Abstract
There is a need for vaccines that can protect broadly across all influenza A strains. We have produced a pseudotyped influenza virus based on suppression of the A/PR/8/34 hemagglutinin signal sequence (S-FLU) that can infect cells and express the viral core proteins and neuraminidase but cannot replicate. We show that when given by inhalation to mice, S-FLU is nonpathogenic but generates a vigorous T cell response in the lung associated with markedly reduced viral titers and weight loss after challenge with H1 and H3 influenza viruses. These properties of S-FLU suggest that it may have potential as a broadly protective A virus vaccine, particularly in the setting of a threatened pandemic before matched subunit vaccines become available.
INTRODUCTION
Experimental infection and recovery from influenza A virus infection confer partial protection from distantly related A strain viruses, in the absence of detectable cross-neutralizing antibodies (reviewed in references 14 and 20). Protection is typically associated with 10- to 100-fold reduction in viral replication in the lungs, milder pathology, and improved survival, even after challenge with highly pathogenic H5N1 viruses (5). Schulman and Kilbourne coined the term “partial heterotypic immunity” for this phenomenon (46), which was first observed soon after the isolation of human influenza in the 1930s when ferrets that had recovered from swine influenza were immune to the human virus but failed to make detectable cross-neutralizing antibodies (18, 46, 48). Heterotypic (in recent years also called “heterosubtypic”) immunity has since been observed in mice, cotton rats, ferrets, pigs (reviewed in references 13, 14, and 20), and nonhuman primates (67). There is suggestive evidence that it can be induced in humans (reviewed in references 6, 9, 14, 20, and 50). However, it may be short-lived (15, 31, 37, 50), whereas strain-specific immunity mediated by antibody is long-lived (54).
Much effort has gone into defining the mechanisms involved, without a complete resolution (reviewed in references 6, 14, and 20). In general, in experimental settings without adjuvants, heterotypic immunity is efficiently induced by infections of the lung with live influenza A virus (28, 38). Likewise, recombinant vectors capable of expressing the conserved viral proteins in host cells, particularly in the lung, tend to be efficient inducers of heterotypic protection (42, 49). The specificity of the protective effect correlates with the conserved viral core antigens recognized by T cells (reviewed in references 12, 14, 20, 29, and 68), and protection can be transferred with core protein-specific T cells, particularly class I restricted cytotoxic T lymphocytes (32, 59, 70). These results suggest that the conserved viral antigens, including NP, M, and the viral polymerases presented via the endosomal and cytosolic antigen presentation pathways, play a key part in heterotypic immunity through the induction of a coordinated protective T cell response in the lung (1, 25, 28, 38).
A vaccine that could induce this form of immunity in humans would be particularly useful for mitigating the first wave of a new pandemic, before matched vaccines could be produced. Cold-adapted live attenuated influenza vaccines have been shown to be safe and effective in prevention of seasonal influenza in humans (3, 4). In addition, they can induce cross-reactive T cell responses (24) and provide a better level of protection against drifted seasonal strains than subunit vaccines. However, they may not be appropriate as prepandemic vaccines, as they can potentially reassort a novel hemagglutinin (HA) vRNA into circulating seasonal influenza viruses.
In recent years, it has become possible to make pseudotyped influenza viruses that are prevented from replication through inactivation of the vRNA encoding the hemagglutinin (33, 34). The defective virus can replicate through multiple cycles in appropriate cell lines transfected with cDNAs encoding an HA of choice, which complements the defect by providing HA protein for the lipid envelope of the budding virus particles. These pseudotyped influenza viruses appear indistinguishable from the wild type (WT) in electron micrographs and can infect cells that lack expression of HA, synthesize viral RNA, and express all of the viral proteins except for HA. But they cannot spread from cell to cell and do not contain a viable HA vRNA that could reassort productively with seasonal influenza. To date, they have been produced as safe alternatives to their virulent counterparts for measurement of neutralizing antibodies in low-level containment facilities (34).
However, these properties also make pseudotyped influenza particles attractive as candidate vaccines for the induction of heterotypic immunity in the lung. They should infect as efficiently as wild-type influenza to give a rigorously contained infection with intracellular expression of the complete set of conserved proteins of the virus, which is ideal for recognition by cytotoxic T lymphocytes (63, 64).
To assess this potential, we have produced pseudotyped influenza based on a subline of A/PR/8/34 (H1N1 Cambridge) (69) that is particularly virulent for mice (21). The vRNA encoding HA was inactivated by suppression of the HA signal sequence to produce S-FLU. We show that this preparation has lost all virulence for mice but can induce solid heterotypic protection with prevention of weight loss and suppression of viral replication in the lungs after challenge with high doses of homologous H1N1 and heterologous H3N2 A viruses. Protection was associated with a vigorous influenza-specific T cell response in the lungs in the relative absence of neutralizing antibodies detected in serum. Our results suggest that appropriate pseudotyped influenza viruses should be considered for development as vaccines for the induction of heterotypic immunity to influenza A viruses.
MATERIALS AND METHODS
Design of S-FLU (Fig. 1).
Fig 1.
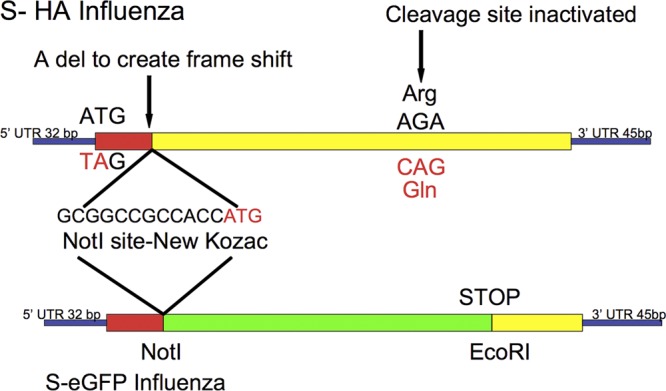
Schematic diagram of the design for the inactivation of the hemagglutinin signal sequence to produce S-HA FLU. For eGFP expression, the S-HA sequence was replaced with eGFP between the NotI site and a unique EcoR I site at position 1268, beyond the HA cleavage site. Viruses were named according to their genotype between square brackets, followed by the identity of the HA in the viral coat obtained from the transfected cells in which replication occurred. For example [S-H1(PR8)/N1(PR8)] H1(Eng195) is S-H1 and N1 vRNAs from A/PR/8/34, coat HA from A/Eng/195/2009 (pandemic H1).
The HA sequence derived from the vRNA of A/PR/8/34 Cambridge (69) (GenBank CAA24272.1) was altered by replacing the original ATG start codon with TAG (bases A33T and T34A) to suppress translation of the signal sequence. A single base at the end of the signal sequence at position 83 was removed to ensure that if the original ATG was reconstituted by back mutation, it would read out of frame. Nucleotide 83 was replaced with the sequence GCGGCCGCCACCATG that contains an idealized Kozac sequence containing a NotI site to initiate protein synthesis beyond the signal sequence and to provide a cloning site for insertion of any desired sequence. Finally, as a further safety mutation, the Arg codon (positions 1063 to 1065, AGA) was changed to CAG to encode Gln and inactivate the HA cleavage site (8). This S-HA sequence was synthesized by GeneArt.
The S-enhanced green fluorescent protein (S-eGFP) form was made by replacing the HA sequence between the introduced NotI site and an EcoRI site at position 1268 with eGFP (Clontech), ending with a stop codon to prevent expression of downstream HA sequence but preserving all of the sequences required for packaging the vRNA (33, 34).
An additional pseudotyped S-eGFP virus was produced based on the pandemic H1N1 strain A/Eng/195/2009 in which the N1 (GenBank accession no. GQ166659.1) vRNA and surface H1 HA (GenBank accession no. ACR15621.1) from A/Eng/195/2009 replace those of A/PR/8/34. Finally, a version of A/PR/8/34 with the wild-type HA from the Cambridge strain (GenBank accession no. CAA24272.1) was also made.
Virus production.
The required cDNA copies of vRNA sequences with SapI sites positioned at both ends were synthesized by GeneArt and cloned into pPolISapIT between the appropriate SapI sites as described previously (16, 53). Recombinant pseudotyped viruses were produced by standard transfection into 106 293T cells in a 2-ml volume with Lipofectamine (Invitrogen) with the addition of the expression plasmid pCDNA 3.1 containing a full-length coding sequence (without the 3′ and 5′ untranslated regions [UTRs]) for the required HA composed of humanized codons to optimize expression and complement for lack of HA expression by the virus. The full-length humanized HA sequences were synthesized by GeneArt.
Pseudotyped viruses from the initial 293T transfections were propagated in MDCK-SIAT1 cells (35) stably transduced with the lentiviral vector pHR-SIN (10) engineered to express the full-length humanized HA from A/PR/8/34 or A/Eng/195/2009 and lacking 3′ and 5′ UT regions. Transduced cells were stained with HA-specific monoclonal antibodies and sorted with a fluorescence-activated cell sorter (FACS) to achieve maximal expression of HA. One milliliter of virus supernatants from the 293T transfections was added to 5 × 105 cells in 1 well of a 6-well plate for 1 h, 2 ml of Dulbecco's modified Eagle's medium (DMEM)-0.1% bovine serum albumin (BSA) containing penicillin and streptomycin and 1 μg/ml of TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (Sigma T-1426) were added, and the virus containing supernatant (s/n) was collected after 48 h of incubation at 37°C. Control plates containing eGFP-expressing viruses were inspected under UV at 48 h for evidence of spreading plaques (Fig. 2). Virus stocks were produced by seeding 0.5 hemagglutinating units (HAU)/106 cells (multiplicity of infection [MOI] of 1:100) on appropriate HA-expressing MDCK-SIAT1 cells in the presence of trypsin as described above.
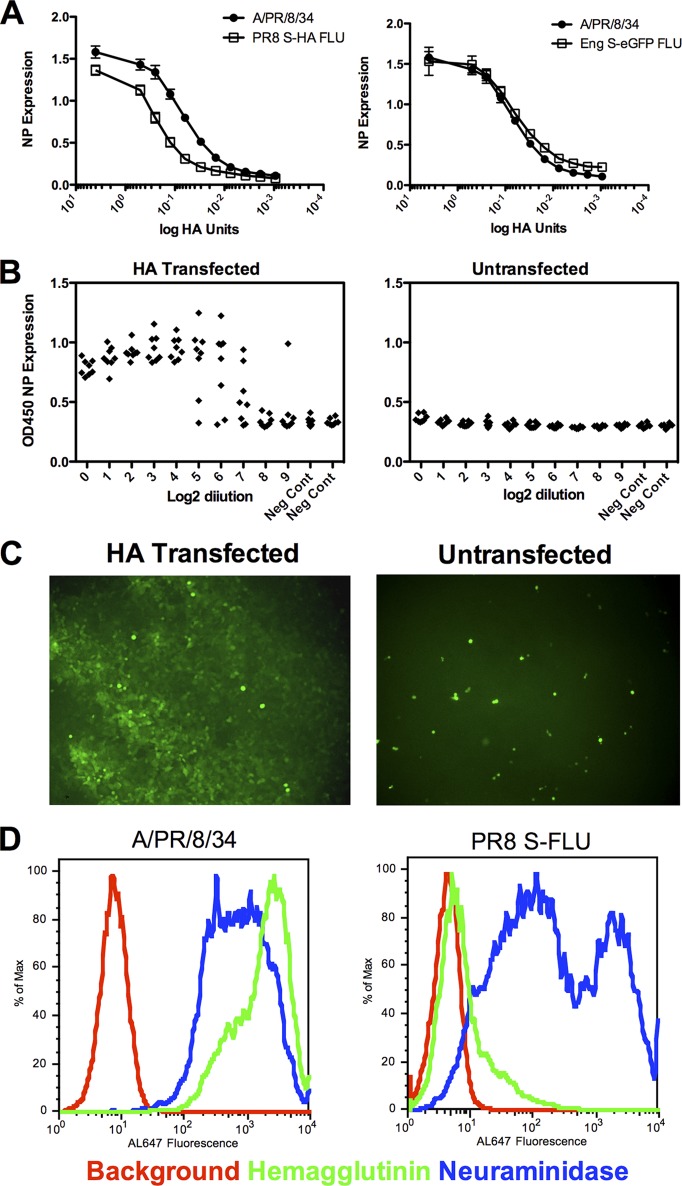
Fig 2.
S-FLU infections in vitro. (A) Infection of MDCK-SIAT1 monolayer with [S-HA(PR8)/N1(PR8)] H1(PR8) or [S-eGFP/N1(Eng195)]H1(Eng195) generates levels of NP expression similar to that of wild-type A/PR/8/34. MDCK-SIAT1 monolayers in replicates of 8 in 96-well plates were infected overnight with 4 and 0.5 HAU/well and then doubling dilutions of virus. (B) Clonal expansion (detected by NP expression) of S-FLU requires the presence of hemagglutinin expressed by the MDCK-SIAT1 cells and added trypsin. Virus [S-eGFP/N1(Eng195)] H1(Eng195) prepared at 1:400 of an HA unit/well and then serially diluted 2-fold in replicates of 8 in 96-well plates was grown in the presence of trypsin for 48 h. (C) The dependence of clonal expansion on HA expression in the monolayer shown by eGFP expression by S-FLU. (D) Expression of neuraminidase but not hemagglutinin at the surface of cells infected with PR8 S-FLU. MDCK-SIAT1 cells were infected overnight with [S-H1(PR8)/N1(PR8)/N1(PR8)] H1(PR8) or A/PR/8/34 and then labeled by indirect immunofluorescence with monoclonal antibodies to hemagglutinin H9-D3 Cb13 or neuraminidase NA2-1C1. Experiments were repeated at least twice.
Nomenclature.
In order to distinguish the various combinations of genotype and surface HA of the pseudotyped viruses, we refer to the HA and neuraminidase (NA) genotypes between square brackets, followed by the origin of the surface HA. Three pseudotyped viruses were produced for this paper: [S-HA(PR8)/N1(PR8)] H1(PR8), [S-eGFP/N1(PR8)] H1(PR8), and [S-eGFP/N1(Eng195)] H1(Eng195), as well as the control A/PR/8/34 with the HA from the Cambridge strain (69).
Virus titration (TCID50 and hemagglutination).
Virus titrations were based on standard procedures (36, 44, 66) with minor modifications. Determination of 50% tissue culture infective dose (TCID50) on the MDCK-SIAT1 cells in 96-well flat-bottomed plates was performed by serial dilution of virus containing s/n in groups of 8 wells in 200-μl volumes onto 3 × 104 MDCK-SIAT1 cells. Virus growth was detected by staining the fixed (10% [vol/vol] formalin) and permeabilized (0.5% Triton X-100) monolayer for NP expression after 48 h at 37°C, with murine (Abcam AA5H) or human (2-8C antibody, produced in-house) monoclonal anti-NP primary antibodies and Dako horseradish peroxidase (HRP)-labeled anti-mouse (P0447) or anti-human (P0241) Ig, as described previously (36), replacing the wash solution with phosphate-buffered saline (PBS)-0.1% BSA. Wells were developed with Roche PM Blue substrate (catalog no. 11-484-281-001) and read at 450 nm. Positive wells were defined as giving a signal of >4 standard deviations (SD) above the mean of 16 uninfected control wells. For titration of virus from infected mouse lung homogenates, detection was by hemagglutination of 50 μl 1% human red cells. TCID50 was calculated as described by Reed and Muench (43).
HA titer was measured as described previously (66) by adding 50 μl 1% (vol/vol) human red cells (adjusted so that a 1:2 dilution gave an optical density at 600 nm [OD600] of 1.8 ± 0.05) to dilutions of virus in 50 μl PBS in V-bottomed 96-well plates. Hemagglutination was read at 1 h by loss of teardrop formation after tilting the plate. Pseudotyped viruses grown in 3-ml volumes in 6-well plates of confluent transduced MDCK-SIAT1 cells routinely gave HA titers of 16 to 32 HAU/50 μl and TCID50 values of 0.5 to 2 ×107/ml. Wild-type A/PR/8/34 (Camb) grew to 10- to 20-fold higher titers.
Indirect immunofluorescence staining of infected cells.
Cell lines (MDCK-SIAT1) were washed once in PBS and infected at an MOI of ∼5:1 for 1 h and then incubated overnight in complete medium (DMEM-10% fetal calf serum [FCS] with Pen/Strep). Harvested cells were stained by indirect immunofluorescence with monoclonal antibodies to hemagglutinin H9-D3 Cb13 or neuraminidase NA2-1C1 kindly provided by J. Yewdell.
Microneutralization assay.
Microneutralization assay was based on Rowe et al. (44) with minor modifications. Viruses were diluted in virus growth medium (VGM; DMEM-penicillin-streptomycin-0.1% BSA [Sigma A0336] without trypsin) and titrated to give plateau expression of NP in 3 × 104 MDCK-SIAT1 cells after overnight infection (1 to 4 HAU = ∼2 × 104 to 8 × 104 TCID50 per well; see Fig. 2) in 96-well flat-bottomed plates. Mouse antisera were heat inactivated at 56°C for 30 min, filtered through a 0.22-μm-pore-size filter, and pooled in equal volumes from groups of 4 to 6 animals. Dilutions of heat-inactivated mouse sera in 50 μl starting at 1:20 were added to 50 μl of virus and incubated for 1 to 2 h at 37°C. A total of 3 × 104 indicator MDCK-SIAT1 cells were then added in 100 μl VGM without trypsin and incubated overnight at 37°C. The monolayer was formalin fixed and permeabilized with Triton X-100 as described above and stained with human anti-NP IgG1 monoclonal antibody 2-8C (produced in-house) and HRP-labeled second-layer anti-human Ig (Dako P0241) as described above. Titers were defined as the final dilution of serum that caused >50% reduction in NP expression (44). Neutralization titers obtained with pseudotyped virus [S-eGFP/H1 (PR8)] H1(PR8) were regularly within one doubling dilution of titers obtained with WT PR8 virus.
T cell ELISPOT assay.
Enzyme-linked immunosorbent spot assays (ELISPOT) were performed according to standard procedures similar to that done in humans (40) but using a mouse ELISPOT kit (Mabtech, Necka Strand, Sweden). Briefly, titrated numbers of spleen or lung single-cell suspensions were incubated with 2 μM (final concentration) peptide solutions in R10 (RPMI [Sigma] and 10% [vol/vol] FCS, 100 U/ml penicillin and 100 μg/ml streptomycin [both Sigma], and 2 mM glutamine). After 18 to 24 h, cells were discarded and plates washed and developed by the addition of anti-IFN-γ-biotin antibody followed by streptavidin alkaline phosphatase and then an alkaline phosphatase substrate kit (Bio-Rad, Hemel Hempstead, United Kingdom). Spots were read using a CTL ELISPOT plate reader (CTL, Shaker Heights, OH). Negative controls were irrelevant peptide (KK-restricted HA epitope) and media. Positive controls used concanavalin A (Sigma, Poole, United Kingdom).
Animals and immunization schedules.
Mice were bred at the BMS, University of Oxford, or purchased from Harlan (Shaw Farm, Bicester, United Kingdom). BALB/c or C57BL/6 females were used at 6 to 8 weeks of age. All procedures were done under the authority of the appropriate personal and project licenses issued by the United Kingdom Home Office. Mice were immunized intranasally (i.n.) twice with S-FLU or eGFP influenza in 50 μl under anesthesia at two weekly intervals followed by at least 14 days before challenge with 32 HAU PR8, X31, or B/Lee viruses. A humane endpoint of weight loss and clinical score was used for mice that would otherwise have succumbed to infection. Animals were assessed for clinical score in terms of mobility, appearance, and breathing intensity. Mice reaching 20% weight loss and/or a morbid clinical score were euthanized.
Production of recombinant HA.
Recombinant HA constructs were based on the design of Stevens et al. (52). cDNA encoding the HA from A/PR/8/34 Cambridge (69) (GenBank accession no. CAA24272.1) or A/Eng/195/2009 (GenBank accession no. ACR15621.1) with human optimized codons was synthesized by GeneArt. HA sequences up to codon 176 of HA2 (H3 numbering) were linked to the T4 fibritin trimerization sequence and hexahistidine tag for purification as described previously (52) and subcloned into the murine retroviral vector pQCXIX (Clontech) containing an eGFP expression cassette beyond an internal ribosome entry site (IRES) as described previously (45). Packaging of retrovirus, transduction, and sorting of 293T cells and HA protein purification was done as previously described with yields of 1 to 5 mg/liter (45). The purified HA0 gave a single dominant band of ∼75 kDa on Coomassie-stained reduced SDS polyacrylamide gels.
ELISA for antibodies to HA.
The enzyme-linked immunosorbent assay (ELISA) was done as described previously (on page 564 in reference 22). Briefly, flat-bottomed 96-well ELISA plates (Falcon 353915) were exposed for 2 h to 20 μg/ml purified HA, washed 2 times in PBS, and blocked with 3% BSA (22). After being washed 2 times in PBS, mouse sera were pooled (4 to 7 animals), heat inactivated at 56°C for 30 min, filtered using a 0.22-μm-pore-size filter, and diluted from 1:20. Fifty-microliter volumes were added to the plates for 1 to 2 h, washed 4 times with PBS, bound with 50 μl Dako anti-mouse HRP antibody (P0447; 1:2,000), washed 4 times, developed with 50 μl Roche PM Blue substrate (catalog no. 11-484-281-001), and read at 450 nm. Titers were expressed as the last dilution to give >50% of the plateau positive signal.
RESULTS
Generation of S-FLU and growth properties in vitro.
We designed two versions of S-FLU based on suppression of the hemagglutinin signal sequence (Fig. 1). The first prevents expression of full-length hemagglutinin (HA) but allows for the expression of a signal-deleted form of the protein (S-HA) in the cytosol that we have previously shown is rapidly degraded and can be presented to class I-restricted cytotoxic T lymphocytes (62) via the cytoplasmic pathway of antigen presentation (63, 64). This version was made to assess whether the addition of HA epitopes might enhance the effectiveness of S-FLU as a vaccine. This was compared to a second version in which most of the HA sequence was replaced with eGFP (S-eGFP), which is expressed in the cytoplasm of infected cells and can be visualized under UV light (33, 34).
We first established that S-FLU viruses could infect target cells and express the viral NP as efficiently as the wild-type A/PR/8/34 virus. Figure 2A (left) shows that [S-HA(PR/8)/N1(PR8)] H1(PR8) infects MDCK-SIAT1 cells at an MOI of >1:1 (1 TCID50:1 cell) in the absence of trypsin and expresses NP to levels equivalent to that of wild-type PR8. The same was found for the S-eGFP versions based on PR/8 or the pandemic H1N1 virus A/Eng/195/2009 (Fig. 2A, right). Figure 2B shows that replication of S-FLU is limited to MDCK-SIAT1 cells that express an appropriate hemagglutinin protein. The left panel shows that clonal expansion of [S-eGFP/N1(Eng)] H1(Eng) titrated to beyond limiting dilutions (MOI of 1: 3 × 104) occurred only in HA-transfected cells in the presence of trypsin. Figure 2C shows this visually for the [S-eGFP/H1(PR8)] H1(PR8) virus, which formed spreading fluorescent plaques in HA-transfected cells but only single-cell infections in untransfected cells. Replication was also dependent on the presence of trypsin (not shown).
Finally, we compared surface expression of HA and NA after overnight infection of MDCK-SIAT1 cells with the S-FLU viruses and wild-type A/PR/8/34. Figure 2D shows that infection with S-FLU achieves surface expression of NA similar to that of the wild type, but as previously shown (62), suppression of the HA signal sequence prevents expression of the folded HA protein at the cell surface. Together, these results established that S-FLU can infect cells and express viral proteins but is not able to spread from cell to cell in vitro without a source of complete hemagglutinin protein in the infected cell membrane and trypsin.
S-FLU is not pathogenic.
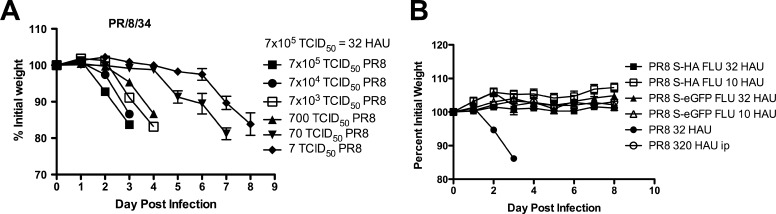
We next compared the pathogenicity of S-FLU with wild-type A/PR/8/34 expressing the Cambridge hemagglutinin (69) in BALB/c mice. The HA from the Cambridge strain of PR/8 confers markedly increased virulence for mice (21) through ∼50× greater viral replication in the lung. Figure 3A shows that as little as 7 TCID50 (∼0.00032 HAU) given in 50 μl into the nose of anesthetized mice is enough to initiate an infection of the lung which results in clinical symptoms and weight loss. As the dose of virus is increased, weight loss occurs earlier after the challenge. The route of administration of A/PR/8/34 is crucial for virulence (17), as is shown in Fig. 3B, where as much as 320 HAU given into the peritoneum does not cause any symptoms.
Fig 3.
Pathogenicity of S-FLU compared to A/PR/8/34 (Cambridge) for BALB/c mice. Anesthetized BALB/c mice were infected intranasally with various doses of wild-type A/PR/8/34 Cambridge strain HA sequence (32 HAU is equivalent to 7 × 105 TCID50) (n = 4 mice per group) (A) or the related pseudotypes S-HA FLU [S-H1(PR8)/N1(PR8)] H1(PR8) or the eGFP-expressing version S-eGFP FLU [S-eGFP/N1(PR8)] H1(PR8) and monitored for clinical symptoms and weight loss (n = 6 mice per group) (B). Mice judged moribund were killed. Data are from a representative experiment repeated three times. Values are means ± standard errors of the means (SEM).
In contrast, the S-FLU viruses can be given in the maximum dose of 32 HAU into the nose of anesthetized mice without causing any clinical symptoms or weight loss (Fig. 3B).
Immune responses to S-FLU: T cell responses.
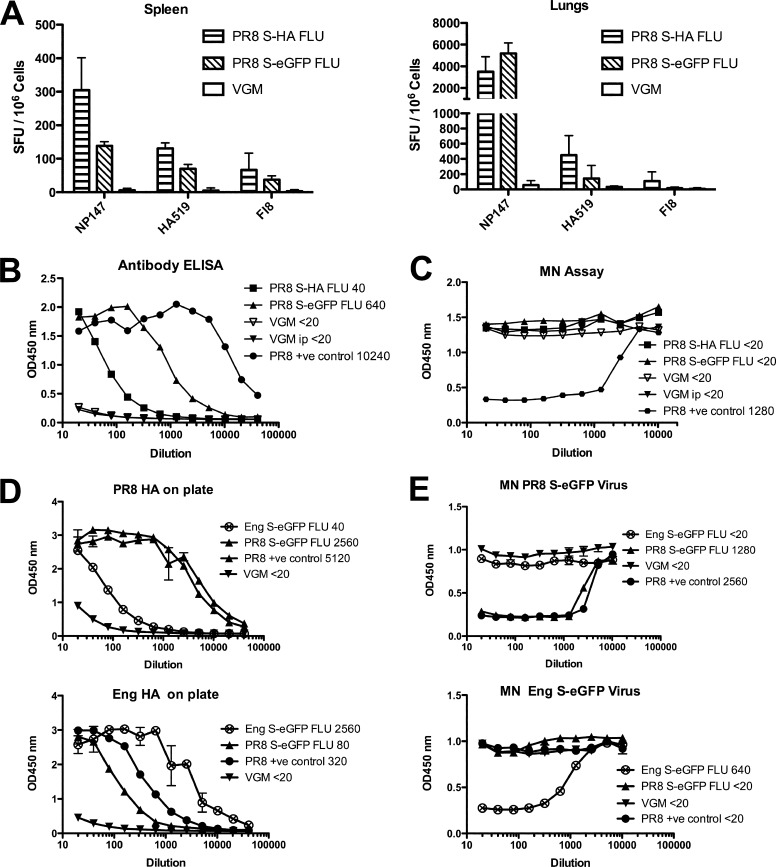
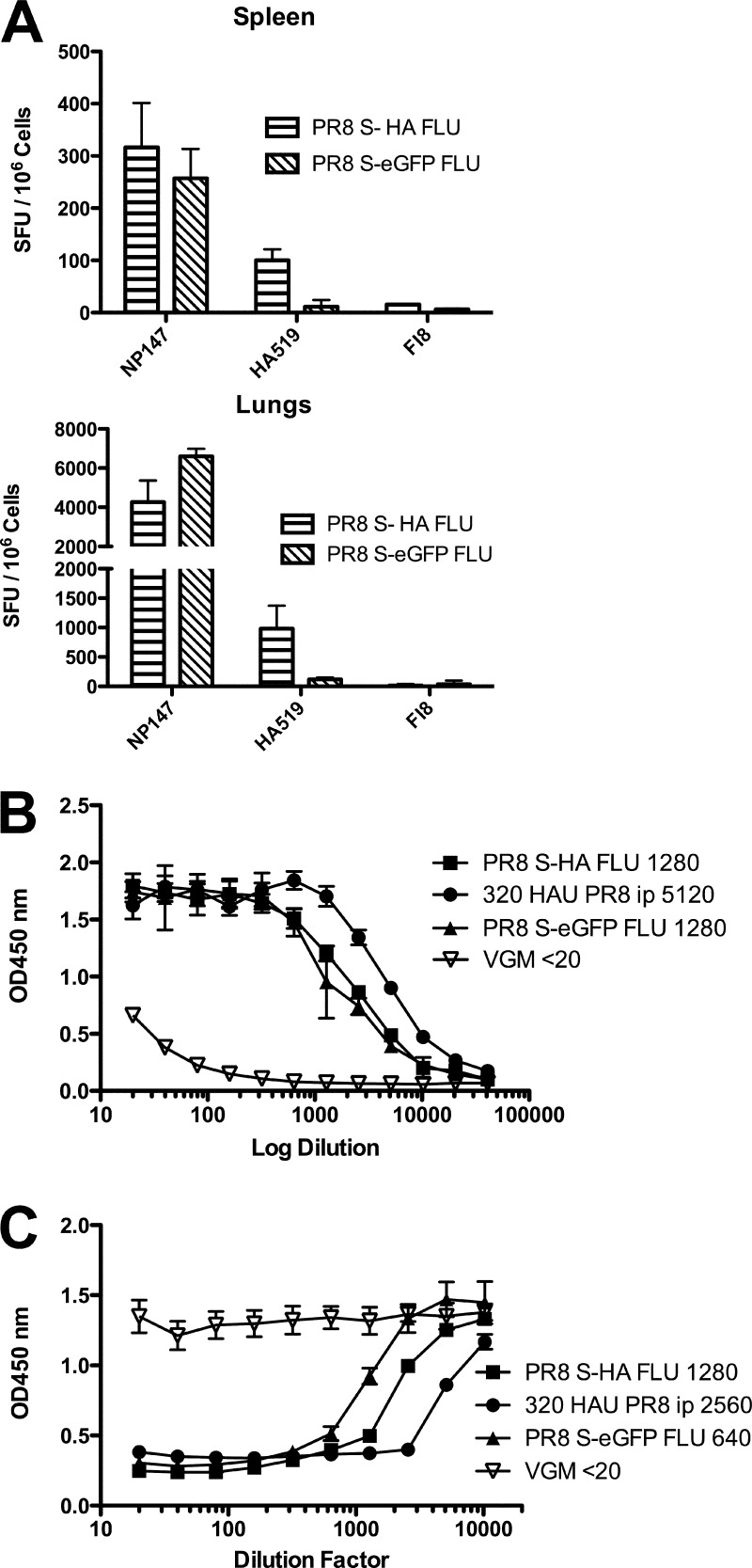
Having established that S-FLU was nonpathogenic in doses up to 32 HAU given i.n. to anesthetized mice, we decided on a vaccination regimen of two doses separated by 2 weeks. The induction of heterotypic immunity by influenza correlates well with the induction of cytotoxic T cell responses in the lower respiratory tract as opposed to the spleen or other sites (1, 28, 38). We therefore measured the level of major histocompatibility complex class I (MHC-I)-restricted T cells in the lung and spleen specific for the conserved H2-Kd-restricted NP peptide 147-158 (60) induced by two doses of 32 HAU of S-FLU given i.n. in 50 μl to anesthetized mice. Figure 4A shows that both versions of S-FLU induced strong NP-specific responses in the lung, with about 10-fold less detectable in the spleen on a per-cell basis. Similar results (see Fig. S1A in the supplemental material) were obtained in C57BL/6 mice responding to the H2-Db-restricted NP peptide 366-374 (64). The version expressing the S-HA molecule did induce some HA-specific T cells to the 518-526 peptide in BALB/c mice, but to levels about 10-fold less than NP.
Fig 4.
Immune responses to S-FLU in mice. BALB/c mice were infected intranasally with 32 HAU PR8 S-HA FLU, [S-H1(PR8)/N1(PR8)] H1(PR8); 32 HAU PR8 S-eGFP FLU, [S-eGFP/N1(PR8)] H1(PR8); or VGM on day 0 and day 14 and culled 21 days later when serum, spleens, and lungs were collected. (A) CTL specific for NP and HA detected in lung and spleen after i.n. inoculation. Pooled spleen or lung single-cell suspensions were frozen before analysis by ELISPOT. (B) Antibody response detected in serum ELISA after 32 HAU i.n. two times. (C) Neutralizing antibody not detected after 32 HAU i.n. two times. (D, E) BALB/c mice were immunized by intraperitoneal (i.p.) injection with 320 HAU Eng S-eGFP FLU, [S-eGFP/N1(Eng195)] H1(Eng195); PR8 S-eGFP FLU, [S-eGFP/N1(PR8)] H1(PR8); WT PR/8/34 Camb virus; or VGM. (D) Antibody response after 320 HAU i.p. two times measured by ELISA. (E) Neutralizing antibody response after 320 HAU i.p. two times measured by MN assay. Data shown are from pooled spleen or lung cells or pooled serum and are representative experiments with n = 6 mice, repeated at least twice with similar results. Assay determinations were triplicates (A), duplicates (B, C), or quadruplicate data (D, E) shown as means ± SD. Titers shown are the last dilution to give >50% of the plateau value for ELISA and <50% NP expression for microneutralization.
Antibody responses to S-FLU.
We next measured antibody levels in response to vaccination both by HA-specific ELISA (Fig. 4B) and by microneutralization (Fig. 4C). In BALB/c mice, an HA-specific response to vaccination i.n. was detected by ELISA, but this was not associated with neutralizing activity at concentrations of serum up to 1:20. In C57BL/6 mice, we also detected HA-specific antibody by ELISA but with minimal neutralization at 1:20 only (see Fig. S1B and C in the supplemental material). In contrast, pooled sera from mice immunized with a single dose of 320 HAU of A/PR/8/34 intraperitoneally (i.p.), followed by challenge with 32 HAU i.n. gave a neutralizing antibody titer of 1:1,280 (Fig. 4C).
To see if the S-FLU preparations could induce neutralizing antibody when given in larger doses, we gave two doses of 320 HAU (in 0.5 ml), separated by 2 to 3 weeks, into the peritoneum and harvested serum after three or more weeks. Figure 4D (top graph) shows that at this dose and route of administration, the S-FLU preparations could induce an HA-specific ELISA titer to 1:2,560, within 2- to 4-fold of that induced by A/PR/8/34 in the same dose. Figure 4E (top graph) shows that this was associated with neutralization titers between 1:640 and 1:1,280.
To extend this result, we compared the pandemic H1 version based on A/Eng/195/2009 with the A/PR/8/34-based version (Fig. 4D, lower). At this higher dose and route, each pseudotyped virus induced a largely strain-specific antibody to HA detected by ELISA, with low levels of cross-reaction (estimated at ∼3% from comparing the titration curves). This was associated with neutralizing antibodies to titers to 1:640 to 1:1,280 that were strain specific with no cross-neutralization (Fig. 4E, bottom). Strain-specific neutralization was also found with antibody raised to the wild-type A/PR/8/34 after two doses of 320 HAU i.p. (Fig. 4E). It is noteworthy that after i.p. administration of these doses, the neutralizing titer induced by the S-FLU-pseudotyped viruses (that cannot synthesize fresh surface HA after infection) are only 2- to 4-fold less than the titer induced by two doses of 320 HAU i.p. of wild-type A/PR/8/34 (Fig. 4E). In several experiments at both doses, we did not detect any reproducible improvement in HA-specific or neutralizing antibody induction by viruses that expressed the S-HA molecule compared to those expressing S-eGFP.
Vaccination with S-FLU induces homotypic and heterotypic immunity. (i) Homotypic immunity.
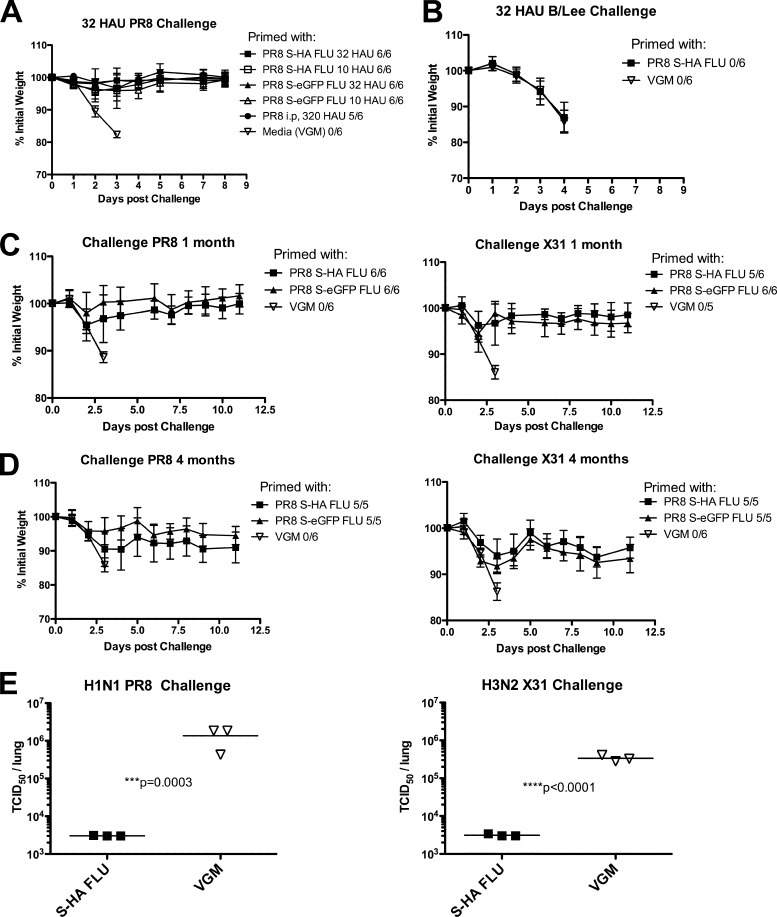
To assess the protective effect of vaccination with S-FLU, mice were given two doses of 32 HAU or one-third of this dose (∼10 HAU) 2 weeks apart i.n. under anesthesia and challenged with the highest dose of 32 HAU wild-type A/PR/8/34 (H1N1) or X31 (H3N2) between 2 weeks to 4 months later. Figure 5A shows that both doses of S-FLU protected mice from weight loss (and clinical scores) after challenge with 32 HAU of A/PR/8/34. No difference in protective effect was seen with the virus that expressed the S-HA compared to S-eGFP. The mice vaccinated with S-FLU showed a level of protection similar to that of mice given a single dose of 320 HAU of the wild-type A/PR/8/34 i.p.
Fig 5.
Protection from challenge after vaccination with S-FLU. BALB/c mice were immunized with PR8 S-HA FLU, [S-H1(PR8)/N1 (PR8)] H1(PR8); PR8 S-eGFP FLU, [S-eGFP/N1(PR8)] H1(PR8); or VGM at day 0 and day 14. Mice were then challenged with 32 HAU wild-type virus, and weight loss and clinical score were monitored for at least 10 days. (A) Mice were primed with 32 or 10 HAU S-HA FLU or S-eGFP FLU virus i.n. Mice were challenged with 32 HAU PR8 on day 28 (A), 32 HAU B/Lee on day 46 (B), 32 HAU PR8 or X31 viruses on day 40 (C), or 32 HAU PR8 or X31 after 4 months (D). In panel D, 4 mice died prechallenge, one from anesthetic overdose, one had a large tumor, and 2 were unexplained. These were the only unexplained deaths in a total of 196 immunizations in 10 experiments. (A to D) Number that were not judged moribund over number in group is shown in the legends. (E) Day 39 challenged mice were culled at day 3 postchallenge, and lungs were collected and snap-frozen in liquid N2. TCID50 titers were then determined using MDCK-SIAT1 cells, and statistical differences are determined by Student's t test comparing log-transformed data. Experiments were repeated at least twice with similar results, and representative data are shown. Percentage initial weights shown are means ± SD.
(ii) Heterotypic immunity to S-FLU.
These results were extended to protection by S-FLU from the H3N2 strain X31 in Fig. 5C. The protective effect of vaccination has been repeated up to 4 months postvaccination with less than 10% weight loss after challenge (Fig. 5D). The specificity of the protective effect for A but not B viruses is shown in Fig. 5B. As classically described (46), weight loss induced by infection with influenza B/Lee/40 is not influenced by heterotypic immunity induced to influenza A viruses.
The replication of virus in the lungs of challenged mice on day 3 of infection is shown in Fig. 5E. The titer of the heterologous virus X31 in the lungs of S-FLU-vaccinated mice was reduced by at least 2 orders of magnitude, whereas homologous virus was reduced by ∼3 orders of magnitude. This is typical of heterotypic immunity, which does not prevent infection by a second A strain virus but reduces viral replication and the associated pathology (46).
Postchallenge immune responses of vaccinated mice.
The levels of NP- and HA-specific T cells in the lungs of vaccinated mice several weeks after homotypic challenge were similar in pattern and extent to those 2 to 4 weeks postvaccination (Fig. 6A). Again, the animals receiving the pseudotype that encoded the S-HA molecule induced a higher level of HA-specific T cells. In contrast, the HA-specific antibody response posthomotypic challenge showed a rise in titer suggestive of a secondary response and was associated with the appearance of neutralizing antibody to titers of 1:640 to 1:1,280. Priming with the lower dose of 10 HAU S-FLU (which was fully protective) also resulted in postchallenge neutralizing titers of 1:160 to 1:320. There was no reproducible difference in the postchallenge titers of animals vaccinated with the S-HA version compared to S-eGFP version of S-FLU.
Fig 6.
Immune responses postimmunization and challenge BALB/c mice were immunized with 32 HAU S-HA FLU, [S-H1(PR8)/N1(PR8)] H1(PR8); S-eGFP FLU, [S-eGFP/N1(PR8)] H1(PR8); or VGM at day 0 and day 14. Mice were challenged at day 28, monitored for recovery, and then culled on day 45 when spleen, lungs, and serum were collected. (A) Pooled spleen and lung single-cell suspensions from 6 animals were tested in ELISPOT assays. (B, C) Pooled sera from 6 animals were tested in an ELISA (B) and microneutralization assay (C). Data shown are triplicates (A) and quadruplicates (B) ±SD, representative of at least two experiments which had similar results. Titers shown are the last dilution to give >50% of plateau value for ELISA and <50% NP expression for microneutralization.
Taken together, our results show that S-FLU, a nonreplicating, nonpathogenic pseudotyped influenza virus, is capable of inducing a strong local T cell response in the lung associated with classical cross-strain heterotypic immunity. In addition, S-FLU can induce a strain-specific neutralizing antibody response if given in sufficient dose.
DISCUSSION
The threat of pandemic influenza persists and has been emphasized by recent evidence that relatively few mutations are required in highly pathogenic H5N1 viruses to render them competent to transmit between ferrets (23, 26). From a practical point of view, the critical period of susceptibility for humans is between the beginning of the first wave of infection with a new pandemic strain and the distribution of matched vaccines, which at the moment is likely to be several months. A cross-protective vaccine to mitigate this first wave of a pandemic would be particularly useful.
Heterotypic immunity across serologically distinct influenza A strains, that at least prevents severe illness and reduces viral replication, has been observed repeatedly in convalescent experimental animals since the earliest period of influenza research (6, 14, 18, 20, 46, 48, 67). Evidence that a degree of cross-protection can be generated in humans that have been recently exposed to influenza A is indirect but persuasive (reviewed in references 6, 9, 14, 20, 50, and 68). In general, this form of postexposure immunity correlates better with cross-reactive T cell responses to the core proteins (6, 14, 20, 68) than with antibody to the glycoproteins and can be transferred between animals with T cells (32, 59, 70). This is not to say that the final effector mechanisms in the lung are solely mediated by T cells. It seems likely that T cells play a key part in coordinating a complex web of responses in the lung that ultimately reduce viral replication and control the innate inflammatory response (11, 27, 56).
On this basis, we reasoned that a pseudotyped influenza virus that is related as closely as possible to the original would reproduce the conditions needed to induce heterotypic immunity. Because pseudotyped influenza can infect but not replicate, immune responses can be induced with a rigorously controlled infection of the lung. Pseudotypes can be made rapidly by standardized techniques (33, 34), can incorporate any HA in the envelope, and are self-replicating in vitro for bulk production. S-FLU is based on inactivation of the vRNA encoding HA so does not contain a viable HA vRNA that could reassort with seasonal strains. Finally, given the complete control over replication, administration by small-droplet aerosol to the lung might be a viable and efficient way to immunize (7, 49).
We have shown that doses of the A/PR8/34 H1N1 S-FLU of 32 and 10 HAU given twice via the nose to anesthetized mice provided solid protection from weight loss and viral replication after challenge with the homologous highly virulent parental H1N1 virus (21, 69) and the heterologous H3N2 strain A-X31. The high dose of the challenge and the increased virulence contributed by the Cambridge hemagglutinin (21) were deliberately chosen to provide a convincing test for vaccination. The S-FLU vaccine at doses greater than 104-fold that of the wild-type virus that induced severe weight loss did not cause any symptoms. At the time of challenge, the S-FLU-vaccinated mice had a brisk cross-reactive T cell response detectable in the lung but at this dose did not make detectable neutralizing antibody in serum. This pattern of immunity was associated with marked suppression of replication of both challenge viruses in the lung and prevented major weight loss 4 months postvaccination. The success of this experiment in mice suggests that further trials of S-FLU in other species challenged with highly pathogenic influenza viruses are warranted.
The design of S-FLU allowed for the expression of either a signal-deleted form of HA or eGFP which replaced most of the HA coding sequence. The S-HA was included to see if presentation of HA T cell epitopes added anything to the protective effect of the vaccine, because in the earliest T cell transfer experiments, polyclonal T cells from animals immunized by influenza infection appeared to give somewhat better protection when transferred to animals challenged with HA-matched strains of influenza (70). Although the S-HA version did induce some HA-specific T cells detected by ELISPOT, we did not detect any obvious benefit from this, either in protection from weight loss, levels of T cells induced in the lung, or priming for an HA-specific antibody response. In the future, it may be wise to use versions of S-FLU that lack most of the HA sequence in their genome to minimize any chance of reversion to the wild type.
The heterotypic protective response induced to S-FLU that we have demonstrated here is comparable to the response in mice to various influenza preparations attenuated by cold adaptation (41, 55, 58), NS deletions (57), HA cleavage site mutations (51), packaging site changes (2), and gamma irradiation (1, 19). Each of these induces heterotypic immunity with doses in the order of 106 or less influenza infectious units. In contrast, the response to intranasal administration of recombinant nonreplicating adenoviruses, capable of expressing NP and M2 proteins of influenza (42), required a dose of ∼1 × 1010 particles of each recombinant adenovirus to minimize weight loss in the vaccinated animals. This large difference in the dose required to achieve protection suggests that influenza particles may have properties that favor the induction of the heterotypic immune response. These might include expression of all of the core proteins in the cytosol to induce efficient antigen presentation and thus a broader repertoire of T cells. In the adenovirus experiment, immunization with either NP or M2 separately resulted in significant weight loss after challenge compared to the combination (42). In addition, the influenza-based vaccines will introduce influenza RNA into the endosome and cytosol to initiate an innate response, which could promote a favorable adjuvant effect.
The dose of S-FLU given i.n. that promoted heterotypic immunity was also sufficient to induce a specific antibody response to hemagglutinin detectable in ELISA but not by neutralization (Fig. 4B). Assuming ∼1,000 HA spikes per virion, we calculate that this represents a dose of HA in the total inoculum of less than 1 ng. The neutralizing response to a 10-fold-higher dose i.p. shows that the HA in the pseudotyped virion is in a configuration favorable to the induction of neutralizing antibody, albeit highly strain specific. In addition, the strong neutralizing response following challenge of immunized mice suggests that the small i.n. dose may have primed for a secondary strain-specific neutralizing response to hemagglutinin. These results were not expected, as the amounts of HA available in the S-FLU virion are limited and cannot be amplified after infection. They suggest that this type of particle may be highly immunogenic for B cells as well as T cells.
Broadly neutralizing monoclonal antibodies to influenza hemagglutinin can be isolated from mice (39) and humans (47), and cross-exposures to serologically distinct strains of influenza can induce these antibodies (30, 61, 65). Classical heterotypic immunity is unlikely to be mutually exclusive to the induction of a cross-neutralizing antibody response. In principle, a vaccine that induces both should be possible. Given that the HA in the envelope of pseudotyped influenza can be changed easily and is highly immunogenic, a regimen of exposures to pseudotypes with an appropriate range of variant hemagglutinins might be capable of priming for broadly neutralizing antibodies while also inducing T cell-dependent heterotypic immunity in the lung.
In summary, we have shown that a pseudotyped, nonreplicating influenza virus based on suppression of the hemagglutinin signal sequence, S-FLU, can induce heterotypic immunity and protect mice from challenge with a highly pathogenic strain of A/PR/8/34 and the heterologous H3N2 virus A-X31. These results suggest that pseudotyped influenza viruses should be considered for development as broadly protective vaccines for influenza.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jon Yewdell for provision of the antineuraminidase and antihemagglutinin antibodies, John McCauley for provision of X31 and B/Lee viruses, and Lisa Schimanski and Pramila Rijal for experimental assistance. Thanks are also due to the anonymous blood donors and other members of the MIG who discussed ideas and data.
These studies were funded by the Townsend-Jeantet Charitable Trust (registered charity no. 1011770) and Oxford University.
A.R.M.T. and T.J.P. conceived the study, planned the experiments, undertook the experiments, analyzed the data, and wrote the paper. E.F. and J.S. supervised production of recombinant viruses. J.D.S. performed experiments and analyzed data.
A.R.M.T., T.J.P., J.S., and E.F. are named on patents concerning the use of S-FLU as a vaccine.
Footnotes
Published ahead of print 26 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alsharifi M, et al. 2009. Intranasal flu vaccine protective against seasonal and H5N1 avian influenza infections. PLoS One 4:e5336 doi:10.1371/journal.pone.0005336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anhlan D, Hrincius ER, Scholtissek C, Ludwig S. 2012. Introduction of silent mutations into the NP gene of influenza A viruses as a possible strategy for the creation of a live attenuated vaccine. Vaccine 30:4480–4489 [DOI] [PubMed] [Google Scholar]
- 3. Belshe RB, Ambrose CS, Yi T. 2008. Safety and efficacy of live attenuated influenza vaccine in children 2–7 years of age. Vaccine 26(Suppl 4):D10–D16 [DOI] [PubMed] [Google Scholar]
- 4. Belshe RB, Coelingh K, Ambrose CS, Woo JC, Wu X. 2010. Efficacy of live attenuated influenza vaccine in children against influenza B viruses by lineage and antigenic similarity. Vaccine 28:2149–2156 [DOI] [PubMed] [Google Scholar]
- 5. Bodewes R, et al. 2011. Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J. Virol. 85:2695–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown LE, Kelso A. 2009. Prospects for an influenza vaccine that induces cross-protective cytotoxic T lymphocytes. Immunol. Cell Biol. 87:300–308 [DOI] [PubMed] [Google Scholar]
- 7. Castro JF, et al. 2005. Evaluation of immunogenicity and side effects of triple viral vaccine (MMR) in adults, given by two routes: subcutaneous and respiratory (aerosol). Vaccine 23:1079–1084 [DOI] [PubMed] [Google Scholar]
- 8. Chen J, et al. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417 [DOI] [PubMed] [Google Scholar]
- 9. Cowling BJ, et al. 2010. Protective efficacy of seasonal influenza vaccination against seasonal and pandemic influenza virus infection during 2009 in Hong Kong. Clin. Infect. Dis. 51:1370–1379 [DOI] [PubMed] [Google Scholar]
- 10. Demaison C, et al. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803–813 [DOI] [PubMed] [Google Scholar]
- 11. De Santo C, et al. 2008. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J. Clin. Invest. 118:4036–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doherty PC, Kelso A. 2008. Toward a broadly protective influenza vaccine. J. Clin. Invest. 118:3273–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doherty PC, Turner SJ, Webby RG, Thomas PG. 2006. Influenza and the challenge for immunology. Nat. Immunol. 7:449–455 [DOI] [PubMed] [Google Scholar]
- 14. Epstein SL, Price GE. 2010. Cross-protective immunity to influenza A viruses. Expert Rev. Vaccines 9:1325–1341 [DOI] [PubMed] [Google Scholar]
- 15. Ferguson NM, Galvani AP, Bush RM. 2003. Ecological and immunological determinants of influenza evolution. Nature 422:428–433 [DOI] [PubMed] [Google Scholar]
- 16. Fodor E, et al. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Francis T, Magill TP. 1935. Immunological studies with the virus of influenza. J. Exp. Med. 62:505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Francis T, Shope RE. 1936. Neutralization tests with sera of convalescent or immunized animals and the viruses of swine and human influenza. J. Exp. Med. 63:645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Furuya Y, et al. 2010. Cytotoxic T cells are the predominant players providing cross-protective immunity induced by {gamma}-irradiated influenza A viruses. J. Virol. 84:4212–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grebe KM, Yewdell JW, Bennink JR. 2008. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 10:1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grimm D, et al. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Herfst S, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoft DF, et al. 2011. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 204:845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hogan RJ, et al. 2001. Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J. Immunol. 166:1813–1822 [DOI] [PubMed] [Google Scholar]
- 26. Imai M, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kok WL, et al. 2012. Pivotal advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J. Leuk. Biol. 91:357–368 [DOI] [PubMed] [Google Scholar]
- 28. Lau YF, Wright AR, Subbarao K. 2012. The contribution of systemic and pulmonary immune effectors to vaccine-induced protection from H5N1 influenza virus infection. J. Virol. 86:5089–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee LY, et al. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li GM, et al. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. U. S. A. 109:9047–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang S, Mozdzanowska K, Palladino G, Gerhard W. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653–1661 [PubMed] [Google Scholar]
- 32. Lin YL, Askonas BA. 1981. Biological properties of an influenza A virus-specific killer T cell clone. Inhibition of virus replication in vivo and induction of delayed-type hypersensitivity reactions. J. Exp. Med. 154:225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marsh GA, Hatami R, Palese P. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81:9727–9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Sobrido L, et al. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. 2003. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77:8418–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McMichael AJ, Gotch FM, Dongworth DW, Clark A, Potter CW. 1983. Declining T-cell immunity to influenza, 1977-82. Lancet ii:762–764 [DOI] [PubMed] [Google Scholar]
- 38. Nguyen HH, et al. 1999. Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8(+) cytotoxic T lymphocyte responses induced in mucosa-associated tissues. Virology 254:50–60 [DOI] [PubMed] [Google Scholar]
- 39. Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Powell TJ, et al. 2012. Identification of H5N1-specific T-cell responses in a high-risk cohort in Vietnam indicates the existence of potential asymptomatic infections. J. Infect. Dis. 205:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Powell TJ, et al. 2007. Priming with cold-adapted influenza A does not prevent infection but elicits long-lived protection against supralethal challenge with heterosubtypic virus. J. Immunol. 178:1030–1038 [DOI] [PubMed] [Google Scholar]
- 42. Price GE, et al. 2010. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2, and H1N1 viruses. PLoS One 5:e13162 doi:10.1371/journal.pone.0013162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. (Lond.) (27):493–497 [Google Scholar]
- 44. Rowe T, et al. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schimanski LM, et al. 2009. In vitro binding of HFE to the cation-independent mannose-6 phosphate receptor. Blood Cells Mol. Dis. 43:180–193 [DOI] [PubMed] [Google Scholar]
- 46. Schulman JL, Kilbourne ED. 1965. Induction of partial specific heterotypic immunity in mice by a single infection with influenza A virus. J. Bacteriol. 89:170–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simmons CP, et al. 2007. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 4:e178 doi:10.1371/journal.pmed.0040178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith W, Andrewes CH, Liadlaw PP. 1933. A virus obtained from influenza patients. Lancet ii:66–68 [Google Scholar]
- 49. Song K, et al. 2010. Genetic immunization in the lung induces potent local and systemic immune responses. Proc. Natl. Acad. Sci. U. S. A. 107:22213–22218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. 1985. Cross-subtype protection in humans during sequential, overlapping, and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. J. Infect. Dis. 151:81–88 [DOI] [PubMed] [Google Scholar]
- 51. Stech J, Garn H, Wegmann M, Wagner R, Klenk HD. 2005. A new approach to an influenza live vaccine: modification of the cleavage site of hemagglutinin. Nat. Med. 11:683–689 [DOI] [PubMed] [Google Scholar]
- 52. Stevens J, et al. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870 [DOI] [PubMed] [Google Scholar]
- 53. Subbarao K, et al. 2003. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology 305:192–200 [DOI] [PubMed] [Google Scholar]
- 54. Subbarao K, Murphy BR, Fauci AS. 2006. Development of effective vaccines against pandemic influenza. Immunity 24:5–9 [DOI] [PubMed] [Google Scholar]
- 55. Suguitan AL, Jr, et al. 2006. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 3:e360 doi:10.1371/journal.pmed.0030360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun J, Madan R, Karp CL, Braciale TJ. 2009. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15:277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Talon J, et al. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tannock GA, Paul JA. 1987. Homotypic and heterotypic immunity of influenza A viruses induced by recombinants of the cold-adapted master strain A/Ann Arbor/6/60-ca. Arch. Virol. 92:121–133 [DOI] [PubMed] [Google Scholar]
- 59. Taylor PM, Askonas BA. 1986. Influenza nucleoprotein-specific cytotoxic T-cell clones are protective in vivo. Immunology 58:417–420 [PMC free article] [PubMed] [Google Scholar]
- 60. Taylor PM, Davey J, Howland K, Rothbard JB, Askonas BA. 1987. Class I MHC molecules rather than other mouse genes dictate influenza epitope recognition by cytotoxic T cells. Immunogenetics 26:267–272 [DOI] [PubMed] [Google Scholar]
- 61. Thomson CA, et al. 2012. Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front. Immunol. 3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Townsend AR, Bastin J, Gould K, Brownlee GG. 1986. Cytotoxic T lymphocytes recognize influenza haemagglutinin that lacks a signal sequence. Nature 324:575–577 [DOI] [PubMed] [Google Scholar]
- 63. Townsend AR, Gotch FM, Davey J. 1985. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell 42:457–467 [DOI] [PubMed] [Google Scholar]
- 64. Townsend AR, et al. 1986. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell 44:959–968 [DOI] [PubMed] [Google Scholar]
- 65. Wang TT, et al. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 6:e1000796 doi:10.1371/journal.ppat.1000796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Webster RG, Cox N, Stohr K. 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, Geneva, Switzerland [Google Scholar]
- 67. Weinfurter JT, et al. 2011. Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates. PLoS Pathol. 7:e1002381 doi:10.1371/journal.ppat.1002381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilkinson TM, et al. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 18:274–280 [DOI] [PubMed] [Google Scholar]
- 69. Winter G, Fields S, Brownlee GG. 1981. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature 292:72–75 [DOI] [PubMed] [Google Scholar]
- 70. Yap KL, Ada GL. 1978. The recovery of mice from influenza A virus infection: adoptive transfer of immunity with influenza virus-specific cytotoxic T lymphocytes recognizing a common virion antigen. Scand. J. Immunol. 8:413–420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.