Abstract
The 80 trimeric, glycoprotein spikes that cover the surface of alphavirus particles are required for mediating viral entry into a host cell. Spike assembly is a regulated process that requires interactions between five structural proteins, E3, E2, 6K and its translational frameshift product TF, and E1. E3 is a small, ∼65-amino-acid glycoprotein that has two known functions: E3 serves as the signal sequence for translocation of the E3-E2-6K-E1 polyprotein into the endoplasmic reticulum (ER), and cleavage of E3 from E2 is essential for virus maturation. Nonetheless, when E3 is replaced with an ER signal sequence, spikes do not form and infectious particles are not assembled, suggesting an additional role(s) for E3 in the viral life cycle. To further investigate the role of E3 in spike assembly, we made chimeric viruses in which E3 from one alphavirus species is replaced with E3 from another species. Our results demonstrate that when E3 is interchanged between alphavirus species that belong to the same virus clade, viral titers and particle morphologies and compositions are similar to what are observed for the parental virus. In contrast, for chimeras in which E3 is derived from a different clade than the parental virus, we observed reduced titers and the formation of particles with atypical morphologies and protein compositions. We further characterized the E3 chimeras using a combination of structure-function and revertant analyses. This work revealed two specific interactions between E3 and its cognate E2 glycoprotein that are important for regulating spike assembly.
INTRODUCTION
Alphavirus particles are characterized by the presence of 80 trimeric, glycoprotein spikes that protrude from the surface of the virion (5, 49). The spikes, which are composed of trimers of E2 and E1 heterodimers, are embedded in a host-derived lipid membrane and are required to initiate infection; E2 is responsible for receptor binding (4, 18, 39) and E1 mediates fusion between the viral and host cell membranes during entry (11, 32, 47). Internal to the viral membrane is the nucleocapsid core. The core consists of 240 copies of the capsid protein, which encapsidate the single-stranded, positive-sense RNA genome (5, 49). The glycoprotein spikes and the nucleocapsid core are linked through interactions between the cytoplasmic tails of E2 and the capsid protein (10, 12, 27, 35). Thus, misfolded or misarranged spikes on the particle surface or aberrant interactions between the spikes and the nucleocapsid core may affect viral entry into a new host cell.
Alphavirus spike assembly is a highly regulated process that requires interactions between both viral and host cell factors. The viral structural proteins are translated as a polyprotein, capsid-E3-E2-6K-E1 (or capsid-E3-E2-TF), from a subgenomic RNA (8, 43). Early in translation, capsid is autoproteolytically cleaved from the rest of the polyprotein (30). Following capsid cleavage, E3 serves as the signal sequence to translocate E3-E2-6K-E1 (or E3-E2-TF) into the endoplasmic reticulum (ER) (2, 3, 8, 23). Multiple cleavage and oligomerization steps within the host ER and secretory system are necessary to form functional spikes. In the ER, the structural polyprotein is processed by the cellular enzyme signalase to yield E3-E2 (also referred to as pE2 or p62), 6K, and E1 (23, 51). After processing, pE2 and E1 form heterodimers, which is followed by the oligomerization of three heterodimers to form the immature, nonfusogenic spike complex (25, 26, 31). Once formed, the immature spikes are transported to the plasma membrane through the host secretory system (6). In the trans-Golgi network, the cellular enzyme furin cleaves E3 from E2, which renders the spikes fusogenic (17, 37, 38, 46, 51). Lateral spike-spike interactions at the plasma membrane arrange the spikes into hexagonal arrays, which are hypothesized to serve as the sites for E2 and nucleocapsid core interactions to initiate budding from the host cell (44). Improper regulation of spike assembly at any of these steps could have drastic effects on particle formation and infectivity.
The observations from several independent lines of work suggest that E3 may function as a chaperone, possibly to mediate E2 folding, during spike assembly. First, when E3 is replaced with an ER signal sequence, the structural proteins are translated; however, spikes do not form and E1 and E2 are retained in the ER (26). Second, when the furin cleavage site is mutated, uncleaved pE2 is incorporated into the released virions, but particle infectivity is severely attenuated. The reduced infectivity is presumably due to the inability of the E2-E1 heterodimers to dissociate during viral entry (17, 45). Third, distinct assembly defects were observed when single amino acid mutations were made in Sindbis virus (SINV) and Ross River virus (RRV) E2, indicating that E2 folding and function are species dependent, possibly due to interactions with a virus-specific chaperone (40). Fourth, in the atomic structure of the Chikungunya virus (CHIK) pE2-E1 heterodimer, E3 contacts E2 only and does not interact directly with E1 (45). There are eight predicted interaction sites between E3 and E2 (through either hydrogen bonds or salt bridges) that are distributed throughout the CHIK E3-E2 interface, but none of these are near the E2 receptor-binding site (4, 18, 39, 45). It is hypothesized that these contact sites are necessary to stabilize the pE2-E1 heterodimer during assembly of the viral spike complex.
To investigate if E3 regulates folding of its cognate E2 glycoprotein and subsequent pE2 and E1 heterodimerization, we generated chimeric viruses in which E3 from one virus species is replaced with E3 from another species. Our results demonstrate that when E3 is interchanged between virus species that belong to the same virus clade, there are minimal effects on viral titer, particle morphology, and particle composition. In contrast, we observed attenuated virus growth and defects in particle morphology and composition for chimeras in which E3 was derived from a different clade than the parental virus. Furthermore, second-site revertants map to two binding sites between E3 and E2, demonstrating that the maintenance of a small number of specific E3-E2 contacts can restore E3 function in chimeras that are defective in particle assembly. Taken together, our results indicate that E3 makes specific interactions with its cognate E2 glycoprotein and these interactions are important for regulating spike assembly.
MATERIALS AND METHODS
Viruses and cells.
The virus strains used in this work were the TE12 strain of SINV (29), the T48 strain of RRV (19), the pSP6-SFV4 infectious clone of Semliki Forest virus (SFV) (24), and the BeAR 10315 strain of Aura virus (AURA) (36). BHK-21 cells (American Type Culture Collection, Manassas, VA) were grown in minimal essential medium (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), nonessential amino acids, l-glutamine, and penicillin-streptomycin. BHK-21 cells were grown at 37°C in the presence of 5% CO2. C6/36 cells (American Type Culture Collection, Manassas, VA) were grown in identical medium but at 28°C in the presence of 5% CO2.
Generation of chimeric and mutant viruses.
All chimeric and mutant viruses of the TE12 cDNA clone and the T48 cDNA clone were generated using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). No additional changes were introduced into the chimeras during cloning. For each construct, the region corresponding to the E3 glycoprotein and/or the E2 glycoprotein was sequenced. Wild-type, chimeric, and mutant cDNA clones were linearized with SacI and in vitro transcribed with SP6 polymerase (33).
Electroporation of BHK-21 cells was performed as previously described (34). Briefly, approximately 107 BHK-21 cells were trypsinized, washed three times with phosphate-buffered saline (PBS), and resuspended with PBS to a final volume of 500 μl. The cells were mixed with in vitro-transcribed RNA in a 2-mm-gap cuvette and pulsed once at 1.5 kV, 25 μF, and 200 Ω using a Bio-Rad Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA). Following a 10-min recovery at room temperature, the cells were diluted 1:10 in BHK cell medium and incubated at 37°C in the presence of 5% CO2. At the indicated time points, virus was harvested and the titer was determined using a standard plaque assay procedure (33).
Electroporation of C6/36 cells was performed as previously described (40). Briefly, approximately 107 C6/36 cells were washed twice with PBS and resuspended in PBS to a final volume of 500 μl. The cells were mixed with in vitro-transcribed RNA in a 2-mm-gap cuvette and pulsed twice at 0.4 kV, 25 μF, and 800 Ω using the Bio-Rad Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA). Following a 1-min recovery at room temperature, the cells were diluted 1:10 in C6/36 cell medium and incubated at 28°C in the presence of 5% CO2. At 25 h postelectroporation, virus was harvested and the titer was determined on BHK-21 cells using a standard plaque assay procedure (33).
Quantitation of genome copies by RT and qRT-PCR.
Quantitation of genome copies was performed as previously described (40). Briefly, BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At 25 h postelectroporation, the medium was harvested and spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium containing either wild-type or chimeric virus (5 μl) was mixed with 500 ng of both nsP1 and either E1 or E2 reverse transcription (RT) primers (SINV nsP1 [5′-AACATGAACTGGGTGGTG-3′] and SINV E1 [5′-ATTGACCTTCGCGGTCGGATTCAT-3′] or RRV nsP1 [5′-GCTCTGGCATTAGCATGG-3′] and RRV E2 [5′-GAACATCATGACCAGCCATA-3′]). The samples were incubated at 94°C for 5 min and then 70°C for 5 min and transferred to ice. Once on ice, an RT mixture containing 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 10 mM dithiothreitol, 3 mM MgCl2, 0.5 mM deoxynucleoside triphosphates, RNase inhibitor (GenScript, Piscataway, NJ), and ImProm-II reverse transcriptase (Promega, Madison, WI) was added to each sample. Following addition of the RT mixture, the samples were incubated at 25°C for 5 min, 42°C for 45 min, and 75°C for 15 min.
The cDNA prepared as described above (2 μl) was mixed with 1× Brilliant SYBR green III reagent (Stratagene, La Jolla, CA) and 250 nM forward and reverse detection primers to a final volume of 25 μl per well in a 96-well plate. Multiple quantitative RT-PCR (qRT-PCR) measurements were made for each sample. Standard curves were generated by serially diluting wild-type SINV or RRV cDNA clones. All data sets included paired standard curves, which were generated by plotting the observed threshold cycle (CT) value using an exponential regression. The number of genome copies present within each qRT-PCR mixture was calculated using the observed CT values for each sample and the equation obtained from the standard curve.
TEM.
Transmission electron microscopy (TEM) analysis was performed as previously described (40). Briefly, medium containing either wild-type or chimeric virus was spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium was overlaid onto a 27% sucrose cushion and spun at 185,000 × g for 2.5 h at 4°C. Pelleted virus particles were resuspended in HNE buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.1 mM EDTA). Following purification, 3 μl of virus was applied to a Formvar- and carbon-coated 400-mesh copper grid (Electron Microscopy Sciences, Hatfield, PA) and stained with 1% uranyl acetate. The stained grids were analyzed using a JEOL 1010 transmission electron microscope (Tokyo, Japan) operating at 80 kV. Images were recorded using a Gatan UltraScan 4000 charge-coupled-device camera (Pleasanton, CA).
Production of radiolabeled virus and purification using a step gradient.
Production and purification of radiolabeled virus were performed as previously described (40). Briefly, BHK-21 cells were electroporated with in vitro-transcribed wild-type, chimeric, or mutant viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At 15 h postelectroporation, the medium was replaced with l-methionine- and l-cysteine-free Dulbecco's minimum essential medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). The cells were incubated at 37°C in the presence of 5% CO2 for 15 min. Following the 15-min incubation, 50 μCi/ml [35S]cysteine-methionine (PerkinElmer, Waltham, MA) was added to the medium and the cells were incubated for an additional 24 h.
Following virus production, medium containing either radiolabeled wild-type, chimeric, or mutant virus was spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium was then overlaid onto a 27% and 60% sucrose step gradient and spun at 250,000 × g for 2.5 h at 4°C. Following ultracentrifugation, virus present at the 27% and 60% sucrose step interface was extracted. Radiolabeled virus was solubilized in reducing SDS sample buffer and analyzed by SDS-PAGE.
Pulse-chase analysis of viral glycoprotein expression.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At 15 h postelectroporation, the medium was replaced with l-methionine- and l-cysteine-free DMEM (Invitrogen, Carlsbad, CA) supplemented with 5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 2 μg/ml actinomycin D (Invitrogen, Carlsbad, CA). The cells were incubated at 37°C in the presence of 5% CO2 for 15 min. After the 15-min incubation, 50 μCi/ml [35S]cysteine-methionine (PerkinElmer, Waltham, MA) was added to the medium and the cells were incubated for an additional 15 min. After radiolabeling, the cells were either immediately lysed with BHK cell lysis buffer (10 mM Tris, pH 7.4, 20 mM NaCl, 0.4% deoxycholic acid, 1% NP-40, and 1 mM EDTA) (0-h-chase time point) or the medium was replaced with unlabeled BHK cell medium. The cells were chased in unlabeled medium for 1 h and then lysed with BHK cell lysis buffer (1-h-chase time point). The cell lysates were solubilized in reducing SDS sample buffer and analyzed by SDS-PAGE.
Biotinylation analysis of glycoprotein cell surface expression.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At 15 h postelectroporation, the medium was replaced with l-methionine- and l-cysteine-free DMEM (Invitrogen, Carlsbad, CA) supplemented with 5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). The cells were incubated at 37°C in the presence of 5% CO2 for 15 min. Following the 15-min incubation, 50 μCi/ml [35S]cysteine-methionine (PerkinElmer, Waltham, MA) was added to the medium and the cells were incubated for an additional 2 h. After radiolabeling, the cells were washed with PBS and cell surface proteins were biotinylated for 30 min at room temperature with 2.5 mM sulfosuccinimidyl-6-[biotin-amido]hexanoate (sulfo-NHS-LC-biotin; Pierce, Rockford, IL). The biotin solution was prepared in PBS immediately before use. The biotinylated cells were lysed with BHK cell lysis buffer (10 mM Tris, pH 7.4, 20 mM NaCl, 0.4% deoxycholic acid, 1% NP-40, and 1 mM EDTA), and cell lysates were incubated with streptavidin resin (Pierce, Rockford, IL) for 1 h at room temperature with rocking. Following incubation, the resin was pelleted and washed three times with PBS. Biotinylated proteins were eluted from the resin by adding reducing SDS sample buffer and boiling at 95°C for 10 min. The eluted proteins were analyzed by SDS-PAGE.
Screening for second-site revertants of the interclade RRV chimeras.
Chimeric viruses were serially passaged on BHK-21 cells, and the presence of second-site revertants was monitored by plaque size. Large plaques observed at passage 3 were isolated and used to infect BHK-21 cells. The infected cells were lysed at 8 h postinfection with TRIzol reagent (Invitrogen, Carlsbad, CA), and cytoplasmic RNA was isolated using phenol-chloroform extraction. The coding sequence of the structural polyprotein was amplified from the viral RNA using RT-PCR. The region corresponding to the structural polyprotein was sequenced to identify the location of second-site mutations. To verify that the large plaque phenotypes were due to the mutation identified in sequencing, the revertant sites were introduced back into the chimeric viruses and growth kinetics and plaque sizes were analyzed.
RESULTS
The interclade E3 chimeras produce fewer infectious particles than wild-type virus.
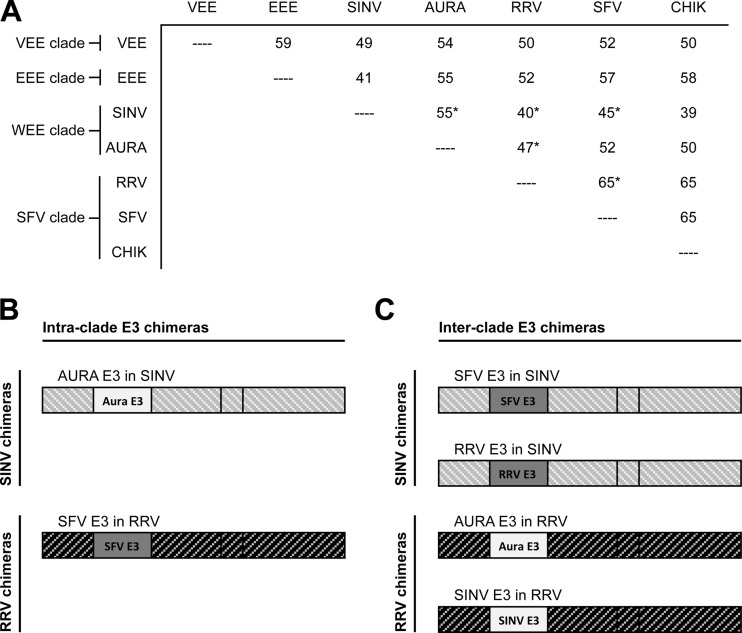
Phylogenetic analysis of the entire viral genome divides alphaviruses into four clades: the Venezuelan equine encephalitis (VEE) clade, the Eastern equine encephalitis (EEE) clade, the Western equine encephalitis (WEE) clade, and the Semliki Forest virus (SFV) clade (Fig. 1A) (9). Pairwise analysis of amino acid sequence identities shows that E3 glycoproteins derived from the same clade have, in general, comparatively higher percent identities than E3 glycoproteins derived from different clades (Fig. 1A) (13, 20).
Fig 1.
Design of SINV and RRV chimeras. (A) Matrix of amino acid percent identities calculated pairwise between different alphavirus E3 glycoproteins. The phylogenetic clade to which each alphavirus is classified is indicated to the left of the matrix (9). Percent identities calculated between the E3 glycoproteins interchanged in this work are indicated (*). Percent identities were calculated using the ClustalW program (13, 20). (B and C) Schematics of the intra- and interclade E3 chimeras, respectively.
To determine how interchangeable E3 is between virus species, we made and characterized E3 chimeric viruses. E3 was interchanged between viruses in the same clade and different clades. If E3 does not play a specific chaperone role with E2 during spike assembly, E3 should be interchangeable between different alphaviruses, including those from different clades; however, if specific contacts between E3 and E2 that promote folding and oligomerization of the viral spike complex are made, the chimeric viruses may have assembly defects that are not observed for wild-type virus. Using Sindbis virus (SINV) and Ross River virus (RRV) as the parental viruses (members of the WEE clade and the SFV clade, respectively), we designed two sets of E3 chimeras (Fig. 1B and C): intraclade chimeras in which E3 is derived from the same phylogenetic clade as the parental virus (AURA E3 in SINV and SFV E3 in RRV) and interclade chimeras in which E3 is derived from a different phylogenetic clade than the parental virus (SFV E3 in SINV, RRV E3 in SINV, AURA E3 in RRV, and SINV E3 in RRV).
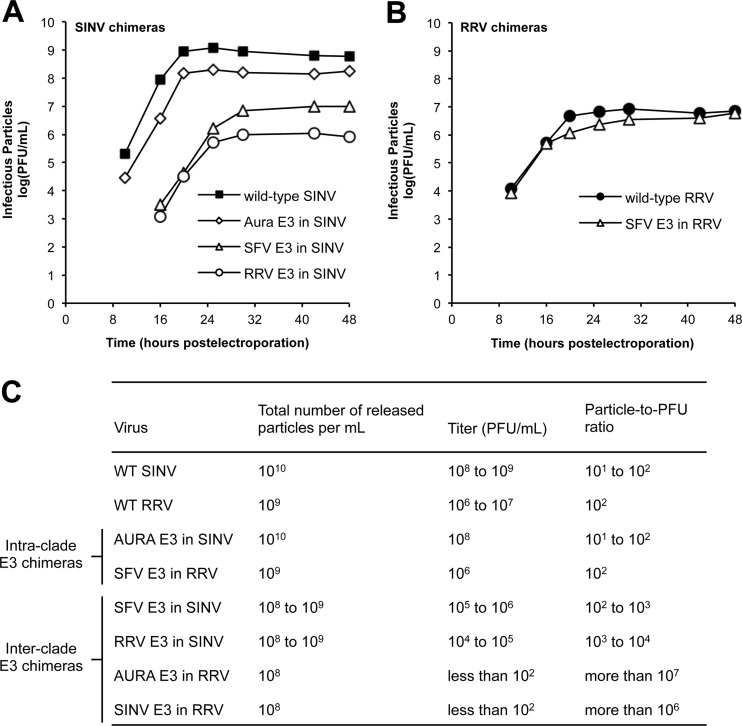
The SINV and RRV chimeras were initially characterized to determine the amount of infectious virus produced over time following electroporation (Fig. 2A and B). The intraclade E3 chimeras produced infectious virus at the same rate and to similar levels as the parental viruses, either wild-type SINV or RRV. Plaque sizes for these chimeras were also similar to those for the parental viruses (data not shown). In contrast, the interclade E3 chimeras produced smaller amounts of infectious virus. The interclade SINV chimeras (SFV E3 in SINV and RRV E3 in SINV) were reduced in titer by 2 to 3 log units, respectively, compared to those in wild-type SINV (Fig. 2A). The interclade RRV chimeras (AURA E3 in RRV and SINV E3 in RRV) displayed a more severe phenotype, producing at most 102 PFU/ml at 48 h postelectroporation. As expected, plaque sizes for each of the interclade E3 chimeras were smaller than those for the parental viruses (data not shown). We observed similar patterns of infectivity and similar differences in plaque size when the chimeric viruses were grown in the C6/36 mosquito cell line derived from Aedes albopictus (data not shown), indicating that the capacity of the E3 chimeras to assemble infectious virions is not cell type specific.
Fig 2.
Infectivities of SINV and RRV chimeras. (A and B) Production of infectious virus by SINV and RRV chimeras, respectively. BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At the indicated time points, the medium was harvested and titers were determined by a standard plaque assay procedure (33). The experiments to obtain growth curves were performed twice for each E3 chimera. The results from one representative experiment are shown. (C) Quality of the released SINV and RRV chimera particles. BHK-21 cells were electroporated with in vitro-transcribed wild-type (WT) or chimeric viral RNA. At 25 h postelectroporation, the medium was harvested and the particle number was quantitated using qRT-PCR. Virus titer was determined using a standard plaque assay procedure (33). For each virus, the experiments used to obtain the total number of particles released and titer were performed on three independent samples. The range of values obtained from these experiments is shown.
The lower titers observed for the interclade E3 chimeras could be due to an overall reduction in the total number of particles produced. qRT-PCR was used to quantify the number of viral RNA genome copies (or total number of particles) present in the medium of infected cells (41, 55). As expected, the intraclade E3 chimeras did not produce fewer particles than wild-type virus and had particle-to-PFU ratios that were similar to those of wild-type virus. For each of the interclade E3 chimeras, we observed only a 1- to 2-log-unit decrease in the total number of particles produced compared to that for the parental viruses; however, the differences in titer between the SINV and RRV interclade E3 chimeras led to significantly different particle-to-PFU ratios (Fig. 2B and C). The interclade SINV chimeras (SFV E3 in SINV and RRV E3 in SINV) had particle-to-PFU ratios that were only 1 to 2 log units higher than those of wild-type SINV. Alternatively, the particle-to-PFU ratios determined for the AURA E3 in RRV and SINV E3 in RRV were 4 to 5 log units higher than those determined for wild-type RRV. Taken together, the elevated particle-to-PFU ratios observed for the interclade E3 chimeras demonstrate that these viruses assemble particles that are not as competent to infect a new cell as the particles assembled by the parental viruses.
The interclade E3 chimeras produce particles with atypical morphologies and protein compositions.
The interclade RRV chimeras showed only a moderate reduction in the total number of particles produced but had significantly elevated particle-to-PFU ratios (Fig. 2C). These observations suggest that the virions formed by these chimeras have severe defects in terms of particle assembly, including the incorporation of misfolded or misarranged spikes or the incorporation of improperly processed spike proteins, such as uncleaved pE2 (E3-E2), into the released particles.
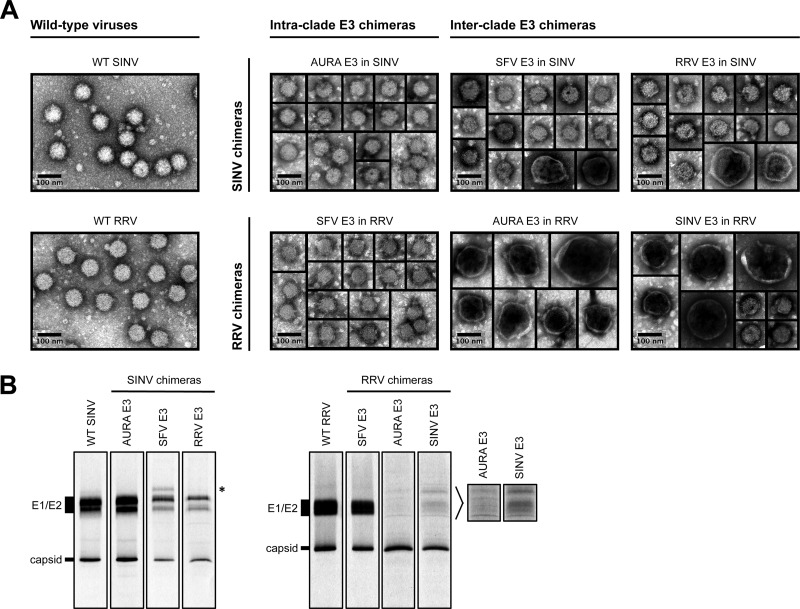
To examine the morphology of the released E3 chimera particles, virus particles present in the medium of infected cells were pelleted through a 27% sucrose cushion, resuspended in HNE buffer, and analyzed by negative-stain TEM (Fig. 3A). Wild-type SINV and RRV assembled spherical particles that were 70 nm in diameter, which is consistent with previous findings (5, 49). The intraclade E3 chimeras also produced homogeneous, spherical particles that were approximately 70 nm in diameter (Fig. 3A).
Fig 3.
Characterization of released SINV and RRV chimera particles. (A) TEM images of pelleted E3 chimera particles. Wild-type and chimeric viruses were pelleted from the medium of infected cells, resuspended in HNE buffer, and stained on a Formvar- and carbon-coated 400-mesh copper grid. The stained grids were imaged at ×40,000 magnification. (B) Composition of radiolabeled E3 chimera particles. [35S]cysteine-methionine-labeled wild-type and chimeric viruses were harvested from the medium of infected cells, purified using a two-step sucrose gradient, solubilized in reducing SDS sample buffer, and analyzed by SDS-PAGE. For each virus, 107 to 108 particles were loaded onto the gel. Particle number was quantitated using qRT-PCR. Migration of E1/E2 and capsid bands is indicated to the left of the images. The band that migrated slower than E1/E2 in SFV E3 in SINV is indicated with an asterisk to the right of the SINV chimera images. Longer exposures of the indicated regions for AURA E3 in RRV and SINV E3 in RRV are shown to the right of the RRV chimera images.
In contrast, we observed mixed morphologies for the interclade E3 chimeras (Fig. 3A). The interclade RRV chimeras produced only a small percentage of particles that were similar in size and morphology to those of wild-type RRV. The majority of the particles assembled by AURA E3 in RRV and SINV E3 in RRV had a large, pleomorphic phenotype. The defects in particle morphology observed for the interclade SINV chimeras were not as severe as those observed for the interclade RRV chimeras. SFV E3 in SINV and RRV E3 in SINV predominantly produced particles that were similar in size to wild-type SINV particles, with only a small percentage of particles displaying a large, pleomorphic phenotype. These results are consistent with the growth analysis and the particle-to-PFU data, in which the interclade RRV chimeras had more deleterious phenotypes than the interclade SINV chimeras (Fig. 2).
To examine the protein composition of the released particles, [35S]cysteine-methionine-labeled virus was purified from the medium using a two-step (27% and 60%) sucrose gradient. Virus present at the 27% and 60% sucrose step interface was extracted and analyzed by SDS-PAGE (Fig. 3B). This purification method was used because it is gentler than pelleting of the virus, which can disrupt unstable particles. As expected, capsid and E1/E2 were the predominant proteins present in wild-type SINV, wild-type RRV, and the intraclade E3 chimeras (AURA E3 in SINV and SFV E3 in RRV). Furthermore, the relative levels of capsid to E1/E2 were similar for each of these viruses. These observations are consistent with the intraclade SINV chimeras predominantly forming particles that are morphologically similar to those of the parental viruses (Fig. 3A). The interclade SINV chimeras (SFV E3 in SINV and RRV E3 in SINV) also showed discrete capsid and E1/E2 bands, though the relative amounts of each band appeared to be slightly different from those for wild-type SINV. Interestingly, SFV E3 in SINV incorporated an additional protein species that migrated where uncleaved pE2 (E3-E2) would be expected to run on an SDS-polyacrylamide gel (Fig. 3B; migration of the extra band is indicated with an asterisk to the right of the SINV chimera images).
In contrast, the interclade RRV chimeras (AURA E3 in RRV and SINV E3 in RRV) did not have discrete bands corresponding to E1 and E2. At longer exposure times, it appeared that multiple bands were present, possibly indicating different folded forms of E1/E2 (Fig. 3B; longer exposures of the glycoprotein bands are indicated to the right of the RRV chimera images). Furthermore, these chimeras had an abundance of capsid relative to E1/E2, which is consistent with the TEM images that show potentially multicored particles (Fig. 3A). Additionally, if the interclade RRV chimeras were damaged during step gradient purification, these particles may have lost their viral envelope. Thus, the purified particles would be expected to have reduced levels of E1/E2 relative to capsid, which is consistent with the findings presented in Fig. 3B. This interpretation would support our conclusion that the interclade RRV chimeras are less stable than either wild-type RRV or the intraclade RRV chimeras.
Thus far, our results demonstrate that when E3 is interchanged between virus species that are classified in the same clade, there are minimal effects on particle assembly. The intraclade E3 chimeras had titers and particle-to-PFU ratios that were comparable to those of the parental viruses (Fig. 2). Furthermore, TEM and SDS-PAGE analysis of purified particles indicated that the intraclade E3 chimeras assembled virions that were similar in size, morphology, and protein composition to the parental viruses (Fig. 3). In contrast, the interclade E3 chimeras had reduced infectivity (Fig. 2) and assembled particles with atypical morphologies and protein compositions (Fig. 3). Interestingly, the interclade RRV chimeras showed consistently more severe assembly defects than the interclade SINV chimeras.
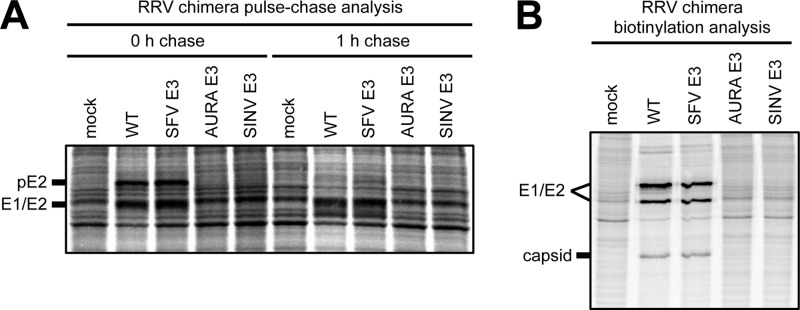
The interclade RRV chimeras have reduced levels of intracellular and cell surface spike glycoprotein expression within infected cells.
The small amounts of E1/E2 relative to capsid in the released interclade RRV chimera particles (Fig. 3B; see AURA E3 in RRV and SINV E3 in RRV) suggest that there is either reduced intracellular or reduced cell surface expression of the spike glycoproteins during infection. Using pulse-chase and biotinylation analyses, we investigated what stage(s) of assembly, either spike protein expression or spike transport to the cell surface, is affected.
We first examined the total amounts of E1/E2 expressed in infected cells (Fig. 4A, 0-h chase). For the interclade RRV chimeras, pE2 and E1/E2 were present at lower levels in cell lysates than those for either wild-type RRV or SFV E3 in RRV. We examined the levels of pE2 and E1/E2 after a 1-h chase to determine the lifetime of the spike proteins in the intra- and interclade RRV chimeras. Wild-type RRV and SFV E3 in RRV showed a decrease of pE2 levels, presumably because of cleavage of E3 from E2. The interclade RRV chimeras. showed reduced levels of E1/E2, consistent with what was observed at the 0-h chase. These results demonstrate that a mismatched E3 affects the folding and stability of the spike glycoproteins within infected cells.
Fig 4.
Intracellular and cell surface spike glycoprotein expression in cells infected with RRV chimeras. (A) Pulse-chase analysis probing for intracellular spike glycoprotein expression. BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At 15 h postelectroporation, the cells were labeled with [35S]cysteine-methionine for 15 min. After radiolabeling, the cells were either immediately lysed (0-h-chase time point) or chased for 1 h in unlabeled medium and then lysed (1-h-chase time point). Cell lysates were solubilized in reducing SDS sample buffer and analyzed by SDS-PAGE. Migration of spike glycoprotein bands is indicated to the left of the image. (B) Biotinylation analysis probing for cell surface spike glycoprotein expression. BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At 15 h postelectroporation, the cells were labeled with [35S]cysteine-methionine medium for 2 h. After incubation, cell surface proteins were biotinylated with 2.5 mM sulfo-NHS-LC-biotin. Following biotinylation, the cells were lysed with BHK cell lysis buffer and biotinylated proteins were purified from cell lysates using streptavidin resin and eluted by adding reducing SDS sample buffer and boiling at 95°C for 10 min. The eluted proteins were analyzed by SDS-PAGE. Migration of spike glycoprotein and capsid bands is indicated to the left of the image.
To address if E1/E2 are transported to the plasma membrane, cell surface proteins were biotinylated, purified with streptavidin resin, and analyzed by SDS-PAGE (Fig. 4B). As expected for SFV E3 in RRV, the spike proteins were present at the cell surface at levels comparable to those of wild-type RRV. In contrast, small amounts of E1/E2 were present at the plasma membrane in cells infected with either of the interclade RRV chimeras, consistent with the overall small amounts of E2 and E1 present during an infection (Fig. 4A). For wild-type RRV and SFV E3 in RRV, we also observed low levels of capsid protein, which is consistent with the spike proteins interacting with the nucleocapsid core at the plasma membrane prior to budding. The results of immunofluorescence experiments probing for cell surface glycoprotein expression were consistent with the biotinylation data (data not shown). Taken together, the reduced intracellular and cell surface spike glycoprotein expression observed for the interclade RRV chimeras is consistent with overall spike misfolding.
Deletion of the CXXC motif from RRV E3 does not affect the production of infectious virus.
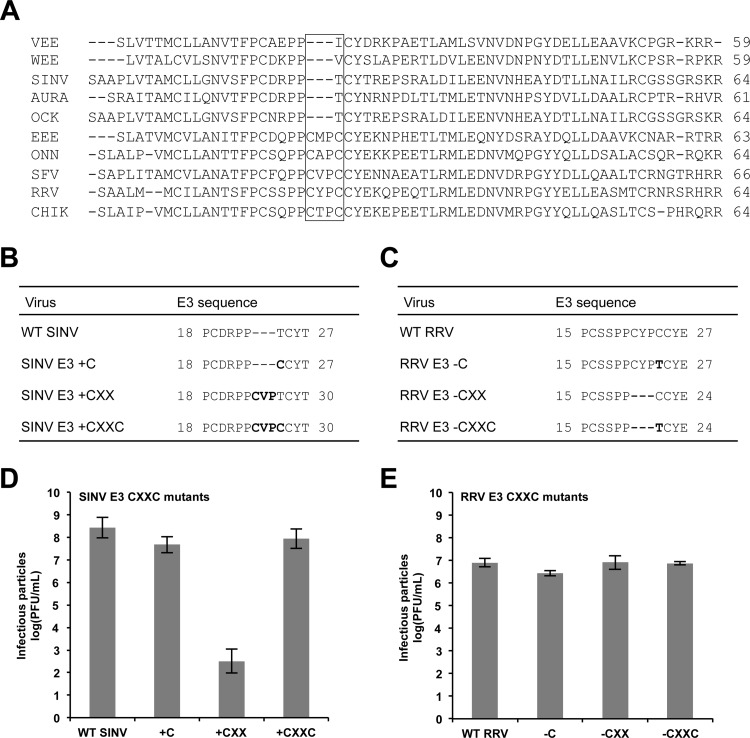
To investigate why E3 is not interchangeable between different clades, we compared the amino acid sequences of a representative group of E3 glycoproteins. This analysis revealed a subset of E3 glycoproteins that contain an extra CXXC motif (Fig. 5A). This motif is characteristic of many redox proteins, such as protein disulfide isomerase, and proteins that undergo disulfide reshuffling during folding (48, 50). Interestingly, the E3 proteins of both RRV and SFV (members of the SFV clade) contain this motif, whereas the E3 proteins of SINV and AURA (members of the WEE clade) do not. In the atomic structure of CHIK E3, the CXXC motif is part of an extended loop that does not make direct contacts with E2 (45).
Fig 5.
Characterization of the RRV and SINV E3 CXXC mutants. (A) Amino acid sequence alignments of a representative group of alphavirus E3 glycoproteins. The E3 CXXC motif is indicated with a box. VEE, Venezuelan equine encephalitis virus; WEE, Western equine encephalitis virus; SINV, Sindbis virus; AURA, Aura virus; OCK, Ockelbo virus; EEE, Eastern equine encephalitis virus; ONN, o'nyong nyong virus; SFV, Semliki Forest virus; RRV, Ross River virus; CHIK, Chikungunya virus. (B and C) Amino acid sequences of SINV and RRV E3 CXXC mutants, respectively. (D and E) Infectivities of SINV and RRV E3 CXXC mutants, respectively. BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA. At 25 h postelectroporation, the medium was harvested and titers were determined by a standard plaque assay procedure (33). For each virus, the mean titer determined from three independent experiments is shown. Error bars indicate standard deviations.
To determine if the CXXC motif plays a role in the clade specificity observed for the E3 chimeras, we inserted all or parts of the CXXC motif into SINV E3 (Fig. 5B). We inserted CXXC, CXX, and C alone (+CXXC, +CXX, and +C, respectively) into wild-type SINV, so SINV E3 was more similar in amino acid sequence to RRV E3. SINV E3 +CXX was the only mutant that produced less infectious virus than wild-type SINV, which is consistent with previous findings (Fig. 5D) (34).
Next, we designed mutants in which all or parts of the CXXC motif were deleted from RRV E3 (Fig. 5C). We deleted CXXC, CXX, and C alone (−CXXC, −CXX, and −C, respectively) from wild-type RRV, so RRV E3 was more similar in amino acid sequence to SINV E3. At 25 h postelectroporation, each of the RRV E3 CXXC mutants produced infectious virus at levels comparable to those of wild-type RRV, demonstrating that this motif is not required for RRV assembly (Fig. 5E). Together, these results suggest that the different phenotypes observed for the inter- and intraclade E3 chimeras are likely not a result of the absence or presence of the CXXC motif.
Restoring proper interactions at the E3-E2 interface enhances assembly of the RRV E3 in SINV chimera.
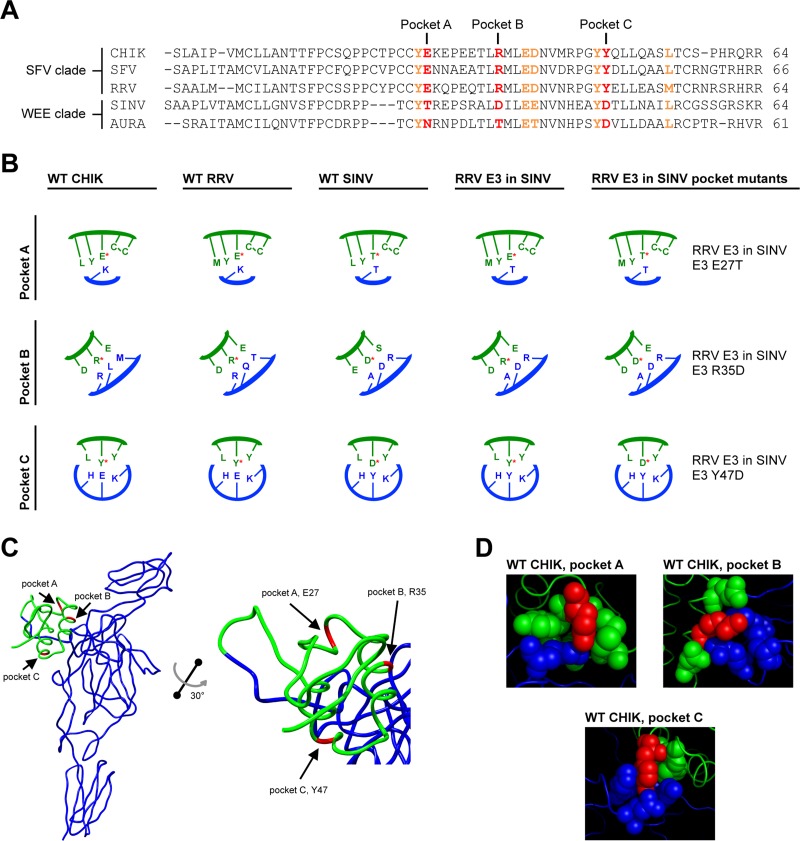
To understand how E3 may affect E2 folding and function, we examined the atomic structure of the pE2-E1 heterodimer of CHIK (a member of the SFV clade). In this structure, there are eight residues in E3 that interact with E2 through either hydrogen bonds or salt bridges (Fig. 6A; residues colored in red and orange) (45). Sequence alignments show that three of the eight amino acids are significantly different in terms of charge and/or size between the SFV and WEE clades (Fig. 6A; residues colored in red). Furthermore, the three E3 contact residues fall into three distinct pockets that are distributed throughout the E3-E2 interface (Fig. 6B and C). We termed these pockets A, B, and C (Fig. 6A to D). To test if one or more of the three E3 pocket residues are important for assembly, we mutated amino acids RRV E3 E27, R35, and Y47 (pockets A, B, and C, respectively) in the RRV E3 in SINV chimera by changing these residues in RRV E3 to the residues that are found at the corresponding positions in SINV E3 (Fig. 6A and B). If any of the three E3 contacts are required for particle formation, these changes were predicted to increase the titer of the RRV E3 in SINV chimera. If multiple sites are important, making double or triple mutations should increase the production of infectious virus.
Fig 6.
E3 contact residues mutated within RRV E3 in SINV. (A) E3 amino acid sequence alignments. E3 residues that interact with E2 through hydrogen bonds or salt bridges are in either red or orange bold type (45). The residues mutated within E3 of RRV E3 in SINV and the corresponding residues in CHIK, SFV, AURA, and SINV E3 are in red bold type. (B) Schematics of E3-E2-interacting pockets A, B, and C. E3 residues are represented in green, and E2 residues are represented in blue. The residues mutated within E3 of RRV E3 in SINV (E27, R35, and Y47) (RRV numbering) are indicated with a red asterisk. (C) Atomic structure of the CHIK pE2 glycoprotein and localization of the E3 contact residues mutated within E3 of RRV E3 in SINV (45) (Protein Data Bank accession number 3N40). The E2 glycoprotein is represented in blue, and the E3 glycoprotein is represented in green. The residues mutated within E3 of RRV E3 in SINV are colored in red and indicated with arrows (RRV numbering). (D) Structures of E3-E2-interacting pockets A, B, and C. E3 and E2 residues that comprise pockets A, B, and C are represented as spheres. E3 residues are represented in green, and E2 residues are represented in blue. The residues mutated within E3 of RRV E3 in SINV are colored in red.
We made the single amino acid mutations RRV E3 E27T, R35D, and Y47D as well as double and triple mutations within the RRV E3 in SINV chimera. The titers of the mutant chimeras were analyzed at 25 h postelectroporation (Fig. 7A). RRV in SINV/E3 E27T (pocket A) and RRV E3 in SINV/E3 R35D (pocket B) produced fewer infectious particles than RRV E3 in SINV. The reduced titers suggest that there may be alternative interactions in pockets A and B that are important for particle assembly. In contrast, we observed a 1- to 2-log-unit increase in titer for RRV E3 in SINV/E3 Y47D (pocket C) compared to that for RRV E3 in SINV. We did not observe significant increases in infectivity for any of the double or triple mutants (data not shown). These results demonstrate that the interactions in pocket C are important for E3 function during assembly of the RRV E3 in SINV chimera.
Fig 7.
Characterization of RRV E3 in SINV pocket mutants. (A) Infectivities of RRV E3 in SINV pocket mutants. BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At 25 h postelectroporation, the medium was harvested and titers were determined by a standard plaque assay procedure (33). For each virus, the mean titer determined from three independent experiments is shown. Error bars indicate standard deviations. (B) Protein composition of the released RRV E3 in SINV/E3 Y47D particles. [35S]cysteine-methionine-labeled wild-type and chimeric virus were harvested from the medium of infected cells, purified using a two-step sucrose gradient, solubilized in reducing SDS sample buffer, and analyzed by SDS-PAGE. For each virus, 107 to 108 particles were loaded onto the gel. Particle number was quantitated using qRT-PCR. Migration of E1/E2 and capsid bands is indicated to the left of the images. The band that migrated slower than E1/E2 in RRV E3 in SINV/E3 Y47D is indicated with an asterisk to the right of the images.
To determine if the released RRV E3 in SINV/E3 Y47D particles had protein compositions that were similar to those of wild-type SINV particles, [35S]cysteine-methionine-labeled virions were purified from the medium of infected cells and analyzed by SDS-PAGE (Fig. 7B). Consistent with RRV E3 in SINV (Fig. 3B), we observed discrete E1/E2 and capsid bands in the purified RRV E3 in SINV/E3 Y47D particles.
RRV E3 in SINV/E3 Y47D also contained one extra band, which migrated slower than E1/E2 (Fig. 7B; migration of the extra band is indicated with an asterisk to the right of the images). The additional band, which was also observed for SFV E3 in SINV (Fig. 3B), migrated where uncleaved pE2 (E3-E2) would be expected to run on an SDS-polyacrylamide gel. Incorporating uncleaved pE2 into the released virions is consistent with the reduced infectivity of RRV E3 in SINV/E3 Y47D compared to that of wild-type SINV (Fig. 7A) (17). Alternatively, the additional band could indicate a furin-processed E2 with an alternative glycosylation pattern. To determine if this slow-mobility band in SFV E3 in SINV or the RRV E3 in SINV/E3 Y47D mutant is due to aberrant glycosylation, we treated purified mutant particles with N-glycosidase F (PNGase F), which cleaves N-linked glycosylation. The treated particles still showed three distinct bands, demonstrating that the differences in mobility are not due to N-linked glycosylation (data not shown).
Revertants of the SINV E3 in RRV chimera map to residues at the E3-E2 interface.
Thus far, our results indicate that the interclade RRV chimeras are severely reduced in infectivity (Fig. 2B and C). As a complementary strategy to examine how E3 affects E2 folding and/or function in a clade-specific manner, we used forward genetics to isolate second-site revertants. Toward this end, we serially passaged the SINV E3 in RRV chimeras. At passage 3, we isolated and sequenced plaques that were 3 mm in diameter; in comparison, the plaques that were initially observed for SINV E3 in RRV were <1 mm diameter (data not shown). Sequencing of the large plaque isolates identified two amino acid changes in E2: T171R and E166K (RRV numbering). These residues are found in pockets B and C, respectively.
RRV E2 T171R and E2 E166K were introduced individually and in combination back into the SINV E3 in RRV chimeras, and titers were analyzed at 25 h postelectroporation (Fig. 8B). SINV E3 in RRV/E2 T171R (pocket B) increased infectivity by approximately 4 log units, bringing the virus titer back to wild-type levels. SINV E3 in RRV/E2 E166K (pocket C) enhanced infectivity by only 1 to 2 log units. Interestingly, introducing the second-site changes together into SINV E3 in RRV generated virus that grew to slightly higher titers than wild-type RRV (Fig. 8B).
Fig 8.
Characterization of SINV E3 in RRV revertants. (A) Schematics of E3-E2-interacting pockets B and C. E3 residues are represented in green, and E2 residues are represented in blue. The location of the revertants within SINV E3 in RRV and the corresponding residues in wild-type RRV and SINV are indicated with a red asterisk. (B) Infectivities of SINV E3 in RRV revertants. BHK-21 cells were electroporated with in vitro-transcribed wild-type or chimeric viral RNA. At 25 h postelectroporation, the medium was harvested and titers were determined by a standard plaque assay procedure (33). For each virus, the mean titer determined from three independent experiments is shown. Error bars indicate standard deviations. (C) Protein composition of the released SINV E3 in RRV revertant particles. [35S]cysteine-methionine-labeled wild-type and chimeric viruses were harvested from the medium of infected cells, purified using a two-step sucrose gradient, solubilized in reducing SDS sample buffer, and analyzed by SDS-PAGE. For each virus, 107 to 108 particles were loaded onto the gel. Particle number was quantitated using qRT-PCR. Migration of E1/E2 and capsid bands is indicated to the left of the images.
To determine if the second-site changes also restored the atypical protein compositions observed for the interclade RRV chimeras (Fig. 3B), [35S]cysteine-methionine-labeled virus was purified from the medium of infected cells and analyzed by SDS-PAGE (Fig. 8C). Consistent with the growth data, both SINV E3 in RRV/E2 T171R and SINV E3 in RRV/E2 T171R E166K assembled particles that were similar in protein composition to wild-type RRV particles. In contrast, SINV E3 in RRV/E2 E166K assembled particles that were similar in protein composition to particles for SINV E3 in RRV. Taken together, the data presented here show that maintaining a small number of specific contacts at the E3-E2 interface is necessary to promote efficient and accurate alphavirus assembly.
DISCUSSION
E3 is a small, ∼65-amino-acid glycoprotein that is dispensable for alphavirus attachment and entry into a new cell (17, 37, 38, 46, 51); however, E3 is absolutely required for particle assembly (2, 3, 23, 26). To further investigate a role for E3 in regulating spike assembly, we made and characterized E3 chimeric viruses. We observed defects in infectivity and in particle morphology and composition for chimeras in which E3 was derived from a different clade than the parental virus. In contrast, assembly proceeded in a manner similar to that for wild-type virus for chimeras in which E3 was derived from the same clade as the parental virus.
The location of E3 in the atomic structure of the CHIK pE2-E1 heterodimer has led to the model in which E3 acts as a clamp to stabilize the interactions between E2 and E1 during assembly (21, 45). Specifically, uncleaved E3 prevents premature disassembly or fusion of the heterodimer as the spikes are exposed to changes in pH in the endoplasmic reticulum and the host secretory system. When E3 is cleaved from E2 in the trans-Golgi network (17, 37, 38, 46, 51), the spikes transition from a stable conformation to a metastable conformation (16, 21, 45). This transition is necessary so that the E2-E1 heterodimer readily dissociates upon exposure to low pH and mediates fusion between the viral and host cell membrane during viral entry (17). Previous work has shown that a second-site revertant of a furin cleavage-deficient strain of SFV maps to E2 R250 (SFV numbering) (52). This residue is located in pocket B and interacts with E3 through a putative hydrogen bond (Fig. 6 and 8) (45). The second-site reversion is predicted to destabilize the E3-E2 contacts in pocket B, allowing E2 and E1 to dissociate more readily during entry. For the interclade E3 chimeras studied in this work, we propose that nonoptimal interactions at the E3-E2 interface prevent accurate spike assembly. The mismatched E3 glycoproteins are likely unable to stabilize the pE2-E1 heterodimer during spike formation and transport to the plasma membrane. Consequently, the heterodimers fold into multiple conformations (Fig. 3) that either are targeted for degradation by the host cell (Fig. 4) or oligomerize to form labile spike complexes. Alternatively, the interclade E3 chimeras may assemble spikes that are more susceptible to premature fusion with cellular membranes during transport in the host secretory system.
Amino acid size and charge are likely to be key factors in maintaining proper protein-protein interactions at the interface between E3 and E2. In pocket A, a putative salt bridge interaction is made between the side chains of E3 E28 and E2 K10 (CHIK numbering). Pocket B is not as well-defined structurally; E2 residues in this pocket are derived from mostly unstructured and loop regions. Nonetheless, the peptide backbone of E3 R36 and the side chain of E2 R251 (CHIK numbering) interact through a putative hydrogen bond. Within pocket C of CHIK, the side chains of E3 Y48 and E2 E166 (CHIK numbering) associate through a putative hydrogen bond.
Using both reverse and forward genetic approaches, we identified mutations in pockets B and C that rescued assembly of the interclade E3 chimeras. For RRV E3 in SINV, the RRV E3 Y47D change increased the viral titer. This mutation likely restored proper E3-E2 contacts in pocket C (Fig. 6 and 7). For SINV E3 in RRV, second-site reversions mapped to E2 residues in pockets B and C (Fig. 8). The T-to-R mutation at RRV E2 171 (pocket B) changes the RRV residue to what is found at the corresponding position in wild-type SINV, thus possibly restoring a putative interaction found naturally in SINV. The change from E to K at RRV E2 166 (pocket C) is likely removing a repelling charge and could be introducing a stabilizing salt bridge. The second-site mutations in SINV E3 in RRV increased the viral titer (Fig. 8B) and restored the presence of E1/E2 in the released virus particles (Fig. 8C).
The E2 residues that comprise pockets B and C (Fig. 6 to 8) are all part of the E2 acid-sensitive region (ASR). During viral entry, exposure to low pH causes large conformational changes in the E2 ASR, which contribute to heterodimer disassembly (21, 45). Recently, it has been shown that mutating a single residue in the E2 ASR improved the yield of virus-like particles generated from CHIK, demonstrating that E3-E2 and E2-E1 interactions in this region are required for assembly (1). Our mutational results are consistent with this finding (Fig. 6 to 8). It is important to note that pockets A, B, and C were identified from the atomic structure of the CHIK pE2-E1 heterodimer in its fully folded conformation (45). There are potentially more E3-E2 interactions that are required for assembly; however, the residues that make these contacts may not be in close proximity to each other in the mature structure.
Similar to the alphavirus E3 glycoprotein, cleavage of the flavivirus prM protein into the pr peptide and the M protein is a critical step in particle maturation (7, 42). Uncleaved prM interacts with the envelope (E) protein to prevent premature conformational changes from occurring within the spikes (14, 15). It has also been proposed that prM functions as a chaperone to aid in folding of the E protein (28). This chaperone activity is mediated through a small number of interactions at the prM-E interface (22, 53, 54). Taken together, many viruses express small, indispensable viral proteins that are required for assembly but whose precise functions are not clearly understood. These proteins, including the alphavirus E3 glycoprotein, may regulate multiple stages of the viral life cycle.
ACKNOWLEDGMENTS
We thank members of the Indiana University virology group for helpful discussions.
Anthony J. Snyder was supported on Indiana University Genetics, Cellular and Molecular Sciences training grant T32-GM007757.
Footnotes
Published ahead of print 3 October 2012
REFERENCES
- 1. Akahata W, Nabel GJ. 2012. A specific domain of the Chikungunya virus E2 protein regulates particle formation in human cells: implications for alphavirus vaccine design. J. Virol. 86:8879–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonatti S, Blobel G. 1979. Absence of a cleavable signal sequence in Sindbis virus glycoprotein PE2. J. Biol. Chem. 254:12261–12264 [PubMed] [Google Scholar]
- 3. Bonatti S, Migliaccio G, Blobel G, Walter P. 1984. Role of signal recognition particle in the membrane assembly of Sindbis viral glycoproteins. Eur. J. Biochem. 140:499–502 [DOI] [PubMed] [Google Scholar]
- 4. Byrnes AP, Griffin DE. 1998. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 72:7349–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng RH, et al. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Curtis I, Simons K. 1988. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci. U. S. A. 85:8052–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elshuber S, Allison SL, Heinz FX, Mandl CW. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183–191 [DOI] [PubMed] [Google Scholar]
- 8. Firth AE, Chung BY, Fleeton MN, Atkins JF. 2008. Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol. J. 5:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forrester NL, et al. 2012. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J. Virol. 86:2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gahmberg CG, Utermann G, Simons K. 1972. The membrane proteins of Semliki Forest virus have a hydrophobic part attached to the viral membrane. FEBS Lett. 28:179–182 [DOI] [PubMed] [Google Scholar]
- 11. Garoff H, Frischauf AM, Simons K, Lehrach H, Delius H. 1980. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature 288:236–241 [DOI] [PubMed] [Google Scholar]
- 12. Garoff H, Simons K, Renkonen O. 1974. Isolation and characterization of the membrane proteins of Semliki Forest virus. Virology 61:493–504 [DOI] [PubMed] [Google Scholar]
- 13. Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 doi:10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guirakhoo F, Bolin RA, Roehrig JT. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191:921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 72(Pt 6):1323–1329 [DOI] [PubMed] [Google Scholar]
- 16. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain SK, DeCandido S, Kielian M. 1991. Processing of the p62 envelope precursor protein of Semliki Forest virus. J. Biol. Chem. 266:5756–5761 [PubMed] [Google Scholar]
- 18. Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhn RJ, Niesters HG, Hong Z, Strauss JH. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430–441 [DOI] [PubMed] [Google Scholar]
- 20. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 21. Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L, et al. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834 [DOI] [PubMed] [Google Scholar]
- 23. Liljestrom P, Garoff H. 1991. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liljestrom P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lobigs M, Wahlberg JM, Garoff H. 1990. Spike protein oligomerization control of Semliki Forest virus fusion. J. Virol. 64:5214–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lobigs M, Zhao HX, Garoff H. 1990. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol. 64:4346–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorenz IC, Allison SL, Heinz FX, Helenius A. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76:5480–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lustig S, et al. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Melancon P, Garoff H. 1987. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 61:1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mulvey M, Brown DT. 1996. Assembly of the Sindbis virus spike protein complex. Virology 219:125–132 [DOI] [PubMed] [Google Scholar]
- 32. Omar A, Koblet H. 1988. Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology 166:17–23 [DOI] [PubMed] [Google Scholar]
- 33. Owen KE, Kuhn RJ. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parrott MM, et al. 2009. Role of conserved cysteines in the alphavirus E3 protein. J. Virol. 83:2584–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rice CM, Bell JR, Hunkapiller MW, Strauss EG, Strauss JH. 1982. Isolation and characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. J. Mol. Biol. 154:355–378 [DOI] [PubMed] [Google Scholar]
- 36. Rumenapf T, Strauss EG, Strauss JH. 1994. Subgenomic mRNA of Aura alphavirus is packaged into virions. J. Virol. 68:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salminen A, Wahlberg JM, Lobigs M, Liljestrom P, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sjoberg M, Lindqvist B, Garoff H. 2011. Activation of the alphavirus spike protein is suppressed by bound E3. J. Virol. 85:5644–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith TJ, et al. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A. 92:10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snyder AJ, Sokoloski KJ, Mukhopadhyay S. 2012. Mutating conserved cysteines in the alphavirus e2 glycoprotein causes virus-specific assembly defects. J. Virol. 86:3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sokoloski KJ, et al. 2012. Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res. 167:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stadler K, Allison SL, Schalich J, Heinz FX. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. von Bonsdorff CH, Harrison SC. 1978. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 28:578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voss JE, et al. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 46. Wahlberg JM, Boere WA, Garoff H. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63:4991–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wahlberg JM, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woycechowsky KJ, Wittrup KD, Raines RT. 1999. A small-molecule catalyst of protein folding in vitro and in vivo. Chem. Biol. 6:871–879 [DOI] [PubMed] [Google Scholar]
- 49. Zhang R, et al. 2011. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30:3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang RM, Snyder GH. 1989. Dependence of formation of small disulfide loops in two-cysteine peptides on the number and types of intervening amino acids. J. Biol. Chem. 264:18472–18479 [PubMed] [Google Scholar]
- 51. Zhang X, Fugere M, Day R, Kielian M. 2003. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 77:2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X, Kielian M. 2004. Mutations that promote furin-independent growth of Semliki Forest virus affect p62-E1 interactions and membrane fusion. Virology 327:287–296 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y, et al. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y, et al. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zybert IA, van der Ende-Metselaar H, Wilschut J, Smit JM. 2008. Functional importance of dengue virus maturation: infectious properties of immature virions. J. Gen. Virol. 89:3047–3051 [DOI] [PubMed] [Google Scholar]