University of Maryland, Baltimore. Slides were deparaffinized in xylenes and rehydrated in an ethanol series. Endogenous peroxidase activity was quenched by incubating slides in 0.3% hydrogen peroxide in methanol for 30 min at room temperature (RT). Antigen retrieval was performed by incubating slides for 20 min in IHC select citrate buffer, pH 6.0 (Millipore, Billerica, MA). Tissue sections were then preincubated in 10% goat serum in PBS for 1 h at RT. Rabbit polyclonal Fizz1 antibody (Abcam, Cambridge, MA) was used at a concentration of 1:100 diluted in 10% goat serum in PBS. Rabbit polyclonal YM1 antibody (StemCell Technologies, Vancouver, BC, Canada) was used at a concentration of 1:200 diluted in 10% goat serum in PBS. Sections were incubated in primary antibody for 30 min at RT. The slides were washed before incubation with biotinylated secondary antibody from the ABC Elite Rabbit kit (Vector Laboratories, Burlingame, CA) for 30 min at RT. The slides were washed again prior to addition of ABC reagent (Vector Laboratories, Burlingame, CA) for 30 min at RT. After 3 changes of PBS, the slides were developed using DAB (3,3-diaminobenzidine) for 2 min per slide. Development was stopped using a 5-min incubation in distilled water (dH2O). A 10-min incubation in 0.5% methyl green in 0.1 M sodium acetate solution, pH 4.2, was used for counterstaining. The counterstain was cleared using 2 changes of PBS, followed by 2 changes of N-butanol for 2 min each. Slides were dehydrated using 2 5-min incubations in xylenes before being mounted.
STAT1 immunohistochemistry.
Slides with 5-μm sections of mouse lung were obtained from the Histology Core at the University of Maryland, Baltimore. The slides were deparaffinized using two changes of xylenes for 5 min each change. An ethanol series was then used to rehydrate the sections. The slides were then incubated in MOM mouse IgG blocking reagent from a MOM kit purchased from Vector Laboratories (Burlingame, CA) for 1 h at RT. The slides were washed in PBS and then incubated with MOM protein diluent for 5 min at RT. Anti-STAT1 primary antibody (BD Transduction Laboratories, San Jose, CA) was diluted at 1:100 in MOM protein diluent. The slides were incubated in primary antibody for 30 min at room temperature. The slides were washed in PBS before being incubated in MOM biotinylated anti-mouse IgG antibody provided in the MOM kit for 10 min at RT and washed in 2 changes of PBS. The biotin signal was amplified using a TSA Plus Biotin kit (Perkin Elmer, Waltham, MA). The slides were incubated in working-strength biotin amplification solution made at 1:25 for 5 min at RT and then washed in 2 changes of PBS. An ABC-Alkaline Phosphatase (AP) kit (Vector Laboratories, Burlingame CA) was then used. The slides were incubated in ABC-AP for 30 min at RT and then developed using an alkaline phosphatase kit (Vector Laboratories, Burlingame, CA) by incubating the slides for 60 min in AP. The slides were then counterstained using methyl green solution before being decolorized, dehydrated in xylenes, and mounted.
Isolation of DNA from intraperitoneal macrophages and peripheral blood.
Peritoneal macrophages were obtained by injecting mice with 3 ml of sterile 3% thioglycolate broth (Remel, Lenexa, KS) intraperitoneally. Three days after injection, cells were harvested by lavage of the intraperitoneal cavity. The cells were washed in DMEM and pelleted for further analysis by reverse transcription-PCR. Peripheral blood was collected by terminal cardiac puncture. DNA was isolated using a DNA/RNA Allprep kit from Qiagen Laboratories (Valencia, CA). The DNA was then used for PCR and analyzed on a 1% agarose gel. The template was amplified by reverse transcription-PCR using forward (5′-GCATGACATGATCAGCATTGC-3′) and reverse (5′-ACTGACGTCAACCAAGCCTG-3′) primers.
RESULTS
rMA15 infection is cleared from both C57BL/6 and STAT1−/− mice.
To evaluate the role of STAT1 in SARS-CoV infection in mice, 10-week-old WT C57BL/6 and C57BL/6 STAT1−/− mice were infected with 1 × 105 PFU of rMA15. rMA15 is a mouse-adapted SARS-CoV containing 6 amino acid mutations from the WT Urbani strain of SARS-CoV that produces lethal disease in BALB/c mice but does not kill C57BL/6 mice (57). The mice were weighed daily over a 9-day course of infection (Fig. 1A), during which lung tissue was harvested at days 2, 5, and 9 for further characterization. WT C57BL/6 mice lost roughly 15% of their body weight during the first 4 days postinfection (p.i.) and then achieved complete recovery of their starting weight by 9 days p.i. In contrast, STAT1−/− mice continued to lose weight throughout the course of infection, losing more than 20% of their starting weight by 9 days p.i.
Fig 1.
C57BL/6 STAT1−/− mice are highly susceptible to rMA15. (A) Weight loss in WT and STAT1−/− mice infected with either rMA15 or PBS control (n = 15 for each strain). The error bars indicate standard deviations. (B) rMA15 lung titers in WT and STAT1−/− mice (n = 5 for each time point). The asterisk indicates a P value of <0.5, and the dashed line indicates the level of detection. (C) H&E staining of WT and STAT1−/− mice infected with rMA15 or mock infected with PBS at 9 days p.i. (D) Histological scoring of lung sections of WT and STAT1−/− mice infected with rMA15 or mock infected with PBS at 9 days p.i.
Viral titers were analyzed using a plaque assay to compare WT to STAT1−/− mouse infections (Fig. 1B). WT mice displayed peak viral titers at 2 days p.i., with titers of 4.75 × 107 PFU/g, after which they decreased by 5 days p.i. to 1.4 × 104 PFU/g. Viral titers were below detectable levels by 9 days p.i. STAT1−/− mice displayed a similar trend, with peak titers of 8.75 × 107 PFU/g at 2 days p.i. STAT1−/− mice showed a 2-log-unit increase in titer at 5 days p.i. compared to WT mice but still cleared virus by 9 days p.i. to below detectable levels (Fig. 1B). Interestingly, despite the differences in weight loss, we found that WT and STAT1−/− mice on the C57BL/6 background were able to clear rMA15 virus from their lungs during infection, although STAT1−/− mice had higher virus titers at 5 days p.i.
Induction of prefibrotic lesions in STAT1−/− mice after virus clearance.
Histological analysis was performed on lungs harvested at 2, 5, and 9 days p.i. in C57BL/6 WT and STAT1−/− mice (Fig. 1C and D). As previously reported, WT mice infected with rMA15 show bronchiolar sloughing at 2 days p.i., with significant cell debris in the bronchioles. A peribronchiolar and periarteriolar mixed inflammatory infiltrate consisting of eosinophils and neutrophils was observed. By 5 days p.i., airways were generally clear of cell debris and ciliated epithelial cells were present in the bronchioles. Inflammation was still present in the WT lungs, presenting with mainly eosinophils and macrophages in focal regions, primarily surrounding small airways. By 9 days p.i., we observed few small focal aggregates predominantly comprised of lymphocytes cuffing around vessels and larger airways; however, in general, inflammatory infiltrates were minimal, and the lungs had returned to their preinfection state.
In C57BL/6 STAT1−/− mice we found a more severe lung pathology that persisted throughout the course of infection. At 2 days p.i., STAT1−/− mice showed lung pathology similar to that seen in WT mice. Lungs showed evidence of bronchiolar sloughing, with mixed inflammatory infiltrates throughout the lung, but most notably displayed cuffing around large airways and blood vessels. At 5 days p.i., STAT1−/− mice did not have the same recovery as WT mice, and we found a continued increase in levels of mixed inflammatory infiltrates comprised of neutrophils and mononuclear cells, macrophages, and lymphocytes. These infiltrates continued to cluster around large airways and vessels and began to fill the alveolar space. By 9 days p.i., there was continued progression in the severity of lung pathology and there was more pronounced neutrophilia and increased macrophage numbers found in the lung interstitium. Lung epithelial cells exhibited squamous metaplasia, which is a characteristic reactive response to damage commonly seen in incidents of ALI, especially in organizing DAD. Interestingly, there was also proliferation of interstitial fibroblasts in both the subpleural and central regions of the lung. This resembled a prefibrotic-like state sometimes seen following extensive lung damage. H&E-stained slides were blinded and scored by pathologist Lindsay Goicochea (UMB). The slides were scored for perivascular, peribronchiolar, and interstitial inflammation. The scores were tallied as described in Materials and Methods and are graphed in Fig. 1D. While pulmonary inflammation was still found in WT mice at 9 days p.i., we observed that at 9 days p.i. STAT1−/− mice had statistically significant inflammation throughout the lung tissue. Collectively, this lung pathology suggests an inappropriate immune response continuing even after virus had been cleared to below detectable levels.
Alternatively activated macrophages are induced during rMA15 infection of STAT1−/− mice.
We observed an expansion of total macrophages during infection in our histological sections of STAT1−/− mice after rMA15 infection. Since we had previously reported the induction of AA macrophage phenotypes in STAT1−/− mice (79), immunohistochemistry was performed to look for the presence of proteins commonly associated with AA macrophages. YM1 and FIZZ1 are both expressed from AA macrophages in the lung during diseases such as allergic airway inflammation models and respiratory syncytial virus (RSV) infection and used as the standard marker of AA macrophage induction in mice (54, 55). To identify whether AA macrophages were present in WT and STAT1−/− mice, lung sections were stained with antibodies for YM1 and FIZZ1 to analyze their expression during rMA15 infection (Fig. 2). Wild-type and STAT1−/− mice were found to express FIZZ1 specifically along the lung epithelia in PBS-inoculated controls; however, at 9 days after infection, FIZZ1 protein is highly enriched both in the lung parenchyma and in macrophages of STAT1−/− mice (Fig. 2). This is in contrast to what is observed in WT mice. In WT mice, we found FIZZ1 expression solely associated with the lung epithelium even at 9 days p.i. (Fig. 2). Minimal induction of FIZZ1 was identified at days 2 and 5 postinfection of WT or STAT1−/− mice (Fig. 2). YM1 expression in PBS-inoculated control mice was associated with alveolar macrophages with little to no expression in the ciliated lung epithelium of the large airways in both wild-type and STAT1−/− mice (Fig. 2). However, at 9 days p.i., STAT1−/− mice showed evidence of YM1 still associated with alveolar macrophages, but now they displayed diffuse staining in the lung parenchyma in addition to increased staining in the ciliated lung epithelial cells. Minimal induction of YM1 was identified at days 2 and 5 postinfection of WT or STAT1−/− mice. This increase in both YM1 and FIZZ1 levels in the lung suggests an increase in AA macrophages in the SARS-CoV-infected STAT1−/− mice.
Fig 2.
Alternatively activated macrophages are induced during rMA15 infection of STAT1−/− mice. Immunohistochemistry was performed in lung sections from WT and STAT1−/− mice after either PBS inoculation or rMA15 infection at 2, 5, and 9 days p.i. The sections were stained for the alternatively activated macrophage markers FIZZ1 and YM1. The images showing 10× resolution focused on airway labeling and those at 40× resolution focused on the pulmonary interstitium to highlight the extent of signal.
The induction of FIZZ1 from cell types other than macrophages has been reported in systems of allergic airway inflammation (8). To confirm the presence of macrophages at sites of FIZZ1 and YM1 expression in these mice, we stained with a panmacrophage marker, F4/80, using immunohistochemistry (Fig. 3). We detected F4/80-positive cells at the same sites that stained for FIZZ1 and YM1, which strongly supports the finding that the positive cells are truly AA macrophages.
Fig 3.
Immunohistochemical staining of lung sections from WT, STAT1−/−, LysM/STAT1, STAT1/6−/−, and STAT6−/− mice for the macrophage marker F4/80 to identify the locations of macrophages in lung sections. Brown staining is positive staining with antibody. All samples are either PBS or 9-day p.i. (D9) samples.
Induction of TH2 cytokines, IL-4 and IL-13, during rMA15 infection.
Alternatively activated macrophages differentiate in response to the TH2 cytokines IL-4 and IL-13. We hypothesized that we would be able to detect an increase in IL-4 and IL-13 concomitant with the induction of AA macrophages in response to infection in STAT1−/− mice. RNA from lungs of WT and STAT1−/− mice mock infected with PBS or infected with rMA15 was isolated and used for real-time PCR analysis of IL-4 and IL-13 (Fig. 4). We found that in WT mice, there was little detectable IL-4 or IL-13 message in either PBS-mock-infected or rMA15-infected mice. However, in STAT1−/− mice infected with rMA15, we found a significant increase in IL-4 and IL-13 mRNAs at 5 days p.i., which was then reduced by 9 days p.i. The increase at 5 days p.i. correlates with the induction of AA macrophages seen beginning at 5 days p.i. and increasing through 9 days p.i. (Fig. 2). Once the induction of AA macrophages occurs, IL-4 and IL-13 are no longer needed to continue the TH2 polarization in STAT1−/− mice.
Fig 4.
RNA from WT and STAT1−/− mice at 2, 5, and 9 days p.i. was extracted from mouse lungs, converted to cDNA, and used to analyze the induction of IL-4 and IL-13 mRNAs. Three mice were used for each time point and strain. The fold change was calculated by comparison of the total induction for each, normalized to 18S rRNA levels and divided by the PBS control. The error bars indicate standard deviations.
STAT1 is necessary in the hematopoietic lineage to protect from rMA15 pathogenesis.
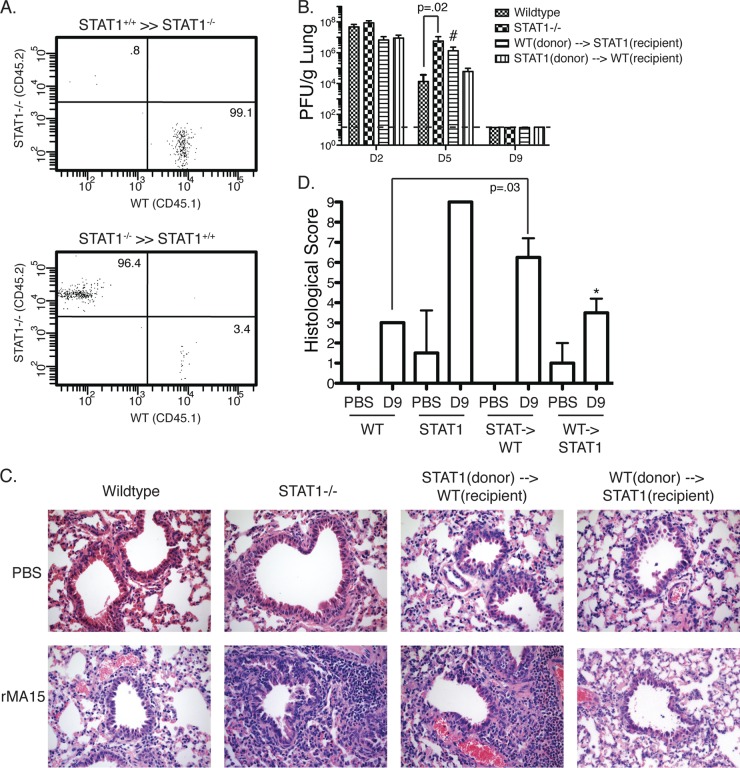
Our data suggest that STAT1−/− mice have a dysregulated response to SARS-CoV infection, leading to a more severe disease phenotype and delayed clearance of virus. Immunohistochemical results suggest that part of this response is related to a change in macrophage phenotype and polarity. We next sought to determine if cells from the hematopoietic lineage were responsible for these changes in outcome following infection. In order to test this, we developed bone marrow chimeric mice by lethally irradiating mice and transferring congenitally marked bone marrow; STAT1−/− bone marrow was injected into wild-type mice, and wild-type bone marrow was injected into STAT1−/− mice. After screening using the CD45 marker on CD4+ cells to confirm successful reconstitution (Fig. 5A), the mice were infected with 1 × 105 PFU of rMA15 and weighed daily for 9 days. Mouse lungs were harvested at days 2, 5, and 9 postinfection for analysis. Quantification of virus titers was performed on lung tissue to evaluate the ability of each mouse to clear virus. We observed that all groups had peak viral titers at 2 days p.i., with titers beginning to diminish by 5 days p.i. By 9 days p.i., virus was below detectable levels (Fig. 5B). The STAT1−/− mice receiving wild-type bone marrow displayed a minor delay in clearance at 5 days p.i., while wild-type mice receiving STAT1−/− bone marrow were indistinguishable from normal wild-type mice.
Fig 5.
STAT1 is necessary in the hematopoietic lineage to protect from rMA15 pathogenesis. Bone marrow chimera experiments between WT and STAT1−/− mice were performed. (A) FACS plot of CD45.1 and CD45.2 CD4+ cells to confirm reconstitution after irradiation. (B) Lung titers from WT, STAT1−/−, and reconstituted mice after infection with rMA15 at 2, 5, and 9 days p.i. (n = 5 for each time point). #, not significant compared to WT day 5. The dashed line indicates the level of detection. (C) H&E staining of WT, STAT1−/−, and bone marrow-reconstituted mice either mock infected with PBS or infected with rMA15 at 9 days p.i. Notice the enhanced inflammation in STAT1−/− and STAT1(donor)→WT(recipient) lungs after infection. (D) Histological scoring of lung sections in panel C. *, not significant compared to WT on day 9. The error bars indicate standard deviations.
Lung sections from bone marrow chimera mice were analyzed for the severity of lung pathology after infection (Fig. 5C). At 9 days p.i., the lung pathology was significantly different between the two chimera groups. In STAT1−/− mice that received wild-type bone marrow, there was damage to the large airways, particularly to the ciliated epithelial cells that line the airways. Cell debris was also still observed in the bronchiolar airspace. These mice also had mixed inflammatory infiltrates that were found cuffing these damaged airways and vessels throughout the lung. Interestingly, these inflammatory infiltrates followed a focal pattern as opposed to the diffuse inflammation seen in the total STAT1−/− animals.
In contrast, WT mice that received STAT1−/− bone marrow exhibited less overall damage to the ciliated epithelial cells of the large airways but displayed enhanced inflammatory infiltrates and both perivascular and peribronchiolar cuffing. There remained foci of reactive squamous metaplasia, but largely, the airways were healthy and intact. We observed a diffuse pattern of inflammation throughout the interstitium comprised of neutrophils and macrophages marked by almost complete collapse of the alveolar space. We also found perivascular neutrophilia, as well as lymphoplasmocytic infiltrates, throughout the lung parenchyma. The inflammation seen in the lungs of these wild-type mice receiving STAT1−/− hematopoietic cells closely resembled that of total STAT1−/− mice, as did their histological scoring (Fig. 5D). This suggests that one or more cell types from STAT1−/− mice are important contributors to the prolonged lung damage after rMA15 infection.
Deleting STAT1 from macrophages and monocytes produces disease similar to that in complete STAT1−/− mice.
After confirming that cells of hematopoietic lineage were predominantly responsible for the increase in pathology in total STAT1−/− mice, we sought to identify potential cell types that could be specifically responsible for this pathology. Together, the hematopoietic link and the increase in AA macrophage markers led us to hypothesize that STAT1 contributes to shaping macrophage responses during SARS-CoV infection and that a change in macrophage polarization was leading to severe lung disease. To test this, we developed 2 mouse strains in which STAT1 is conditionally knocked out. In one strain, we crossed mice containing a STAT1/LoxP integrant with a LysM promoter-driven Cre recombinase to delete STAT1 in monocytes (mice referred to as LysM/STAT1). In the second strain, we crossed mice containing the same STAT1/LoxP integrant with a FoxJ1 promoter-driven Cre recombinase to delete STAT1 in ciliated epithelial cells (77). While we hypothesized that STAT1 was modulating the macrophage response, we created the FoxJ1/STAT1 mouse strain to control for the effect of STAT1 in ciliated epithelial cells, which are the primary target of SARS-CoV in the lung (77). The FoxJ1/Cre mouse has been analyzed and displays specific expression in the ciliated cells of the respiratory epithelium, as well as the choroid plexus, ependyma, oviduct, and testis (77). SARS-CoV specifically infects the respiratory epithelium, so expression in other tissues does not complicate the analysis. To confirm the deletion of STAT1 in monocytes and macrophages in LysM/STAT1 mice, we used thioglycolate to induce intraperitoneal macrophages in WT, STAT1−/−, and LysM/STAT1 mice to compare deletions (Fig. 6A). We confirmed deletion of STAT1 using gene-specific primers and found that in macrophages, a higher-molecular-weight band is found in WT mice than in LysM/STAT1 mice, demonstrating the deletion, and no band is found in the STAT1−/− mice, as is expected with the primers used. In blood, where the LysM promoter is not active, we do not see a deletion in the LysM/STAT1 mice, demonstrating that LysM/Cre is specific to monocytes and macrophages. To verify the FoxJ1/STAT1 deletion in ciliated epithelial cells, we used IHC staining for STAT1 to compare WT, STAT1−/−, and FoxJ1/STAT1 mouse airways (Fig. 6B). We found high levels of expression of STAT1 in WT mice and no expression in any lung cells in STAT1−/− mice. In FoxJ1/STAT1 mice, we found STAT1 expression in the Clara cells of the airways and throughout the alveoli; however, STAT1 expression was lacking in ciliated airway epithelial cells, confirming correct deletion of STAT1 in FoxJ1/Cre-expressing cells.
Fig 6.
Pathogenesis of rMA15 in LysM/STAT1 and FoxJ1/STAT1 mice. (A) PCR confirmation for LysM/STAT1 mice showing STAT1 deletion only in monocytes and macrophages and not in blood of transgenic mice used for the experiments. The arrow indicates the WT STAT1 band, and the arrowhead indicates a deleted STAT1 band. (B) Immunohistochemical labeling of STAT1 in WT, STAT1−/−, and FoxJ1/STAT1 mice to confirm deletion of STAT1 in ciliated epithelial cells. Lung sections were stained with anti-STAT1 antibody. The bright red color is positive staining with antibody. Note in the FoxJ1/STAT1 image that alveolar cells and Clara cells are positive but ciliated epithelial cells are negative for staining. (C) Weight loss curve of mice infected with rMA15 (n = 5 for each time point). The error bars indicate standard deviations. (D) Histological scoring of H&E-stained lung sections in panel E. (E) H&E staining of WT, STAT1−/−, and LysM/STAT1 mouse lungs after infection with rMA15 or mock infection with PBS control at 9 days p.i.
The 10-week-old WT, STAT1−/−, LysM/STAT1, and FoxJ/STAT1 mice were inoculated with 1 × 105 PFU of rMA15 and were weighed daily for 9 days (Fig. 6C). WT mice displayed a 10% starting weight loss by day 3 postinfection, with the mice regaining 100% of their starting weight by 9 days p.i. STAT1−/− mice lost 10% of their starting weight during the first 3 days and then, as before, lost more than 20% of their starting weight through 9 days p.i. Additionally, FoxJ/STAT1 mice had weight loss very similar to that of WT mice. We observed 10% weight loss by 3 days p.i. and then complete recovery by 9 days. Interestingly, LysM/STAT mice displayed early weight loss that closely followed that of the total STAT1−/− mice and exceeded STAT1−/− mouse weight loss by 4 days p.i.; however, they also proceeded to recover their body weight by 9 days postinfection, albeit more slowly than WT mice.
H&E-stained lung sections were analyzed to determine the degree of lung damage and inflammation in the conditional knockout mice following infection with rMA15. As before, at 9 days p.i., we found minimal residual inflammation in WT mice, while STAT1−/− mice displayed overactive and uncontrolled inflammation cuffing the vasculature, bronchi, and throughout the interstitium (Fig. 6E). We observed that LysM/STAT1 mice had persistent inflammation throughout the lung, and specifically, we found cuffing of the large airways, as well as the vessels. The LysM/STAT1 lungs contained inflammatory infiltrates comprised of neutrophils, as well as macrophages, with lymphoid aggregates throughout the interstitial space. Along with these aggregates, there was also evidence of foci of fibroblast proliferation, which is consistent with a prefibrotic-like state seen in total STAT1−/− mice infected with rMA15 (15). In comparison, FoxJ1/STAT1 mice do not display this increased inflammatory response upon infection with rMA15. We found minimal residual inflammation throughout the lungs of infected mice at 9 days p.i. These mice looked comparable to WT mice at the same time point, with repaired ciliated epithelial cells and no observed cuffing of vasculature or bronchi. Histological scoring demonstrated a reduced score in FoxJ/STAT1 mice similar to that of WT mice at 9 days p.i., while LysM/STAT1 mice were found to have a higher score, similar to that of STAT1−/− mice at 9 days p.i. (Fig. 6D). We conclude that the LysM/STAT1 lung pathology is very similar to the full STAT1−/− lung pathology seen during infection and hypothesize that dysregulation of the monocyte and macrophage cell population by deletion of STAT1 specifically in these cells leads to enhanced lung injury and disease after rMA15 infection.
Alternatively activated macrophages are induced in LysM/STAT1 mice after infection with rMA15 similarly to induction in total STAT1−/− mice.
After infection of rMA15 in total STAT1−/− mice, we found induction of AA macrophages in the lungs. Given the above finding that LysM/STAT1 mice have lung pathology similar to that of STAT1−/− mice but that FoxJ1/STAT1 mice have pathology similar to WT mice, we sought to identify whether the induction of AA macrophages correlated with lung disease. In order to examine the activation profile of macrophages following infection, immunohistochemistry was performed on lung sections from mice harvested at 9 days p.i. Mouse lungs were fixed in 4% PFA and embedded in paraffin for sectioning and staining with antibodies for YM1 and FIZZ1, markers for AA macrophage presence (Fig. 7). After staining for YM1 and FIZZ1, we observed that WT mice showed very little detectable YM1 or FIZZ1 in both uninfected and infected mice. In STAT1−/− mice, we found high levels of YM1 expression by 9 days p.i. associated with macrophages throughout the lungs (Fig. 2). FIZZ1 is found highly induced in the airway epithelial cells of STAT1−/− mice after infection, as well as collecting in pools of proteinaceous fluid found in foci throughout the interstitium (Fig. 2). In LysM/STAT1 mice, YM1 levels were found to be low in PBS inoculation control mice, but at 9 days p.i., the levels were increased and protein could be detected throughout the lung tissue, similar to what is seen in STAT1−/− mice (Fig. 7). FIZZ1 levels were also low in uninfected LysM/STAT1 mice; however, similar to YM1, these levels were elevated at 9 days p.i. (Fig. 7). We observed FIZZ1 staining diffusely throughout the lung parenchyma at this time point, and FIZZ1 was again found collecting in fluid-filled foci in the interstitium. Conversely, FoxJ1/STAT1 mice displayed very little induced YM1 or FIZZ1 at 9 days p.i. with rMA15 (Fig. 7). Minor induction of YM1 was found after infection; however, it was found at background levels, as seen in WT mice. Collectively, these data suggest that STAT1 plays a role in macrophage polarization during infection with SARS-CoV and that when macrophages lack STAT1, they are increasingly of an alternatively activated phenotype and are induced during rMA15 infection.
Fig 7.
Alternatively activated macrophages are induced during rMA15 infection of LysM/STAT1−/− mice but not FoxJ/STAT1−/− mice. IHC was performed in lung sections from LysM/STAT1 and FoxJ/STAT1 mice after either PBS inoculation or rMA15 infection at 9 days p.i. The sections were stained for the alternatively activated macrophage markers FIZZ1 and YM1. The images showing 10× resolution focused on airway labeling, and those at 40× resolution focused on the pulmonary interstitium to highlight the extent of signal.
Inhibition of AA macrophages in STAT1−/− mice eliminates severe lung disease.
Our data suggest that AA macrophages are associated with enhanced lung disease after SARS-CoV infection. We hypothesized that these macrophages could be contributing to the exacerbated lung damage, since they have been previously shown to be involved in altered lung repair pathways in fibrosis (48, 69) and asthma (3, 4, 30, 43, 68). Development of AA macrophages is completely dependent on the STAT6 transcription factor (7, 34, 41). In mice lacking STAT6, no AA macrophages are found after treatment with several AA macrophage inducers (7, 34, 41). We created STAT1−/− × STAT6−/− double-knockout mice (called STAT1/6−/−) that contain the STAT1 deletion but lack the ability to produce AA macrophages and thus YM1 and FIZZ1.
As before, we infected these mice and STAT6−/− mice, to control for a phenotype with the loss of STAT6, with rMA15. WT, STAT1−/−, STAT6−/−, and STAT1/6−/− mice were infected with 1 × 105 PFU of rMA15 and weighed daily for 9 days (Fig. 8A). Weight loss for WT and STAT1−/− mice was consistent with what was observed in previous experiments. WT mice lost 10% of their body weight during the first 3 days postinfection and then regained 100% of their body weight by 9 days p.i. Also, STAT1−/− mice lost more than 20% of their body weight by 9 days p.i. STAT6−/− mice displayed weight loss similar to that of WT mice, with early weight loss by day 3, but then regained weight through 9 days p.i. Interestingly, STAT1/6−/− mice also displayed 10% weight loss by day 3, but unlike STAT1−/− mice, the double-knockout mice regained weight and had returned to their starting weight by 9 days p.i.
Fig 8.
Pathogenesis of rMA15 in STAT1/6−/− mice. (A) Weight loss curves of WT and STAT1/6−/− mice infected with rMA15 (n = 5 for each time point). The error bars indicate standard deviations. (B) H&E staining of WT, STAT1−/−, STAT6−/−, and STAT1/6−/− mice either mock infected with PBS or infected with rMA15. (C) Histological scoring of H&E- stained mouse lungs for WT, STAT1−/−, STAT6−/−, and STAT1/6−/− mice at 9 days p.i.
Mouse lungs were harvested and analyzed for histological lesions by H&E staining (Fig. 8B). WT and STAT1−/− mice displayed similar lung pathology, as found previously, with minimal residual inflammation in WT mice at 9 days p.i., but STAT1−/− mice displayed severe inflammatory infiltrates around blood vessels and airways and throughout the interstitium. Consistent with the weight loss data, STAT1/6−/− mice displayed minimal inflammation and lung damage after rMA15 infection, similar to WT mice (Fig. 8B). Histological scoring of lung sections demonstrated reduced lung damage and inflammation in both STAT6−/− and STAt1/6−/− mice, corroborating the weight loss data (Fig. 8C). Together, these data suggest that elimination of AA macrophages in a STAT1−/− background is able to abrogate induced lung disease and inflammation after rMA15 infection.
To confirm that AA macrophage development was inhibited during infection, we again analyzed YM1 and FIZZ1 induction by IHC as markers of AA macrophage levels in lung tissue (Fig. 9). Lung sections from WT, STAT1−/−, STAT6−/−, and STAT1/6−/− mice inoculated with PBS or rMA15 were analyzed. WT and STAT6−/− mice displayed minimal YM1 and FIZZ1 expression in rMA15-infected mice, while STAT1−/− mice displayed high levels of both YM1 and FIZZ1, as observed previously. Concomitant with weight loss and H&E staining, we found low levels of FIZZ1 and YM1 staining in STAT1/6−/− mice with either PBS inoculation or rMA15 infection. The level of staining was similar to that observed in WT mice after infection. This demonstrates that the STAT1/6−/− mice have decreased numbers of AA macrophages and that this reduction correlates with decreased lung pathology. Together, these data demonstrate that in a STAT1−/− mouse model where AA macrophages are eliminated there is reduced lung pathology and disease after rMA15 infection. We interpret this to mean that AA macrophages contribute to the severe lung disease seen in STAT1−/− mice and that even in the presence of total STAT1 deletion, the removal of AA macrophages inhibits disease progression, prefibrotic lesions, and the enhanced inflammatory infiltrate. We hypothesize that the inhibition of the AA macrophage induction pathway could be a therapeutic target for SARS-CoV infection, as well as that of other highly acute respiratory viruses that may induce similar pathways during infection.
Fig 9.
Immunohistochemical staining of lung sections from WT, STAT1−/−, STAT6−/−, and STAT1/6−/− mice for AA macrophage markers YM1 and FIZZ1. Brown staining is positive staining with antibody. All sections are from either PBS-inoculated or rMA15-inoculated mice at 9 days p.i.
DISCUSSION
The interaction between host and pathogen controls the outcome of an infection. From the virus side, many viral proteins from a wide range of viruses directly affect host cell function during infection. From the host side, the response to infection can exacerbate disease and lead to increased damage, causing more destruction than the viral infection alone. In this series of experiments, we have shown that the host response to SARS-CoV infection is able to exacerbate disease and that the host protein STAT1 is important for controlling that response.
The repair of lung tissue after infection or mechanical trauma normally occurs in a rapid, but controlled series of events: inflammation, growth factor secretion, basement membrane repair, and finally resolution of the injured tissue (58). Under normal circumstances, this process proceeds without any problems; however, when the infectious burden overwhelms the host or there is continuous damage, the wound-healing response can become dysregulated, resulting in scarring and fibrosis (39, 61). When scarring develops into ARDS and pulmonary fibrosis, lung function is reduced, which can ultimately lead to death. Both ARDS and pulmonary fibrosis are traditionally treated with corticosteroids, which act to dampen the immune response; however, this course of treatment has not been shown to be particularly effective (38, 53). In cases where the etiology is an infectious agent, such a treatment can actually be harmful to the patient, as it delays the clearance of the pathogen (6, 17, 76).
After injury occurs, there is a cascade of events that progress from initiation of the wound-healing response to recruiting inflammatory cells and fibroblasts to resolution of the wound. Alteration in any of these cascades can drive the host to induce development of fibrosis in the tissue that is in need of repair. Several factors have been shown to be involved in pushing a profibrotic spectrum in animal models; however, these targets have yet to be therapeutically validated in human studies (20, 52, 74). A novel pathway in this process is the STAT1/EGFR signaling axis. Mutations in STAT1 have been found in many cancers throughout the body, especially the lung (21, 71). STAT1 has been shown to activate cell cycle arrest and apoptosis and has demonstrated the properties consistent with a tumor suppressor. Additionally, STAT1−/− mouse embryonic fibroblasts (MEFs) are highly susceptible to uncontrolled cell growth compared to WT MEFs (70), and STAT1−/− mice display increased sensitivity to radiation (12, 22) and bleomycin-induced fibrosis (70).
In viral models of chronic airway infection using Sendai virus as the inducer, STAT1 expression in airway epithelial cells has been shown to be key to protecting against severe lung disease (63). In this model, STAT1−/− mice are highly susceptible to Sendai virus, and using bone marrow chimeras of WT and STAT1−/− mice, it was found that in STAT1−/− mice reconstituted with WT bone marrow, the enhanced susceptibility remained, but not with the converse reconstitution. This result is the opposite of what we found for SARS-CoV infection in the above-mentioned bone marrow chimera experiment and suggests that the intimate and virus-specific interactions between the virus and host can modify the inflammatory and disease states. Using the same model of Sendai virus-induced chronic airway infection, Kim et al. found that even after viral clearance, induction of natural killer T (NKT) cells activates macrophages in the lung (23). These macrophages and NKT cells secrete IL-13, which polarizes macrophages into an alternatively activated macrophage subtype. This activation produces a positive-feedback loop to amplify IL-13 production and alternative activation of macrophages (23). Importantly, the authors found that it is the persistent activation of the AA macrophage pathway that is critical to the continued lung damage and inflammation.
In our previous study, we showed the result of infection by rMA15 in 129/Sv STAT1−/− mice, which display a phenotype different in important ways from that of C57BL/6 STAT1−/− mice (15). We found that 129/Sv wild-type mice infected with a mouse-adapted SARS-CoV (rMA15) lose around 15% of their starting weight but recover that weight over a 9-day course of infection. In 129/Sv STAT1−/− mice infected with the same virus, we observed a similar initial weight loss; however, the mice do not recover and lose >20% of their starting weight by 9 days p.i. and/or succumb to the infection. The STAT1−/− mice develop acute lung injury with the development of pre-pulmonary-fibrosis-like lesions consisting of alternatively activated macrophages, eosinophils, neutrophils, and fibroblasts. In this study, we wanted to identify what the key mediator of the lung lesions was and what cell type necessitated STAT1 for protection.
In this work, we utilized mice on the C57BL/6 genetic background and a STAT1−/− mouse, also on this background (10). We switched to C57BL/6 background mice in this work for several reasons. First, we found that, unlike 129/Sv STAT1−/− mice, C57BL/6 STAT1−/− mice do not die during the infection, allowing us a longer period of pathogenesis for analysis. Second, there are no congenic markers like CD45.1 and CD45.2 in the 129/Sv background to allow the bone marrow chimera experiments to be performed. Third, the FoxJ1/Cre and LysM/Cre mice are only on the C57BL/6 background to allow us to perform the Cre/LoxP experiments. Finally, and most importantly, the C57BL/6 STAT1−/− mice are able to clear rMA15 during the course of infection while still producing the same lung pathology as 129/Sv STAT1−/− mice. This provides us a more realistic model without the issue of continued viral replication potentially altering the results.
Using the C57BL/6 mouse model, we found that mice infected with rMA15 presented with ∼10% weight loss through 4 days of infection and that the mice recovered to their starting weight by 9 days p.i. Importantly, in WT C57BL/6 mice, the rMA15 virus is cleared by 9 days p.i., and it is also cleared in the STAT1−/− mice, unlike what is seen in 129/Sv STAT1−/− mice (15). Even with virus clearance, the STAT1−/− mice still developed pre-pulmonary-fibrosis-like lesions in the lungs by 9 days p.i. This suggests that, as we hypothesized, the STAT1 protein has a role in the development of lung pathology in addition to its role in the innate immune response to infection.
Based on previous data, we predicted that STAT1 was playing a role either in the lung ciliated epithelial cells, which are the only cells in the lung that are infected by SARS-CoV (64), or in macrophages, since we found an altered macrophage response in total STAT1−/− mice infected with rMA15 (79). Using this C57BL/6 model, we produced mice that lacked STAT1 in only specific cell types, either FoxJ1/STAT1 mice (77) or LysM/STAT1 mice (5). FoxJ1/STAT1 mice have STAT1 deleted in the ciliated airway epithelial cells, and LysM/STAT1 mice have STAT1 deleted in monocytes and macrophages. FoxJ1/STAT1 mice infected with rMA15 displayed lung pathology, weight loss, and virus growth kinetics similar to those of WT C57BL/6 mice. LysM/STAT1 mice displayed severe lung pathology and a cytokine response similar to that of total STAT1−/− mice. Interestingly, the weight loss was not significantly different from that of WT mice, suggesting that weight loss and lung pathology may be separate responses during infection. While the lung pathologies and the AA macrophage induction in STAT1−/− and LysM/STAT1 mice are very similar, the weight loss is not, with LysM/STAT1-infected mice regaining their weight through the experiments. We hypothesize that this difference may be due to the role of STAT1 in cytokine induction from host cells and that this induction involves unidentified host response factors affecting morbidity and subclinical symptoms of mice. We are currently using transcriptomics and mathematical modeling to identify pathways contributing to weight loss and pathogenesis in these mice.
The LysM/STAT1 mice developed lung pathology very similar to that of the total STAT1−/− mice. When assayed for the induction of alternatively activated macrophages, which we suspect are critical for the development of these lesions, we found significant upregulation of this macrophage population compared to WT mice and to levels very similar to those in total STAT1−/− mice. Recently, it has been shown that macrophages isolated from patients with idiopathic pulmonary fibrosis (IPF) have an increased number of alternatively activated macrophages, as shown by the human AA macrophage markers of increased CD163 and decreased inducible nitric oxide synthase (iNOS) expression (60). In order to evaluate the role of the AA macrophages more closely, we crossed STAT6−/− mice, which are known to have a blocked AA macrophage phenotype (59), with our STAT1−/− mice. These STAT1/6−/− mice developed disease in a manner that replicated that seen in wild-type mice and also lacked induction of AA macrophage-associated proteins YM1 and Fizz1. This further supports our hypothesis that AA macrophage induction in STAT1−/− mice is responsible for the enhanced disease phenotype.
Together, our findings demonstrate that control of the AA macrophage response is critical during the response to acute lung injury, potentially from a variety of etiologies, and that STAT1 is an important mediator of the development of AA macrophages during infection. We are actively investigating the role of this pathway in other models of chronic inflammation in the lung to identify the role of AA macrophages, STAT1, and the inflammatory response in the induction and maintenance of lung disease. We hypothesize that if we can modulate the host response during infection to skew the classical versus alternatively activated macrophage induction pathways, we may be able to alter the disease outcome in cases of ALI and limit the development of fibrotic-like disease.
ACKNOWLEDGMENTS
We thank David Levy and Skip Virgin for the C56BL/6 STAT1−/− mice and KariAnn Shirey for assistance with macrophage isolation.
This work was supported by NIAID/NIH grant R01AI095569-01 (M.F.) and NIAID/NIH grant T32AI007540 (C.P.).
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Bauer TT, Ewig S, Rodloff AC, Muller EE. 2006. Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin. Infect. Dis. 43:748–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. U. S. A. 93:7673–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byers DE, Holtzman MJ. 2011. Alternatively activated macrophages and airway disease. Chest 140:768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byers DE, Holtzman MJ. 2010. Alternatively activated macrophages as cause or effect in airway disease. Am. J. Respir. Cell Mol. Biol. 43:1–4 [DOI] [PubMed] [Google Scholar]
- 5. Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277 [DOI] [PubMed] [Google Scholar]
- 6. Collard HR, et al. 2007. Acute exacerbations of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 176:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE. 2010. The phenotype of murine wound macrophages. J. Leukoc. Biol. 87:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dasgupta P, Chapoval SP, Smith EP, Keegan AD. 2011. Transfer of in vivo primed transgenic T cells supports allergic lung inflammation and FIZZ1 and Ym1 production in an IL-4Ralpha and STAT6 dependent manner. BMC Immunol. 12:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding Y, et al. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200:282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durbin JE, et al. 2000. Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 164:4220–4228 [DOI] [PubMed] [Google Scholar]
- 11. Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450 [DOI] [PubMed] [Google Scholar]
- 12. Eschrich S, et al. 2009. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int. J. Radiat. Oncol. Biol. Phys. 75:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franks TJ, et al. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34:743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frieman M, et al. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frieman MB, et al. 2010. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6:e1000849 doi:10.1371/journal.ppat.1000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3:23–35 [DOI] [PubMed] [Google Scholar]
- 17. Gross TJ, Hunninghake GW. 2001. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 345:517–525 [DOI] [PubMed] [Google Scholar]
- 18. Han Y, et al. 2003. A follow-up study of 69 discharged SARS patients. J. Tradit. Chin Med. 23:214–217 [PubMed] [Google Scholar]
- 19. Hussain S, Perlman S, Gallagher TM. 2008. Severe acute respiratory syndrome coronavirus protein 6 accelerates MHV infections by more than one mechanism. J. Virol. 82:7212–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jakubzick C, et al. 2003. Therapeutic attenuation of pulmonary fibrosis via targeting of IL-4- and IL-13-responsive cells. J. Immunol. 171:2684–2693 [DOI] [PubMed] [Google Scholar]
- 21. Kaganoi J, et al. 2007. STAT1 activation-induced apoptosis of esophageal squamous cell carcinoma cells in vivo. Ann. Surg. Oncol. 14:1405–1415 [DOI] [PubMed] [Google Scholar]
- 22. Khodarev NN, et al. 2004. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. U. S. A. 101:1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim EY, et al. 2008. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat. Med. 14:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klover PJ, et al. 2010. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia 12:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kobasa D, et al. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323 [DOI] [PubMed] [Google Scholar]
- 26. Kollef MH, Schuster DP. 1995. The acute respiratory distress syndrome. N. Engl. J. Med. 332:27–37 [DOI] [PubMed] [Google Scholar]
- 27. Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ksiazek TG, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 29. Kuiken T, et al. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurowska-Stolarska M, et al. 2009. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J. Immunol. 183:6469–6477 [DOI] [PubMed] [Google Scholar]
- 31. Lau YL, Peiris JS. 2005. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 17:404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee N, et al. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986–1994 [DOI] [PubMed] [Google Scholar]
- 33. Levy DE, Garcia-Sastre A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143–156 [DOI] [PubMed] [Google Scholar]
- 34. Liddiard K, et al. 2006. Interleukin-4 induction of the CC chemokine TARC (CCL17) in murine macrophages is mediated by multiple STAT6 sites in the TARC gene promoter. BMC Mol. Biol. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lillemeier BF, Koster M, Kerr IM. 2001. STAT1 from the cell membrane to the DNA. EMBO J. 20:2508–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopez-Navarrete G, et al. 2011. Th2-associated alternative Kupffer cell activation promotes liver fibrosis without inducing local inflammation. Int. J. Biol. Sci. 7:1273–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mauad T, et al. 2010. Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181:72–79 [DOI] [PubMed] [Google Scholar]
- 38. Meduri GU, Rocco PR, Annane D, Sinclair SE. 2010. Prolonged glucocorticoid treatment and secondary prevention in acute respiratory distress syndrome. Expert Rev. Respir. Med. 4:201–210 [DOI] [PubMed] [Google Scholar]
- 39. Meneghin A, Hogaboam CM. 2007. Infectious disease, the innate immune response, and fibrosis. J. Clin. Invest. 117:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meraz MA, et al. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431–442 [DOI] [PubMed] [Google Scholar]
- 41. Mishra BB, Gundra UM, Teale JM. 2011. STAT6(-)/(-) mice exhibit decreased cells with alternatively activated macrophage phenotypes and enhanced disease severity in murine neurocysticercosis. J. Neuroimmunol. 232:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mora AL, et al. 2006. Activation of alveolar macrophages via the alternative pathway in herpesvirus-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol. 35:466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moreira AP, Hogaboam CM. 2011. Macrophages in allergic asthma: fine-tuning their pro- and anti-inflammatory actions for disease resolution. J. Interferon Cytokine Res. 31:485–491 [DOI] [PubMed] [Google Scholar]
- 44. Moreno JL, Kaczmarek M, Keegan AD, Tondravi M. 2003. IL-4 suppresses osteoclast development and mature osteoclast function by a STAT6-dependent mechanism: irreversible inhibition of the differentiation program activated by RANKL. Blood 102:1078–1086 [DOI] [PubMed] [Google Scholar]
- 45. Narayanan K, et al. 2008. Severe acute respiratory syndrome coronavirus Nsp1 suppresses host gene expression, including type I interferon, in infected cells. J. Virol. 82:4471–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Netland J, et al. 2007. Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J. Virol. 81:11520–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohtsubo M, Takayanagi A, Gamou S, Shimizu N. 2000. Interruption of NFkappaB-STAT1 signaling mediates EGF-induced cell-cycle arrest. J. Cell. Physiol. 184:131–137 [DOI] [PubMed] [Google Scholar]
- 48. Pechkovsky DV, et al. 2010. Alternatively activated alveolar macrophages in pulmonary fibrosis-mediator production and intracellular signal transduction. Clin. Immunol. 137:89–101 [DOI] [PubMed] [Google Scholar]
- 49. Peiris JS, et al. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115 doi:10.1371/journal.ppat.1000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pesce J, et al. 2006. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J. Clin. Invest. 116:2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pierce EM, et al. 2007. Therapeutic targeting of CC ligand 21 or CC chemokine receptor 7 abrogates pulmonary fibrosis induced by the adoptive transfer of human pulmonary fibroblasts to immunodeficient mice. Am. J. Pathol. 170:1152–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pierrakos C, Karanikolas M, Scolletta S, Karamouzos V, Velissaris D. 2012. Acute respiratory distress syndrome: pathophysiology and therapeutic options. J. Clin. Med. Res. 4:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raes G, et al. 2002. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J. Leukoc. Biol. 71:597–602 [PubMed] [Google Scholar]
- 55. Raes G., Van den Bergh R., De Baetselier P., Ghassabeh G. H., Scotton C., Locati M., Mantovani A., Sozzani S. 2005. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol. 174:6561 (Author's reply, 174:6561–6562.) [DOI] [PubMed] [Google Scholar]
- 56. Reiman RM, et al. 2006. Interleukin-5 (IL-5) augments the progression of liver fibrosis by regulating IL-13 activity. Infect. Immun. 74:1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roberts A, et al. 2007. A Mouse-Adapted SARS-Coronavirus Causes Disease and Mortality in BALB/c Mice. PLoS Pathog. 3:e5 doi:10.1371/journal.ppat.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rock JR, Hogan BL. 2011. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27:493–512 [DOI] [PubMed] [Google Scholar]
- 59. Rodriguez-Sosa M, et al. 2002. Chronic helminth infection induces alternatively activated macrophages expressing high levels of CCR5 with low interleukin-12 production and Th2-biasing ability. Infect. Immun. 70:3656–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saleh D, Barnes PJ, Giaid A. 1997. Increased production of the potent oxidant peroxynitrite in the lungs of patients with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 155:1763–1769 [DOI] [PubMed] [Google Scholar]
- 61. Schafer M, Werner S. 2007. Transcriptional control of wound repair. Annu. Rev. Cell Dev. Biol. 23:69–92 [DOI] [PubMed] [Google Scholar]
- 62. Schindler C, Darnell JE., Jr 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621–651 [DOI] [PubMed] [Google Scholar]
- 63. Shornick LP, et al. 2008. Airway epithelial versus immune cell Stat1 function for innate defense against respiratory viral infection. J. Immunol. 180:3319–3328 [DOI] [PubMed] [Google Scholar]
- 64. Sims AC, et al. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511–15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Spiegel M, et al. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stein M, Keshav S, Harris N, Gordon S. 1992. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 176:287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stolfi C, et al. 2011. Interleukin-25 fails to activate STAT6 and induce alternatively activated macrophages. Immunology 132:66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Subrata LS, et al. 2009. Interactions between innate antiviral and atopic immunoinflammatory pathways precipitate and sustain asthma exacerbations in children. J. Immunol. 183:2793–2800 [DOI] [PubMed] [Google Scholar]
- 69. Sun L, et al. 2011. New concepts of IL-10-induced lung fibrosis: fibrocyte recruitment and M2 activation in a CCL2/CCR2 axis. Am. J. Physiol. Lung Cell. Mol. Physiol. 300:L341–L353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walters DM, et al. 2005. Susceptibility of signal transducer and activator of transcription-1-deficient mice to pulmonary fibrogenesis. Am. J. Pathol. 167:1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Watanabe G, et al. 2001. Progression of esophageal carcinoma by loss of EGF-STAT1 pathway. Cancer J. 7:132–139 [PubMed] [Google Scholar]
- 72. Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81:11620–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wong LH, et al. 1997. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J. Biol. Chem. 272:28779–28785 [DOI] [PubMed] [Google Scholar]
- 74. Wynn TA. 2008. Cellular and molecular mechanisms of fibrosis. J. Pathol. 214:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wynn TA, et al. 2011. Quantitative assessment of macrophage functions in repair and fibrosis. Curr. Protoc. Immunol. Chapter 14:Unit14.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu H, et al. 2008. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One 3:e2985 doi:10.1371/journal.pone.0002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang Y, et al. 2007. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am. J. Respir. Cell Mol. Biol. 36:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao J, et al. 2009. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 83:2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zornetzer GA, et al. 2010. Transcriptomic analysis reveals a mechanism for a pre-fibrotic phenotype in STAT1 knockout mice during SARS-coronavirus infection. J. Virol. 84:11297–11309 [DOI] [PMC free article] [PubMed] [Google Scholar]