Abstract
In this study, an antisense luciferase-expressing human immunodeficiency virus type 1 (HIV-1) molecular clone was used to infect primary cells. We found that antisense transcription activity from the 3′ long terminal repeat (LTR) was significantly more abundant in monocyte-derived cells than in activated T lymphocytes. Moreover, by analyzing antisense transcription in infected monocyte-derived dendritic cells (MDDCs), we observed that the majority of HIV-1-infected MDDCs with significant antisense transcription activity did not produce Gag. We also confirmed that the negative-strand-encoded antisense protein (ASP) was expressed in monocyte-derived cells.
TEXT
Human T-cell leukemia virus type 1 (HTLV-1) is the first retrovirus from which the production of antisense transcripts from the 3′ long terminal repeat (LTR) has been clearly demonstrated (11, 17, 19, 22). Antisense transcription in human immunodeficiency virus type 1 (HIV-1) has also been characterized (10, 16, 20). Interestingly, such antisense transcripts can potentially encode proteins (9, 14, 15, 18). We have indeed demonstrated that the antisense protein (ASP) can be expressed in HIV-1-infected cells (12). We focused here on the regulation of HIV-1 antisense transcription in primary cells.
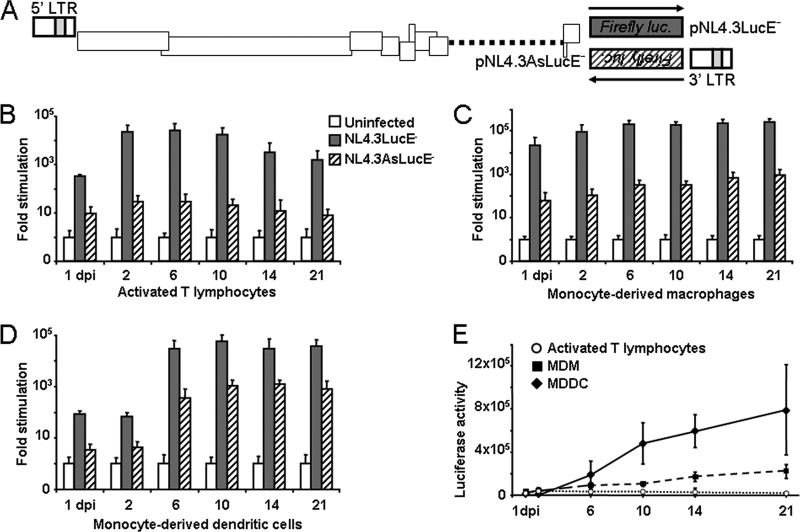
In this study, we used pNL4.3LucE−, a proviral DNA in which the Firefly luciferase gene had been inserted in nef (13). We removed its reporter gene to reinsert the luciferase gene in the same position but in the inverse orientation as already described (16). This vector, pNL4.3AsLucE− (Fig. 1A), permits quantification of antisense transcription from the 3′ LTR, since the reporter gene was placed under the control of the antisense transcription promoter (16). As these proviral DNAs are env deficient, vesicular stomatitis virus envelope (VSVg)-pseudotyped HIV-1 particles were produced. Their decreased requirement for Nef, together with their high infectious titers and their broad host range, made these viruses useful for our experiments, which involved different target cells for infection (1). Moreover, the restriction of monocyte-derived cells to HIV-1 is also observed in this case (2). To produce pseudotyped viruses, HEK-293T cells were cotransfected with proviral DNA (30 μg) and a VSVg expression vector (18 μg). Cells were grown for 2 days before viruses were collected. Virus titers were determined by infecting Jurkat cells with 2-fold serial dilutions of virus stocks. At 2 days postinfection (p.i.), the percentage of infected cells was determined by flow cytometry following intracellular staining of Gag with KC57 antibodies.
Fig 1.
HIV-1 antisense transcription during viral infection in primary cells. (A) Structure of the luciferase reporter vectors pNL4.3LucE− and pNL4.3AsLucE−. The dotted line indicates the untranslated region of env due to the introduction of a stop codon. The dark gray and diagonally striped boxes correspond, respectively, to the Firefly luciferase reporter gene under the control of the 5′ LTR (sense arrow) and the 3′ LTR (antisense arrow) for sense or antisense transcription analyses. (B to D) Analysis of transcriptional sense and antisense activities in primary cells prepared from three different blood donors. Freshly isolated CD4+ T cells (B) were cultured in plates coated with anti-human CD3 and anti-human CD28 antibodies (3, 8), while monocytes were differentiated into macrophages (C) or dendritic cells (D). On day 5, cells were infected with NL4.3LucE− or NL4.3AsLucE− virions pseudotyped with VSVg (MOI = 2). At different days p.i., cell extracts were prepared and normalized for protein content, and Firefly luciferase activity was subsequently measured. Luciferase values are expressed as fold increases relative to values measured in uninfected cells for each day p.i. Values represent means ± standard errors of the means (SEM) (n = 3). Luciferase activities are presented on a logarithmic scale. (E) Comparison of levels of antisense transcription activity in primary cells infected with NL4.3AsLucE− virions. Reported values are the average luminescence levels determined from the three experiments represented in panels B, C, and D. Results are represented as the luciferase activity values normalized for protein content. Values represent means ± SEM. dpi, days postinfection.
Peripheral blood mononuclear cells (PBMC) were isolated from buffy coats, and CD14+ cells were isolated with anti-human CD14-coated magnetic beads (Miltenyi Biotec). Differentiation of CD14+ monocytes into macrophages or dendritic cells was performed as already described (6, 7). CD4+ T cells were isolated from CD14− peripheral blood lymphocytes with a CD4+ T-cell enrichment kit (Stem Cell Technologies). Cell populations were infected at a multiplicity of infection (MOI) of 2 for both virus strains, and Firefly luciferase activity was measured at different days p.i. Infection of activated primary CD4+ T lymphocytes showed a strong activation of sense transcription from the 5′ LTR, while antisense transcription from the 3′ LTR remained low (Fig. 1B and E); levels of antisense activity were about 1,000-fold lower than that of sense transcription. Infection of monocyte-derived macrophages (MDMs) again showed higher sense than antisense transcription levels. However, antisense transcription activity was significantly higher than in T cells (Fig. 1E), increasing steadily and reaching 1,000-fold induction at 21 days p.i. (Fig. 1C). Regulation of antisense transcription was quite different in infected monocyte-derived dendritic cells (MDDCs). During the 2 first days of infection, sense and antisense transcription levels were low, and both increased at 6 days p.i. and remained high at up to 21 days p.i. (Fig. 1D and E). Interestingly, levels of antisense transcription in MDDCs were only 25- to 100-fold lower than that of sense transcription.
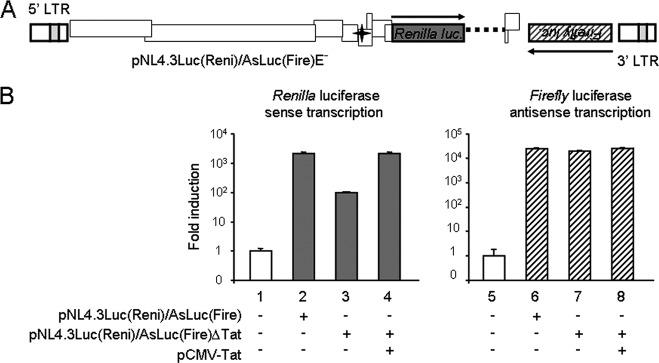
We next studied the effects of the Tat transactivator on antisense transcription activity. Controversial data have been published so far (16, 20, 21). Most results were obtained from cell lines transfected with a Tat expression plasmid and reporter vectors containing only one LTR. We readdressed these results using pNL4.3Luc(Reni)/AsLuc(Fire)E−, corresponding to the NL4.3 molecular clone expressing an antisense Firefly luciferase gene under the control of the 3′ LTR and the Renilla luciferase gene after insertion into the env gene under the control of the Tat-dependent sense transcription (Fig. 2A). In addition, we introduced a mutation into the tat gene (pNL4.3Luc(Reni)/AsLuc(Fire)E−ΔTat), blocking its expression. Tat mutation reduced Renilla luciferase activity, while Firefly luciferase expression was not altered by the absence of Tat (Fig. 2B). When pNL4.3Luc(Reni)/AsLuc(Fire)E−ΔTat was cotransfected with a Tat expression vector, sense transcription, unlike antisense transcription, was stimulated.
Fig 2.
Study of effect of Tat on antisense transcription. (A) Generation of a proviral construct expressing the antisense Firefly luciferase gene under the control of the 3′ LTR (antisense arrow) and the sense Renilla luciferase gene under the control of the 5′ LTR (sense arrow). pNL4.3Luc(Reni)/AsLuc(Fire)E− was derived from pNL4.3AsLucE− as described for Fig. 1A. The NheI-KpnI fragment of the pNL4.3AsLucE− env gene was replaced by the NheI-KpnI Renilla luciferase gene from pRL-TK (Promega) (23). The dark gray box and the star correspond to the Renilla luciferase gene cloned into the env gene and the stop codon introduced in the tat gene, respectively. (B) Effect of Tat on sense and antisense transcription. HEK 293T cells were cotransfected with 1 μg of the dual-luciferase-expressing proviral DNA, 1 μg of pCMV-Tat or empty pCMV-Neo, and 1 μg of pcDNA3.1-lacZ. Cells were grown for 2 days and then lysed in passive lysis buffer (Promega). Cell extracts were normalized for protein content, and Firefly and Renilla luciferase activities were measured using the Genofax A and C reporter assay systems (Yelen Corp., France), respectively. Luciferase activities were normalized to β-galactosidase activity. Luciferase activities are presented on a logarithmic scale. Results are presented as fold induction compared to the control (CTL) data (white columns), which correspond to levels measured in transfected cells without reporter vector. Values represent means ± standard deviations (SD) (n = 3).
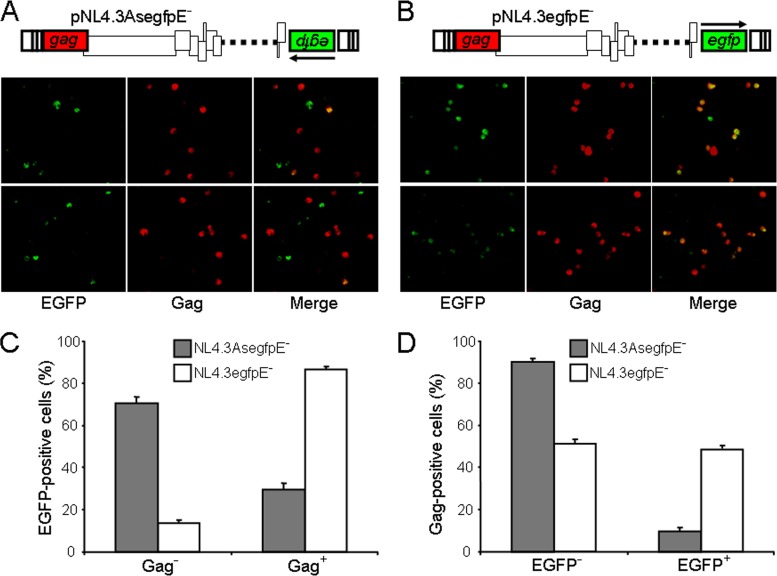
Higher antisense transcription activity was preferentially observed in HIV-1-infected MDDCs at 14 days p.i. However, the question remained whether both transcriptional activities could coexist within the same cell. Therefore, we analyzed the antisense transcription in single infected MDDCs. To identify cells capable of activating antisense transcription from the 3′ LTR, we inserted egfp under the control of the antisense transcription promoter (see Fig. 3A). The novel virus (NL4.3AsegfpE−) was pseudotyped with VSVg to infect MDDCs. As a control, we produced virions from a sense enhanced green fluorescent protein (EGFP)-expressing NL4.3 clone (pNL4.3egfpE−) in which egfp had been inserted in nef (Fig. 3B). At 14 days p.i., transcriptional activity of each infected MDDC was assessed by analyzing the resulting EGFP expression. Expression of gag was also analyzed by intracellular staining of Gag. Few single cells infected with NL4.3AsegfpE− virus were able to express both Gag and EGFP, while that was not the case for the NL4.3egfpE− virus-infected cells (Fig. 3). A total of 70.5% ± 3.1% of the EGFP-positive cells infected with NL4.3AsegfpE− virus did not produce Gag, while 86.5% ± 1.5% of the EGFP-positive cells infected with NL4.3egfpE− virus produced Gag (Fig. 3C). Interestingly, 90.1% ± 1.2% of Gag-expressing cells infected with NL4.3AsegfpE− virus did not express antisense egfp (Fig. 3D). When the same approach was used in the presence of zidovudine (20 μM), Gag could not be detected, confirming that the Gag signal was not derived from the infecting viruses (data not shown).
Fig 3.
Analysis of antisense transcription activity in individual infected MDDCs. Freshly isolated monocytes were differentiated into dendritic cells and then infected with NL4.3AsegfpE− or NL4.3egfpE− virions pseudotyped with VSVg (MOI = 2). pNL4.3AsegfpE− was constructed by replacing the Firefly luciferase gene of pNL4.3LucE− with the egfp reporter gene in the inverse orientation. The molecular organization of the antisense (A) or sense (B) EGFP-expressing NL4.3 clones is depicted. At 14 days p.i., cells were fixed for staining with anti-p24 antibody coupled to phycoerythrin (anti-Gag) before observation under an epifluorescence microscope. Images were acquired using the scan slide command of Metamorph, and analyses were performed using ImageJ software. Cells expressing both Gag (red) and EGFP (green) appear yellow on merged images. A total of 772 and 994 cells infected with NL4.3AsegfpE− and NL4.3egfpE− virions, respectively, were analyzed (n = 3). Graphics represent (C) the percentages (means ± SEM) of Gag-negative or Gag-positive cells among EGFP-expressing cells or (D) the percentages of EGFP-negative or EGFP-positive cells among Gag-expressing cells.
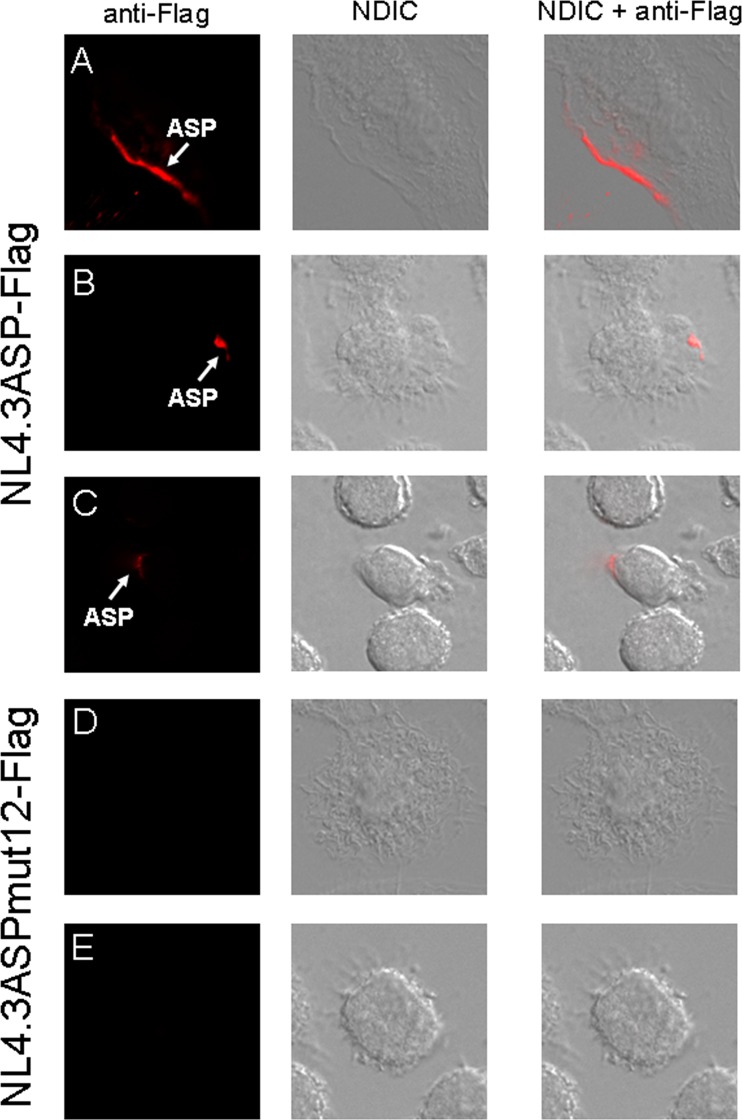
ASP expression was next studied in HIV-1-infected primary cells. Cells were infected with pseudotyped NL4.3ASP-Flag virions, expressing an ASP tagged with the Flag epitope at its C-terminal end (12). While no staining was detected in activated T lymphocytes, we were able to detect ASP expression in MDMs (Fig. 4A) and MDDCs (Fig. 4B and C). No staining was detected in MDMs (Fig. 4D) or MDDCs (Fig. 4E) infected with NL4.3ASPmut12-Flag virions containing a stop codon at amino acid 12 of ASP (12).
Fig 4.
ASP expression in infected monocyte-derived cells. MDM (A and D) or MDDC (B, C, and E) cells were infected with VSVg-pseudotyped virus NL4.3ASP-Flag (A to C) or NL4.3ASPmut12-Flag (D and E) virions. At 14 days p.i., ASP expression was visualized by fluorescence microscopy after immunostaining with a primary anti-Flag antibody and a secondary antibody coupled to Alexa Fluor 568. The morphology of the cells was assessed by Normaski differential interference contrast (NDIC).
We have provided strong evidence for the existence of antisense transcription in infected primary cells and for the production of ASP in monocyte-derived cells. Indeed, HIV-1 antisense transcription is more active in monocyte-derived cells than in activated T cells and unaltered by Tat expression. Moreover, at 14 days p.i., about two-thirds of HIV-1-infected MDDCs showing efficient antisense transcription activity did not express the structural Gag protein. Interestingly, our previous results had already suggested this inverse correlation between Gag and negative-stranded-encoded protein expression in HTLV-1-infected cells (4, 5, 11).
ACKNOWLEDGMENTS
This work was supported by institutional grants from the Centre National de la Recherche Scientifique (CNRS) and the Université Montpellier 1 (UM 1), a grant to J.-M.M. from the CNRS for an international project of scientific cooperation, a grant to J.-M.M. from the Agence Nationale de Recherches sur le Sida et les hépatites virales (ANRS), and a grant to B.B. from The Canadian Foundation for AIDS Research (CanFAR). B.B. holds a Canada Research Chair in Human Retrovirology (Tier 2). S.L. was supported by fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche, and I.C. was supported by ANRS and Sidaction.
Confocal microscopy was performed using Montpellier RIO Imaging (MRI) facilities.
Footnotes
Published ahead of print 3 October 2012
REFERENCES
- 1. Aiken C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arfi V, et al. 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 82:6557–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arpin-André C, Mesnard JM. 2007. The PDZ domain-binding motif of the human T cell leukemia virus type 1 Tax protein induces mislocalization of the tumor suppressor hScrib in T cells. J. Biol. Chem. 282:33132–33141 [DOI] [PubMed] [Google Scholar]
- 4. Barbeau B, Mesnard JM. 2011. Making sense out of antisense transcription in human T-cell lymphotropic viruses (HTLVs). Viruses 3:456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belrose G, et al. 2011. Opposite effects of valproate on Tax and HBZ expressions in T-lymphocytes from HTLV-1 asymptomatic carriers and HAM/TSP patients. Blood 118:2483–2491 [DOI] [PubMed] [Google Scholar]
- 6. Billard E, Cazevieille C, Dornand J, Gross A. 2005. High susceptibility of human dendritic cells to invasion by the intracellular pathogens Brucella suis, B. abortus, and B. melitensis. Infect. Immun. 73:8418–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Billard E, Dornand J, Gross A. 2007. Brucella suis prevents human dendritic cell maturation and antigen presentation through regulation of tumor necrosis factor alpha secretion. Infect. Immun. 75:4980–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Billard E, Dornand J, Gross A. 2007. Interaction of Brucella suis and Brucella abortus rough strains with human dendritic cells. Infect. Immun. 75:5916–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Briquet S, Vaquero C. 2002. Immunolocalization studies of an antisense protein in HIV-1-infected cells and viral particles. Virology 292:177–184 [DOI] [PubMed] [Google Scholar]
- 10. Bukrinsky MI, Etkin AI, Ivanovsky DI. 1990. Plus strand of the HIV provirus DNA is expressed at early stages of infection. AIDS Res. Hum. Retroviruses 6:425–426 [DOI] [PubMed] [Google Scholar]
- 11. Cavanagh M-H, et al. 2006. HTLV-I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology 3:15 doi:10.1186/1742-4690-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clerc I, et al. 2011. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology 8:74 doi:10.1186/1742-4690-8-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944 [DOI] [PubMed] [Google Scholar]
- 14. Gaudray G, et al. 2002. The complementary strand of HTLV-1 RNA genome encodes a bZIP transcription factor that down-regulates the viral transcription. J. Virol. 76:12813–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halin M, et al. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classical bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landry S, et al. 2007. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology 4:71 doi:10.1186/1742-4690-4-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larocca D, Chao LA, Seto MH, Brunck TK. 1989. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem. Biophys. Res. Comm. 163:1006–1013 [DOI] [PubMed] [Google Scholar]
- 18. Larocque E, et al. 2011. Human T-cell lymphotropic virus type 3 (HTLV-3)- and HTLV-4-derived antisense transcripts encode proteins with similar Tax-inhibiting functions but distinct subcellular localization. J. Virol. 85:12673–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li M, Kesic M, Yin H, Yu L, Green PL. 2009. Kinetic analysis of human T-cell leukemia virus type 1 gene expression in cell culture and infected animals. J. Virol. 83:3788–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michael NL, et al. 1994. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J. Virol. 68:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peeters A, Lambert PF, Deacon NJ. 1996. A fourth Sp1 site in the human immunodeficiency virus type 1 long terminal repeat is essential for negative-sense transcription. J. Virol. 70:6665–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Usui T, et al. 2008. Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells. Retrovirology 5:34 doi:10.1186/1742-4690-5-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang H, et al. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]