Abstract
Hantaviruses are important contributors to disease burden in the New World, yet many aspects of their distribution and dynamics remain uncharacterized. To examine the patterns and processes that influence the diversity and geographic distribution of hantaviruses in South America, we performed genetic and phylogeographic analyses of all available South American hantavirus sequences. We sequenced multiple novel and previously described viruses (Anajatuba, Laguna Negra-like, two genotypes of Castelo dos Sonhos, and two genotypes of Rio Mamore) from Brazilian Oligoryzomys rodents and hantavirus pulmonary syndrome cases and identified a previously uncharacterized species of Oligoryzomys associated with a new genotype of Rio Mamore virus. Our analysis indicates that the majority of South American hantaviruses fall into three phylogenetic clades, corresponding to Andes and Andes-like viruses, Laguna Negra and Laguna Negra-like viruses, and Rio Mamore and Rio Mamore-like viruses. In addition, the dynamics and distribution of these viruses appear to be shaped by both the geographic proximity and phylogenetic relatedness of their rodent hosts. The current system of nomenclature used in the hantavirus community is a significant impediment to understanding the ecology and evolutionary history of hantaviruses; here, we suggest strict adherence to a modified taxonomic system, with species and strain designations resembling the numerical system of the enterovirus genus.
INTRODUCTION
Hantaviruses are enveloped, single-stranded negative-sense RNA viruses in the Bunyaviridae family. Their tripartite genome consists of a small (S) segment encoding both the nucleoprotein (N) and a small nonstructural (NSs) protein in an overlapping (+1) open reading frame (ORF), a medium (M) segment that encodes the envelope glycoproteins (Gn and Gc, formerly G1 and G2), and a large (L) segment that encodes the RNA-dependent RNA polymerase (RdRp) (25, 66, 77). Unlike other members of the Bunyaviridae, hantaviruses are not vector-borne but are instead transmitted between their vertebrate hosts through aggressive interactions or the inhalation of excreta (55). While Muridae/Cricetidae rodents have traditionally been considered the reservoir hosts of hantaviruses, novel viruses continue to be described in a wide range of species, including shrews and bats (1, 28, 33, 67, 68, 71).
In the New World, hantaviruses carried by Sigmodontinae rodents have been associated with hantavirus pulmonary syndrome (HPS) since the Four Corners outbreak, which occurred in the United States in 1993 and led to the identification of Sin Nombre virus (48). Since that time, more than 2,000 HPS cases have been reported and more than 30 hantaviruses have been described throughout the New World, the majority in South America (28). HPS cases where an associated virus could be identified have been reported in Argentina (Andes [ANDV], Bermejo [BMJV], Laguna Negra [LANV], Lechiguanas [LECV], Oran [ORNV]), Bolivia (ANDV, BMJV, LANV, Tunari [TUNV]), Brazil (Anajatuba [ANJV], Araraquara [ARQV], Araucaria [ARAUV], Castelo dos Sonhos [CASV], Juquitiba [JUQV], LANV-like), Chile (ANDV), French Guiana (Maripa [MARV]), Paraguay (LANV), and Uruguay (Andes Central Plata [ACPV]) (10, 12, 19, 26, 27, 36, 44, 51, 58, 59, 76). Although many hantaviruses have been described, few meet the criteria for species demarcation suggested by the International Committee on Taxonomy of Viruses (ICTV) (32). According to ICTV, hantavirus species should be delineated based on four criteria: (i) at least 7% amino acid (aa) divergence in the complete N and glycoprotein precursor (GPC) proteins, (ii) a unique reservoir host, (iii) at least a 4-fold difference in a two-way cross neutralization test, and (iv) the absence of naturally occurring reassortants. Despite these clear criteria, publications in this field regularly describe new viruses/strains/genotypes/species/lineages according to a variety of measures, which are neither standardized across the field nor regularly in accordance with those suggested by ICTV (7, 37, 42, 64).
Although hantaviruses have been recognized as important agents of disease in the New World since the late 20th century, they have now been identified in almost all Central and South American countries (28). Nonetheless, the incidence of human hantavirus infection has likely been underestimated, due in part to shared clinical features with a variety of other disease syndromes, including leptospirosis, acute respiratory distress syndrome, and pneumonia (31, 49). Human serosurveys in regions of Brazil with few or no reported HPS cases have repeatedly found evidence of past hantavirus infection in a substantial portion of the population, indicating the existence of undiagnosed or subclinical infections (5, 8, 18, 23, 49). In addition, despite the high case fatality rates often associated with HPS in South America (≥50%), the identity of the etiologic agent is frequently not determined (10, 28). Therefore, although many South American hantaviruses have thus far been identified only in rodents (e.g., Itapua [ITPV], Rio Mamore [RIOMV]), the frequency of undiagnosed infections and the association of closely related viruses with HPS cases (e.g., ARAUV and JUQV are related to ITPV; ANJV and MARV are related to RIOMV) indicates that a potential link to human disease cannot be excluded (3, 7, 8, 28, 76).
The diversity and distribution of hantaviruses in rodents in South America and their contributions to human disease are not yet clearly understood. To further the development of effective methods of diagnosis and treatment, a more complete understanding of these aspects of hantavirus biology is critical. In this study, we sequenced the complete N genes and full genomes of a variety of hantaviruses identified in both HPS cases and Oligoryzomys rodents in Brazil and compared them to all previously described hantaviruses from South America. Using genetic and phylogeographic approaches, we characterized the diversity of South American hantaviruses to add insight into the patterns and processes behind their geographic and host distributions.
MATERIALS AND METHODS
Viral sequencing and genetic analysis.
Samples from 19 HPS cases collected at hospitals in the states of Maranhão, Pará, Mato Grosso, Rondônia, and Amazonas, Brazil, were analyzed along with 24 liver, lung, or heart samples from Oligoryzomys sp. rodents in Mato Grosso, Rondônia, and Amazonas (Fig. 1; see also Table S1 in the supplemental material). Of these, four HPS case samples and four rodent samples have been previously sequenced, but only small (∼400-nucleotide [nt]) fragments of the N gene were used for characterization (45, 74, 75). All samples were preserved in TRIzol reagent (Invitrogen) and extracted according to the manufacturer's specifications. cDNA was synthesized using Superscript III (Invitrogen). PCR products from the S, M, and L genome segments were generated by overlapping consensus PCR and sequenced by Sanger sequencing. In all cases, an attempt was made to amplify the complete genome of representatives from each virus type detected in these samples. In addition, full-length cytochrome b gene sequences of each rodent were amplified and sequenced using primers L14115 and H15288 (43). Sequences generated in this study were deposited in GenBank (accession numbers JX443647 to JX443704).
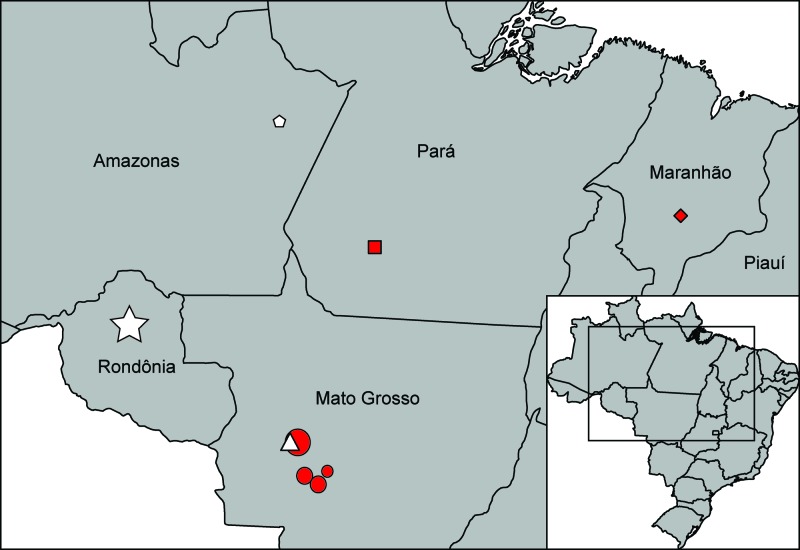
Fig 1.
Map of Brazil indicating the sampling locations of the viruses sequenced in this study. HPS cases are indicated in red, and rodent samples are indicated in white. Viruses identified were ANJV (diamond), CASV (square), CASV-2 (triangle), LANV-2 (circle), RIOMV-3 (pentagon), and RIOMV-4 (star), where the size of the symbol is proportional to the number of samples from a given location.
The hantavirus sequences generated in this study were analyzed for putative glycosylation sites and signal peptidase cleavage sites using the programs NetNGlyc and SignalP-NN, respectively (available at the Center for Biological Sequence Analysis, http://www.cbs.dtu.dk/services/).
Phylogenetic analyses.
Multiple sequence alignments were manually created for each hantavirus segment and gene, using the program Se-Al (http://tree.bio.ed.ac.uk/software/seal). In addition to those viruses sequenced here, each data set included at least two representatives from all Sigmodontinae-borne viruses available on GenBank, whenever possible. Untranslated regions were excluded from this analysis. Maximum likelihood (ML) phylogenies for the N (n = 89, 1,287 nt), GPC (n = 50, 3,444 nt), and RdRp (n = 15, 6,462 nt) genes were inferred using PAUP* (72) with tree bisection-reconnection (TBR) branch swapping and the general time-reversible (GTR) model of nucleotide substitution with an among-site rate heterogeneity parameter (γ) and a proportion of invariant sites (I), as determined by Modeltest version 3.7 (57). Bootstrap values (BSV) were calculated using 1,000 replicate neighbor-joining (NJ) trees and the ML substitution model. Phylogenetic trees were also constructed using the Bayesian Markov chain Monte Carlo (MCMC) method implemented in MrBayes version 3.2 (63). Two independent runs were performed for each data set for at least 10 million generations, with sampling every 10,000 generations, and were terminated after the standard deviation of split frequencies reached ≤0.005. The posterior distribution of trees, Bayesian posterior probabilities (BPP), and model parameters were summarized from the MCMC sampling, and a consensus tree for each data set was created by summarizing the trees from each run after the initial 10% of trees was discarded as burn-in. All trees were rooted based on the position of the included taxa relative to Puumala virus (GenBank accession numbers NC_005224 [N gene] and AB433850 [GPC gene]).
Phylogenetic relationships within the Oligoryzomys genus of rodents were inferred using both ML and Bayesian approaches and the GTR+γ+I model of nucleotide substitution, as described above. This data set consisted of full-length or nearly full-length cytochrome b gene sequences from 20 of the 25 rodent samples in this study, as well as representatives from all Oligoryzomys species available on GenBank (n = 57, 1,140 bp) (see Table S2 in the supplemental material).
Genetic distance.
Pairwise percent nucleotide and amino acid sequence divergence between all taxa for the N, GPC, and RdRp genes was calculated using the pairwise deletion method with the Tamura-Nei (maximum composite likelihood method) and Jones, Taylor, and Thornton (JTT) substitution models in MEGA5, respectively (73).
Phylogeographic analysis.
Hantaviruses, unlike many arthropod-borne or endemic human viruses, have not been well characterized with respect to the environmental and demographic factors that shape their distribution and dynamics. However, the geographic distribution and evolutionary relationships of their hosts (particularly with respect to the rodent-borne viruses) are regularly invoked as critical factors in determining the spatial and evolutionary dynamics of hantaviruses (38, 60, 61). To assess how the geographic distribution of known Sigmodontinae hosts might be used to explore the spatial dynamics of South American hantaviruses, we compared phylogeographic models incorporating host distribution with models that assigned location states according to other metrics (k-means clustering or South American biome type). Spatial analyses were performed using N gene sequences for (i) all South American hantaviruses (SAmC; n = 134) and (ii) all ANDV clade viruses (AVC; n = 89) for which time and location of sampling were known. The spatial dynamics of the LANV and RIOMV clades were not analyzed due to small sample sizes.
For the host distribution model, each sequence was assigned a location state corresponding to the identity of its reservoir host. Because the distance between host ranges is thought to be an important factor for cross-species transmission, published host distributions (www.iucn.org) were mapped onto South America using ArcGIS (17), and the mean distance between them was calculated by finding the centroids of each range using the program R with the geosphere package (22, 24). For the k-means and biome type spatial models, the GPS coordinates associated with each hantavirus sequence were used to map their locations within South America as described above. K-means clustering in R was used to assign each mapped sequence (s) to one of a predefined number of clusters (k) (k = 6, 8, and 13 [SAmC only]), where each s was assigned to the k with the nearest mean (16, 21, 24, 39). Finally, location states were assigned based on the location of each data point within a South American biome: (i) east tropical and subtropical moist broadleaf forest; (ii) west tropical and subtropical moist broadleaf forest; (iii) dry broadleaf forest; (iv) temperate broadleaf and mixed forests; (v) east tropical and subtropical grasslands, savannas, and shrublands; (vi) west tropical and subtropical grasslands, savannas, and shrublands; (vii) temperate grasslands, savannas, and shrublands; (viii) flooded grasslands and savannas; and (ix) Mediterranean forests, woodlands, and shrublands (50, 80). The centroid of each biome type was calculated as described above.
Posterior distributions under a Bayesian phylogeographic model were estimated using the MCMC method implemented in BEAST version 1.7 (14, 35) using BEAGLE (70) to optimize computational efficiency. The model included an uncorrelated, lognormal relaxed molecular clock with a prior informed by the long-term substitution rates previously estimated for hantaviruses (13, 60), a flexible Bayesian skyride coalescent prior (46), and the SRD06 model of nucleotide substitution with two codon position partitions (1st and 2nd positions, 3rd position) and rate heterogeneity, substitution model, and base frequencies unlinked across codon positions. The prior distribution for the rate of movement between location states was based on the assumption of equal rates between all pairs of locations as well as a distance-informed prior calculated as the normalized inverse distance between the centroids of each location state. Bayesian stochastic search variable selection (BSSVS) was used to identify links between location states among the posterior set of trees that explain the most likely movement patterns under each geographic clustering model. The length of the MCMC chain for each run was 100 million iterations, with subsampling every 10,000. Tracer version 1.5 was used to assess convergence of all parameters, with minimum estimated sample size (ESS) values of 200 achieved in all cases. Ten percent of each chain was removed as burn-in. The geographic clustering approach that best fit the data was assessed using a posterior simulation-based analogue of Akaike's information criterion through MCMC (AICM) (2). Bayes factor tests (BF) were used to determine the statistical significance of the movement between location states in the models with the best fit.
RESULTS
Three distinct hantaviruses were recovered from HPS patient samples, each of which has been previously described (see Table S1 in the supplemental material). Analysis of the complete N gene of the single HPS case from Maranhão (H759113) indicated that ANJV virus was the causative agent of disease in this case. The N gene sequence of H759113 was 99% similar at the nucleotide level to previous ANJV isolates from the same location (Fig. 2); unfortunately, it was not possible to sequence the M or L segments of this virus due to template limitations. CASV was recovered from four HPS cases from Amazonas, Pará, and Rondônia, and it was possible to sequence the complete S segment as well as partial M (the C-terminal 2,839-nt) and L (2,428-nt) segments of a single representative (H745332). This virus was greater than 99% similar at the protein level to previously described CASV and formed a monophyletic clade with known CASV in both the N (BSV = 98, BPP = 1) and GPC (BSV = 100, BPP = 1) gene phylogenies (Fig. 2). Viruses similar to LANV were recovered from 14 HPS cases from Mato Grosso state, and complete N genes were sequenced for 10 of these. Very little nucleotide divergence was observed between samples (maximum pairwise nucleotide divergence = 1.3%); therefore, a single representative was selected for full genome sequencing (H731172). Both the N and GPC gene trees revealed that all nine of these viruses form a monophyletic clade with LANV-like virus HMT 08-02 (FJ816031), also from Mato Grosso state (BSV = 100, BPP = 1; Fig. 2). These viruses from Mato Grosso represent a genotype similar to, but distinct from, those of the true LANV viruses identified in Bolivia, Paraguay, and Brazil (Fig. 2) (26, 75). We propose designating this genotype (which would include HMT 08-02) LANV-2; however, sequences from the M and L segments are not available from HMT 08-02, limiting the comparison.
Fig 2.
Maximum likelihood phylogeny of the N (A) and GPC (B) genes from representatives of all available Sigmodontinae-borne hantaviruses, including those described here. Sequences generated in this study are shown in red, and the GPC genes with the WAVSA mutational variant in the signal peptidase cleavage site are indicated with an asterisk. Bootstrap values and Bayesian posterior probabilities are given above and below each node, respectively, for those nodes with BSV of ≥70 or BPP of ≥0.7. The tree is rooted relative to the position of Puumala virus (not shown). The viruses included in the N gene tree are as follows: ACPV (Andes Central Plata; GenBank accession no. EU564715, EU564719), ALPA (Alto Paraguay; DQ345762), ANDV (Andes; AF291702, AF004660), ANJV (Anajatuba; HM238887, HM238889), AAIV (Ape Aime Itapua; GU205340), ARQV (Araraquara; AF307325, EU170208), ARAUV (Araucaria; AY740628, HQ337907), BAYV (Bayou; L36929), BMJV (Bermejo; AF482713), BCCV (Black Creek Canal; L39949), BRV (Blue River; DQ090890), CADV (Cano Delgadito; DQ285566, EU939921), CARV (Carrizal; AB620093), CASV (Castelo dos Sonhos; AF307324, HQ719471), CATV (Catacamas; DQ256126), CHOV (Choclo; DQ285046), CCV (Convict Creek; L33816, U47135), ELMC (El Moro Canyon; U11427, U11429), FCV (Four Corners; U18098), ITPV 16 (Itapua 16; AY515594), ITPV 37 (Itapua 37; AY515595), ITPV 38 (Itapua 38; AY515596), JABV (Jabora; GU205339, JN232079), JUQV (Juquitiba; EF492472, GU213197), JUQV-like (Juquitiba-like; EU564724), LANV (Laguna Negra; AF005727), LANV-like (Laguna Negra-like; FJ816031), LECV (Lechiguanas; AF482714, EU564713), LSCV (Limestone Canyon; AF307322), MACV (Maciel; AF482716), MAPV (Maporal; FJ008979), MARV (Maripa; GQ179973), MTNV (Montano; AB620079, AB620092), MULV (Muleshoe; U54575), BMJV-NEBU (Neembucu; DQ345763), NYV (New York; U09488), ORNV (Oran; AF482715), PARV (Paranoa; EU643619, EU643620), PERV (Pergamino; AF482717), RIOMV (Rio Mamore; FJ532244, U52136), RIOMM (Rio Mearim; DQ451828), RIOSV (Rio Segundo; U18100), SNV (Sin Nombre; L25784, L37904), TUNV (Tunari; JF750417).
Three new genotypes of previously described hantavirus species were recovered from rodent samples: two genotypes of RIOMV (n = 21) and one of CASV (n = 3). Complete N genes were sequenced from 14 of the 21 RIOMV viruses, and the resulting phylogenetic analysis indicated the presence of two distinct genotypes that clustered by sampling location. The first genotype was found in a single sample from Amazonas (AN683313), while the other was recovered from 20 samples from Rondônia (represented by AN693292; Fig. 1 and 2). We tentatively suggest designating these genotypes RIOMV-3 and RIOMV-4, respectively. Further analysis of the complete M and L segments of RIOMV-4 and 2,108 nt of the GPC of RIOMV-3 supports the designation of these viruses as distinct genotypes (Fig. 2; see also Fig. S1 in the supplemental material). RIOMV-3 and RIOMV-4 are the first RIOMV to be identified in Brazil; however, they are closely related to previously described Rio Mearim (RIOMM) and ANJV, found in Maranhão state (76). Phylogenetic analysis of the cytochrome b gene from the host of RIOMV-3 allowed us to identify it as Oligoryzomys microtis, previously sampled throughout Bolivia, Brazil, and Peru (BPP = 0.9; Fig. 3). Interestingly, the 14 cytochrome b sequences recovered from the hosts of RIOMV-4 formed a strongly supported clade (BSV = 100, BPP = 1) that was distinct from O. microtis and more closely related to a newly described rodent, Oligoryzomys sp. RR-2010a from Parque Estadual do Cantão in Tocantins State, Brazil (62). The average percent nucleotide difference between Oligoryzomys sp. RR-2010a and the hosts of RIOMV-4 was 7.1% (±0.7%), while the percent nucleotide difference between these same rodents and O. microtis was 11.4% (±0.5%).
Fig 3.
Maximum likelihood phylogeny of the cytochrome b genes of all available Oligoryzomys rodents. The sequences generated in this study are shown in red.
A new genotype of CASV (henceforth CASV-2) was recovered from three Oligoryzomys utiaritensis rodents from Mato Grosso state (Fig. 1). O. utiaritensis has a range throughout central and southeastern Brazil and is a known carrier of CASV (74). Phylogenetic analysis of the complete genome of a single representative of CASV-2 (AN717313) indicates a close relationship with TUNV from Bolivia (Fig. 2), although the branching order varies by segment and has relatively low branch support. Despite some uncertainty in the phylogenetic positioning within this clade, it is clear that TUNV, CASV, and CASV-2 all cluster together with ANDV and ANDV-related viruses (Fig. 2). The percent amino acid divergences between TUNV, CASV, and CASV-2 are all <7%; therefore, all three of these viruses would be considered a single species (CASV) under the ICTV criteria (see Table S3 in the supplemental material).
Genetic characterization.
All sequenced S segments contained one contiguous open reading frame (ORF) from nt positions 43 to 1326, which could be translated into a predicted 428-aa nucleocapsid protein. An overlapping (+1) ORF, from nt positions 122 to 310 and encoding a putative 63-aa nonstructural (NSs) protein, was also identified. The amino acid sequence of the CASV-2 NSs was identical to that of ANDV (AF004660), while between 5 and 11 aa substitutions distinguished the other NSs proteins sequenced here from ANDV. Work in an in vitro system has indicated that the NSs protein of ANDV is expressed and likely has interferon-modulating functions, similar to other members of the Bunyaviridae family (25, 54, 77).
All sequenced M segments contained a predicted 1,138-aa GPC polyprotein from nt positions 52 to 3465. All conserved N-linked glycosylation sites and cysteine residues previously identified in hantaviruses were also present in these sequences (66). Cleavage of the GPC polypeptide into the Gn and Gc transmembrane proteins is thought to occur at the highly conserved pentapeptide motif WAASA, located just before the amino terminus of G2 (from aa 1990 to 2004) (40). Although the CASV-2, LANV-2, and RIOMV-4 sequences generated in this study did contain the WAASA motif, CASV sample H745332 contained a WAVSA variant that could also be identified in the GPC sequences of ARQV (AF307327, AY970821), CASV (AF307326), Maciel (MACV, AF028027), Paranoa (PARV, EU62116, EU62117, EU62118), and Pergamino (PERV, AF028028) viruses from the New World (Fig. 2), as well as Asama virus (EU929073, EU929074, EU929075) from the shrew mole (Urotrichus talpoides) in Japan (1) and Qiandao Lake virus (GU566022) from the stripe-backed shrew (Sorex cylindricauda) in China.
The sequenced L segments of CASV-2 (AN717313), LANV-2 (H731172), and RIOMV-4 (AN693292) all contained a single ORF from nt positions 36 to 6494, encoding a predicted 2,153-aa RdRp protein. Phylogenetic analysis of the L segment supports the presence of three monophyletic clades of South American hantaviruses: one comprised of ANDV-like viruses (ANDV, CASV, CASV-2), one of LANV-like viruses (LANV and LANV-2), and a third of RIOMV-like viruses (MARV, RIOMV, RIOMV-4); however, only eight full-length L segment sequences are available from South American hantaviruses, limiting the analysis (see Fig. S1 and Table S3 in the supplemental material). Because very few RdRp sequences of substantial length are available, it is unclear whether a robust analysis of all New World hantavirus RdRp genes would affect our understanding of their diversity. Although RdRp is widely regarded as a highly conserved gene and its utility in population-level analyses may be limited, this same characteristic may render it a useful addition to the N and GPC genes when examining deeper phylogenetic or taxonomic relationships within the genus. While the ICTV criteria consider only the N and GPC genes for species delineation, future descriptions of novel hantaviruses should certainly attempt to include sequencing and analysis of the L segment, as attempted here.
Phylogeographic analysis.
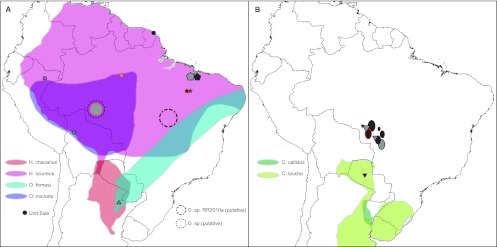
To assess the spatial dynamics of all South American hantaviruses (SAmC) and ANDV clade viruses only (AVC) in the context of rodent host distribution, phylogeographic models incorporating host distribution were compared to models that assigned location states according to other metrics (k-means clustering or South American biome type). According to AICM-based model selection, k-means clustering of the SAmC data into 6 groups (k-means [6]) fit the data better than the model where location states were assigned by host distribution (see Fig. S2a and b in the supplemental material). Adding a distance-based prior to the phylogeographic models resulted in a minutely better fit of both the k-means (6) and host distribution models to the data, but probably insignificantly so. Under the k-means (6) model, a general pattern of migration outward from the central part of the continent emerged for the SAmC data set (see Fig. S2c and d). Three well-supported unidirectional pathways of movement were observed that originated from south-central Brazil, central Bolivia, and Paraguay toward Uruguay/northern Argentina (BF = 9), north-central Brazil (BF = 13), and eastern Brazil (BF = 48) (see Fig. S2d). Significant migration was also observed from north-central Brazil to northeastern Brazil and French Guiana (BF = 78), as well as between Uruguay/northern Argentina and eastern Brazil (BF = 6). No significant migration across the Andes Mountains was detected. Examination of significant movement patterns between location states under the host distribution model for SAmC revealed the presence of unidirectional movement (i) from Necromys benefactus (host of MACV) to Necromys lasiurus (ARQV) (BF = 8), (ii) from O. microtis (RIOMV) to Oligoryzomys fornesi (ANJV) (BF = 8), and (iii) from Akodon azarae (PERV) to N. benefactus (BF = 5) (Fig. 4, 5). Significant bidirectional movement was also observed between the sympatric species Oligoryzomys chacoensis (BMJV and BMJV-NEBU) and Oligoryzomys flavescens (LECV and ACPV) (BF = 10 and 13) (see Fig. S2c). While N. benefactus/N. lasiurus and O. microtis/O. fornesi are congeneric, their published ranges do not overlap (Fig. 4, 5). However, N. lasiurus has been found as far south as Argentina, and both O. microtis and O. fornesi inhabit the contiguous lowlands of Peru, Bolivia, and Paraguay, indicating that contact between these hosts is theoretically possible (Fig. 4, 5) (15, 47). Furthermore, as illustrated by the discrepancies between the published distributions of Calomys laucha and Calomys callidus, and the areas where they have been sampled as hosts of LANV and LANV-2, it is clear that the known ranges of many of the rodent species of South America remain incomplete (Fig. 5).
Fig 4.
Geographic distribution of hantaviruses in the Andes clade and their hosts. Andes clade hantaviruses with sequences available in GenBank (black-outlined shapes) or generated in this study (red-outlined shapes) are shown along with corresponding host distributions from iucn.org. Viruses indicated are ACPV (triangle), ANDV (pentagon), ARAUV (hexagon), ARQV (star), BMJV (rounded square), BMJV-NEBU (oval), CASV (circle), CASV-2 (square), ITPV 37 (chevron), ITPV 38 (inverted triangle), JUQV (sun), LECV (rounded ex), MACV (diamond), ORNV (squared ex), PARV (cross), PERV (six-pointed star), TUNV (octagon), where the size of the symbol is proportional to the number of samples from a given location. Samples from HPS cases are indicated in black, while those from rodents are indicated in gray.
Fig 5.
Geographic distribution of hantaviruses of the Rio Mamore (A) and Laguna Negra (B) clades and their hosts. Hantaviruses with sequences available on GenBank (black-outlined shapes) or generated in this study (red-outlined shapes) are shown, along with corresponding host distributions from iucn.org. Viruses indicated are ALPA (triangle), ANJV (pentagon), MARV (hexagon), RIOMMV (star), RIOMV (ex), RIOMV-3 (square), and RIOMV-4 (circle) (A) and LANV (inverted triangle), LANV-like (sun), and LANV-2 (oval) (B), where the size of the symbol is proportional to the number of samples from a given location. Samples from HPS cases are indicated in black, while those from rodents are indicated in gray.
AICM-based model selection of the AVC data set indicated that the model incorporating location states assigned by host species fit the data significantly better than any other model, with no improvement when distance-informed priors were included (see Fig. S2b in the supplemental material). Adding a distance-informed prior may not have increased the fit of the model in this analysis because of the uncertainty present in the known geographic ranges of these rodents. As these ranges become better described, the relationship between the locations of host ranges and the frequency of cross-species transmission may become better defined. Although AVC viruses did form strongly supported clusters by host species (data not shown), most of the significant movement events identified in the SAmC data set were also reconstructed in the AVC analysis: unidirectional movement from N. benefactus to N. lasiurus (BF = 6) and bidirectional movement between O. chacoensis and O. flavescens (BF = 7 and 8) (see Fig. S2c in the supplemental material). Many of the viruses included in this analysis were represented by only one or a few sequences or may not have been sampled in all relevant host species, perhaps leading to an underestimation of cross-species transmission events. Increasing the amount and diversity of hantavirus sequence data will greatly improve our knowledge about the true geographic and host distributions of hantaviruses in South America.
DISCUSSION
The results of this study indicate that the diversity and distribution of hantaviruses in South America are highly complex. Although the initial identification of hantaviruses in the New World arose from an HPS case in the United States, the greatest diversity of hantaviruses in the Western Hemisphere clearly lies in South America, where more than 25 genotypes have now been described. Based on our phylogenetic analyses, South American hantaviruses form at least three well-supported, monophyletic clades: an Andes clade, a Laguna Negra clade, and a Rio Mamore clade, each classified as a unique species by ICTV (Fig. 2). Bayesian phylogeographic analyses of all South American hantaviruses indicated a general pattern of spread from the south-central part of the continent to the north, south, and east (see Fig. S2d in the supplemental material). While Paraguay, south-central Brazil, and central Bolivia appear to be the origin of much of the current sampled diversity of hantaviruses in South America, this region also contains more genotypes of hantavirus than any other and has been sampled more intensively. Therefore, it is unclear whether this central region represents an origin of hantavirus diversity or whether the patterns reconstructed here are an artifact of sampling. Increasing the amount of available sequence data throughout the range of hantaviruses in South America may clarify this observation. Interestingly, although hantaviruses must have crossed the Andes Mountain range at some point in the past, our analysis could not detect any present-day movement, indicating that the Andes must represent a powerful barrier to dispersal.
Andes clade.
If only those viruses with complete or nearly complete N and GPC gene sequences are considered, the ANDV clade would be comprised of approximately 12 viruses, found in six countries (Argentina, Bolivia, Brazil, Chile, Paraguay, Uruguay) and carried by three genera of rodents (Akodon, Necromys, Oligoryzomys). Furthermore, three well-supported groups (BPP = 1) exist within the ANDV clade: (i) CASV, CASV-2, and TUNV; (ii) PERV, MACV, ARQV, and PARV; (iii) ORNV, BMJV, BMJV-NEBU, LECV, and ACPV. However, the exact relationships between these groups was not well resolved (Fig. 2). Analysis of the spatial dynamics of the ANDV clade revealed evidence of potential cross-species transmission events between closely related (congeneric) and sympatric host species (Fig. 4). Although cross-species transmission events between closely (and distantly) related hosts have been widely reported in hantaviruses, host switching as a primary driving mechanism of hantavirus diversity and distribution is often overlooked in favor of codivergence between virus and host (4, 29, 30, 60, 69). Nonetheless, both host geographic distribution and phylogenetic relatedness is known to influence the host-switching potential of many RNA and DNA viruses, even if their relative importance in shaping these dynamics remains uncertain (20, 41, 81). The cross-species transmission events inferred in the present analysis of the ANDV clade (and all SAmC viruses) indicate that both the geographic proximity of and phylogenetic relatedness between hosts are likely important in shaping the evolutionary dynamics of this group (Fig. 4 and 5).
Both of the Andes clade viruses sequenced in this study (CASV, CASV-2) form a single, well-supported group with TUNV from Bolivia. Of these, CASV and TUNV have both been associated with fatal HPS, but thus far a potential reservoir has been suggested only for CASV (10, 27, 74). Short fragments (421 nt) of the CASV-2 sequences generated here from O. utiaritensis in Mato Grosso were previously reported and identified as CASV, along with viruses from HPS cases taken from Pará (74). Based on these short N gene sequences, it was suggested that the viruses sequenced from rodents in Mato Grosso and HPS cases from Pará were both CASV and that O. utiaritensis is a probable reservoir of CASV in Brazil (74). However, when the full genome sequence of one of these same CASV-2 viruses from O. utiaritensis (AN717313) was analyzed, it became clear that these sequences are not CASV but cluster more closely with TUNV. Therefore, it is uncertain whether both CASV and CASV-2 are carried by O. utiaritensis and if they both contribute to HPS in this region.
Genetic analysis of the GPC protein of the Andes clade revealed the presence of a WAVSA mutational variant in the signal peptidase cleavage site of the Gn and Gc proteins of several viruses. To our knowledge, this is the first time the WAVSA variant has been identified as the GPC cleavage site in New World hantaviruses. In all cases, the alanine to valine amino acid substitution was the result of a C-to-T transition at the second codon position, a substitution that must have occurred multiple times throughout the evolution of these viruses (Fig. 2). Experimental evidence has shown that an amino acid change at the −3 position of the C-terminal portion of the signal peptide cleavage site will not interfere with cleavage, provided that this amino acid is small and uncharged (i.e., alanine, serine, valine, and cysteine) (53, 78, 79). Cleavage of the GPC polypeptide was not impeded in vitro when the WAASA motif was replaced with WAVSG, indicating that the WAVSA motif identified in CASV and others is likely to be effectively cleaved (40).
Laguna Negra clade.
The Laguna Negra clade contains only LANV, originally identified in HPS cases and Calomys laucha from Paraguay and Bolivia, and LANV-2 viruses, identified from HPS patients in Brazil (26, 58). Recently, LANV was reported for the first time in HPS cases in Mato Grosso, expanding the range of this virus into Brazil (75). The authors further suggested the crafty vesper mouse (Calomys callidus) as a potential reservoir for LANV in Brazil. However, as only 435 nt of the S segment was used to characterize these viruses, we further sequenced the full S segment of one of these (H711891, accession no. JQ775503) and determined that it is more closely related to the Brazilian LANV-2 than to LANV. In the original publication, the sequence of H711891 clustered with the Brazilian Calomys callidus-borne LANV, indicating that this rodent is more likely to be the reservoir of LANV-2 virus (see Fig. S3 in the supplemental material). Notably, LANV and LANV-2 share 96 to 97% aa sequence similarity in both the N and GPC proteins, falling below the 7% divergence recommended for species-level distinction (32). However, based on the substantial level of nucleotide divergence observed between these viruses (∼15% in the N and ∼20% in the GPC) and the possibility that they are carried by different hosts, we suggest they be considered distinct genotypes of LANV virus. Both viruses are cocirculating in Mato Grosso state, where they have been associated with HPS since at least 2001 (75). LANV-2 viruses are likely important contributors to HPS in west central Brazil, as they have now been recovered from 18 HPS cases spanning 3 years (75) (this study).
Rio Mamore clade.
The Rio Mamore clade of viruses is comprised of Alto Paraguay (Paraguay), ANJV (Brazil), MARV (French Guiana), RIOMM (Brazil), RIOMV (Bolivia and Peru), and RIOMV-3 and RIOMV-4 (Brazil, this study). Of these, only ANJV and MARV have been associated with HPS; however, the close relationships between these viruses and other members of the Rio Mamore clade imply that an association between HPS and these viruses cannot be excluded. Hantavirus-positive rodents are regularly collected in regions where HPS cases have been reported; however, the absence of direct evidence of hantavirus infection in patient samples limits the association. For example, all RIOMV-4 viruses detected here were recovered from rodents in Rondônia, Brazil, where HPS cases have been reported, yet no etiological agent has been identified (45). Similarly in Peru, serological reactivity to ANDV has revealed past evidence of hantavirus exposure in febrile patients, and fatal HPS has recently been reported, yet no associated hantavirus has been identified (6).
Within the Rio Mamore clade, only RIOMV is currently recognized as a species by ICTV. Despite the characterization of several novel viruses since the initial description of RIOMV in 1997, none are more than 7% divergent (aa) in either the N or GPC proteins, indicating that they should be considered a single species, Rio Mamore virus (32). However, the presence of substantial nucleotide sequence divergence between viruses in all three genome segments (approximately 20%) along with disparate rodent hosts does support the distinction of these viruses as unique genotypes of RIOMV (see Table S3 in the supplemental material). The reservoir of RIOMV in Peru, Bolivia, and Amazonas, Brazil (RIOMV-3, this study), is Oligoryzomys microtis; however, the host of RIOMV-4 was identified as a novel but related species of Oligoryzomys that is similar to Oligoryzomys sp. RR-2010a (Fig. 3). Oligoryzomys sp. RR-2010a has been identified only once, in Tocantins, Brazil (Fig. 5) (62). The host geographic ranges of all viruses in the RIOMV clade overlap, creating opportunities for host switching, as well as reassortment and recombination.
Taxonomic implications.
Despite the explicit criteria for species delineation recommended by the ICTV, hantavirus nomenclature remains somewhat arbitrary, potentially obscuring the true relationships between viruses. The criteria by which hantaviruses are named has a significant impact not only on our understanding of their diversity but also on how their evolutionary history, especially with respect to host relationships, is viewed. For example, if ANDV, BMJV, ORNV, and LECV are considered unique viruses (as suggested by their unique names), we see that ANDV is carried primarily by O. longicaudatus, BMJV by O. chacoensis, ORNV by O. longicaudatus, and LECV by O. flavescens. Therefore, we might postulate a long coevolutionary history of hantaviruses and their hosts. However, if these four viruses are genotypes of ANDV, we must then give an evolutionary history that includes cross-species transmission, such as that inferred in this study, more weight. It is also problematic that many viruses have been characterized based solely on small fragments of the genome. Phylogenetic relationships between viruses can change with the addition of full gene sequences, and taxonomic allocations are likely to change further once the serological and ecological aspects of these viruses are taken into account.
Complicating matters further, it is not clear how effectively the ICTV criteria describe and delineate the diversity present in the genus, particularly with respect to the requirements for percent genetic divergence and a unique host species. It is now clear that cross-species transmission between hosts has occurred multiple times throughout the history of hantaviruses, in both the Old and New Worlds (9, 11, 30, 34, 52, 56, 60, 65). Unique hantaviruses may infect multiple host species, individual hosts may carry multiple viruses, and host-switching events can occur between both closely (congeneric) and distantly (between orders Rodentia and Soricomorpha) related mammals. Therefore, the ICTV requirement for each hantavirus species to infect a unique primary host may be inappropriate, potentially resulting in an artificial taxonomic structure and obscuring the biological context of emergence. In addition, recent work has suggested that a 7% aa difference in the complete N and GPC proteins for species demarcation may be unsuitably small given the high levels of genetic diversity observed within hantaviruses (42). Based on the results of the present study, more stringent criteria for species delineation (such as the >10% aa difference in the N or 12% aa difference in the GPC suggested by Maes et al.) would appear to be more appropriate (42). Further, it is critical that these calculations be performed using complete coding sequence only; the consensus primers routinely used to detect novel hantaviruses necessarily target highly conserved regions of the genome, biasing percent difference calculations (see Table S3 in the supplemental material). Finally, we suggest the adaptation of a numerical taxonomic system to indicate species and genotype relationships, similar to the classification structure already in use for the enterovirus (e.g., EV-D68) and adenovirus (HAdV-12) genera (32). With this approach, a reference genotype would be designated for each novel and known hantavirus species that would ideally correspond to the initial species description (e.g., ANDV, RIOMV). All additional and related viruses that do not meet the criteria for species delineation would then be given sequential numerical genotype designations, akin to the nomenclature used in this study (e.g., RIOMV-3, LANV-2). This type of classification structure would bring important clarification to the relationships between hantaviruses (32).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Health Surveillance Secretariat (Mauro R. Elkhoury, Marilia Lavocat) and the Mato Grosso State Health Secretariat (Aparecido Marques, Alba Via) for their technical support during collection of samples in the field, as well as Maia Rabaa for helpful comments on previous versions of the manuscript and Steve Sameroff for technical assistance. This work was supported by CNPq (CNPq 302987/2008-8 and 301641/2010-2 and CNPq/CAPES/FAPESPA 573739/2008-0), the National Institutes of Health (AI057158; Northeast Biodefense Center-Lipkin), and the Defense Threat Reduction Agency.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Arai S, et al. 2008. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proc. Natl. Acad. Sci. U. S. A. 105:16296–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baele G, et al. 7 March 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. [Epub ahead of print.] doi:10.1093/molbev/mss084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. 1997. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am. J. Trop. Med. Hyg. 57:368–374 [DOI] [PubMed] [Google Scholar]
- 4. Bohlman MC, et al. 2002. Analysis of hantavirus genetic diversity in Argentina: S segment-derived phylogeny. J. Virol. 76:3765–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campos GM, et al. 2003. Serological survey of hantavirus in Jardinopolis County, Brazil. J. Med. Virol. 71:417–422 [DOI] [PubMed] [Google Scholar]
- 6. Castillo Ore RM, et al. 2012. Serologic evidence for human hantavirus infection in Peru. Vector Borne Zoonotic Dis. 12:683–689 [DOI] [PubMed] [Google Scholar]
- 7. Chu YK, Goodin D, Owen RD, Koch D, Jonsson CB. 2009. Sympatry of 2 hantavirus strains, Paraguay, 2003–2007. Emerg. Infect. Dis. 15:1977–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu YK, Owen RD, Gonzalez LM, Jonsson CB. 2003. The complex ecology of hantavirus in Paraguay. Am. J. Trop. Med. Hyg. 69:263–268 [PubMed] [Google Scholar]
- 9. Chu YK, Owen RD, Jonsson CB. 2011. Phylogenetic exploration of hantaviruses in Paraguay reveals reassortment and host switching in South America. Virol. J. 8:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cruz CD, et al. 2012. Novel strain of Andes virus associated with fatal human infection, central Bolivia. Emerg. Infect. Dis. 18:750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dekonenko A, et al. 2003. Genetic similarity of Puumala viruses found in Finland and western Siberia and of the mitochondrial DNA of their rodent hosts suggests a common evolutionary origin. Infect. Genet. Evol. 3:245–257 [DOI] [PubMed] [Google Scholar]
- 12. Delfraro A, et al. 2003. Yellow pigmy rice rat (Oligoryzomys flavescens) and hantavirus pulmonary syndrome in Uruguay. Emerg. Infect. Dis. 9:846–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88 doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eisenberg FJ, Redford HK. 1999. Mammals of the neotropics, volume 3: Ecuador, Bolivia, Brazil. University of Chicago Press, Chicago, Illinois [Google Scholar]
- 16. Espindola A, Alvarez N. 2011. Comparative phylogeography in a specific and obligate pollination antagonism. PLoS One 6:e28662 doi:10.1371/journal.pone.0028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. ESRI 2011. ArcGIS desktop: release 10. Environmental Systems Research Institute, Redlands, CA [Google Scholar]
- 18. Ferrer JF, et al. 1998. High prevalence of hantavirus infection in Indian communities of the Paraguayan and Argentinean Gran Chaco. Am. J. Trop. Med. Hyg. 59:438–444 [DOI] [PubMed] [Google Scholar]
- 19. Figueiredo LT, Moreli ML, Campos GM, Sousa RL. 2003. Hantaviruses in Sao Paulo State, Brazil. Emerg. Infect. Dis. 9:891–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gottschling M, et al. 2011. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol. Biol. Evol. 28:2101–2113 [DOI] [PubMed] [Google Scholar]
- 21. Hartigan JA, Wong MA. 1979. A K-means clustering algorithm. Appl. Stat. 28:100–108 [Google Scholar]
- 22. Hijmans JR, Williams E, Vennes C. 2012. Geosphere: spherical trigonometry. R package, 1.2-28. [Google Scholar]
- 23. Holmes R, Boccanera R, Figueiredo LT, Mancano SR, Pane C. 2000. Seroprevalence of human hantavirus infection in the Ribeirao Preto region of Sao Paulo State, Brazil. Emerg. Infect. Dis. 6:560–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299 [Google Scholar]
- 25. Jaaskelainen KM, et al. 2007. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 79:1527–1536 [DOI] [PubMed] [Google Scholar]
- 26. Johnson AM, et al. 1997. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 238:115–127 [DOI] [PubMed] [Google Scholar]
- 27. Johnson AM, et al. 1999. Genetic investigation of novel hantaviruses causing fatal HPS in Brazil. J. Med. Virol. 59:527–535 [DOI] [PubMed] [Google Scholar]
- 28. Jonsson CB, Figueiredo LT, Vapalahti O. 2010. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 23:412–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang HJ, et al. 2009. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 388:8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R. 2011. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 85:7496–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ketai LH, et al. 1998. Distinguishing hantavirus pulmonary syndrome from acute respiratory distress syndrome by chest radiography: are there different radiographic manifestations of increased alveolar permeability? J. Thorac. Imaging 13:172–177 [DOI] [PubMed] [Google Scholar]
- 32. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, Amsterdam, The Netherlands [Google Scholar]
- 33. Klempa B, et al. 2007. Novel hantavirus sequences in shrew, Guinea. Emerg. Infect. Dis. 13:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klingstrom J, et al. 2002. Rodent host specificity of European hantaviruses: evidence of Puumala virus interspecific spillover. J. Med. Virol. 68:581–588 [DOI] [PubMed] [Google Scholar]
- 35. Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5:e1000520 doi:10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levis S, et al. 2004. Hantavirus pulmonary syndrome in northwestern Argentina: circulation of Laguna Negra virus associated with Calomys callosus. Am. J. Trop. Med. Hyg. 71:658–663 [PubMed] [Google Scholar]
- 37. Levis S, et al. 1998. Genetic diversity and epidemiology of hantaviruses in Argentina. J. Infect. Dis. 177:529–538 [DOI] [PubMed] [Google Scholar]
- 38. Lin XD, et al. 2012. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J. Virol. 86:972–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lloyd S. 1982. Least squares quantization in PCM. IEEE Trans. Inform. 28:129–137 [Google Scholar]
- 40. Lober C, Anheier B, Lindow S, Klenk HD, Feldmann H. 2001. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289:224–229 [DOI] [PubMed] [Google Scholar]
- 41. Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. 2011. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog. 7:e1002260 doi:10.1371/journal.ppat.1002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maes P, et al. 2009. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect. Genet. Evol. 9:813–820 [DOI] [PubMed] [Google Scholar]
- 43. Martin Y, Gerlach G, Schlotterer C, Meyer A. 2000. Molecular phylogeny of European muroid rodents based on complete cytochrome b sequences. Mol. Phylogenet. Evol. 16:37–47 [DOI] [PubMed] [Google Scholar]
- 44. Matheus S, et al. 2010. Hantavirus pulmonary syndrome, French Guiana. Emerg. Infect. Dis. 16:739–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Medeiros DB, et al. 2010. Circulation of hantaviruses in the influence area of the Cuiaba-Santarem Highway. Mem. Inst. Oswaldo Cruz 105:665–671 [DOI] [PubMed] [Google Scholar]
- 46. Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25:1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Musser GG, Carleton MD. 2005. Super family Muroidea. In Wilson DE, Reeder DM. (ed), Mammal species of the world. A taxonomic and geographic reference. Johns Hopkins University Press, Baltimore, MD [Google Scholar]
- 48. Nichol ST, et al. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914–917 [DOI] [PubMed] [Google Scholar]
- 49. Oliveira RC, et al. 2009. Genetic characterization of a Juquitiba-like viral lineage in Oligoryzomys nigripes in Rio de Janeiro, Brazil. Acta Trop. 112:212–218 [DOI] [PubMed] [Google Scholar]
- 50. Olson DM, et al. 2001. Terrestrial ecoregions of the world: new map of life on earth. Bioscience 51:933–938 [Google Scholar]
- 51. Padula P, et al. 2002. Andes virus and first case report of Bermejo virus causing fatal pulmonary syndrome. Emerg. Infect. Dis. 8:437–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Palma RE, Polop JJ, Owen RD, Mills JN. 2012. Ecology of rodent-associated hantaviruses in the Southern Cone of South America: Argentina, Chile, Paraguay, and Uruguay. J. Wildl. Dis. 48:267–281 [DOI] [PubMed] [Google Scholar]
- 53. Perlman D, Halvorson HO. 1983. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J. Mol. Biol. 167:391–409 [DOI] [PubMed] [Google Scholar]
- 54. Plyusnin A. 2002. Genetics of hantaviruses: implications to taxonomy. Arch. Virol. 147:665–682 [DOI] [PubMed] [Google Scholar]
- 55. Plyusnin A, Morzunov SP. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47–75 [DOI] [PubMed] [Google Scholar]
- 56. Plyusnina A, et al. 2011. Genetic evidence for the presence of two distinct hantaviruses associated with Apodemus mice in Croatia and analysis of local strains. J. Med. Virol. 83:108–114 [DOI] [PubMed] [Google Scholar]
- 57. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 58. Raboni SM, et al. 2009. Evidence of circulation of Laguna Negra-like hantavirus in the Central West of Brazil: case report. J. Clin. Virol. 45:153–156 [DOI] [PubMed] [Google Scholar]
- 59. Raboni SM, Probst CM, Bordignon J, Zeferino A, dos Santos CN. 2005. Hantaviruses in Central South America: phylogenetic analysis of the S segment from HPS cases in Parana, Brazil. J. Med. Virol. 76:553–562 [DOI] [PubMed] [Google Scholar]
- 60. Ramsden C, Holmes EC, Charleston MA. 2009. Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol. Biol. Evol. 26:143–153 [DOI] [PubMed] [Google Scholar]
- 61. Richter MH, Hanson JD, Cajimat MN, Milazzo ML, Fulhorst CF. 2010. Geographical range of Rio Mamore virus (family Bunyaviridae, genus Hantavirus) in association with the small-eared pygmy rice rat (Oligoryzomys microtis). Vector Borne Zoonotic Dis. 10:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rocha RG, et al. 2011. Small mammals of the mid-Araguaia River in central Brazil, with the description of a new species of climbing rat. Zootaxa 2789:1–34 [Google Scholar]
- 63. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 64. Rosa ES, et al. 2005. Newly recognized hantaviruses associated with hantavirus pulmonary syndrome in northern Brazil: partial genetic characterization of viruses and serologic implication of likely reservoirs. Vector Borne Zoonotic Dis. 5:11–19 [DOI] [PubMed] [Google Scholar]
- 65. Saasa N, et al. 2012. Ecology of hantaviruses in Mexico: genetic identification of rodent host species and spillover infection. Virus Res. 168:88–96 [DOI] [PubMed] [Google Scholar]
- 66. Schmaljohn C. 1996. Bunyaviridae: the viruses and their replication, 3 ed Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 67. Song JW, Baek LJ, Schmaljohn CS, Yanagihara R. 2007. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg. Infect. Dis. 13:980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Song JW, et al. 2007. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol. J. 4:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Song JW, et al. 2009. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura). J. Virol. 83:6184–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Suchard MA, Rambaut A. 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 25:1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sumibcay L, et al. 2012. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d'Ivoire. Virol. J. 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4.0b. Sinauer Associates, Sunderland, MA [Google Scholar]
- 73. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 74. Travassos da Rosa ES, et al. 2011. Pygmy rice rat as potential host of Castelo dos Sonhos Hantavirus. Emerg. Infect. Dis. 17:1527–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Travassos da Rosa ES, et al. 2012. Molecular epidemiology of Laguna Negra virus, Mato Grosso State, Brazil. Emerg. Infect. Dis. 18:982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Travassos da Rosa ES, et al. 2010. Hantaviruses and hantavirus pulmonary syndrome, Maranhao, Brazil. Emerg. Infect. Dis. 16:1952–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vera-Otarola J, et al. 2012. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J. Virol. 86:2176–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. von Heijne G. 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133:17–21 [DOI] [PubMed] [Google Scholar]
- 79. von Heijne G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wade TG, Riitters KH, Wickham JD, Jones KB. 2003. Distribution and causes of global forest fragmentation. Conserv. Ecol. 7:7 [Google Scholar]
- 81. Wertheim JO, Worobey M. 2007. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 3:e95 doi:10.1371/journal.ppat.0030095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.