Abstract
The rotavirus spike protein domain VP8* is essential for recognition of cell surface carbohydrate receptors, notably those incorporating N-acylneuraminic acids (members of the sialic acid family). N-Acetylneuraminic acids occur naturally in both animals and humans, whereas N-glycolylneuraminic acids are acquired only through dietary uptake in normal human tissues. The preference of animal rotaviruses for these natural N-acylneuraminic acids has not been comprehensively established, and detailed structural information regarding the interactions of different rotaviruses with N-glycolylneuraminic acids is lacking. In this study, distinct specificities of VP8* for N-acetyl- and N-glycolylneuraminic acids were revealed using biophysical techniques. VP8* protein from the porcine rotavirus CRW-8 and the bovine rotavirus Nebraska calf diarrhea virus (NCDV) showed a preference for N-glycolyl- over N-acetylneuraminic acids, in contrast to results obtained with rhesus rotavirus (RRV). Crystallographic structures of VP8* from CRW-8 and RRV with bound methyl-N-glycolylneuraminide revealed the atomic details of their interactions. We examined the influence of amino acid type at position 157, which is proximal to the ligand's N-acetyl or N-glycolyl moiety and can mutate upon cell culture adaptation. A structure-based hypothesis derived from these results could account for rotavirus discrimination between the N-acylneuraminic acid forms. Infectivity blockade experiments demonstrated that the determined carbohydrate specificities of these VP8* domains directly correlate with those of the corresponding infectious virus. This includes an association between CRW-8 adaption to cell culture, decreased competition by N-glycolylneuraminic acid for CRW-8 infectivity, and a Pro157-to-Ser157 mutation in VP8* that reduces binding affinity for N-glycolylneuraminic acid.
INTRODUCTION
Rotaviruses belong to the Reoviridae family and are recognized as a major cause of life-threatening gastroenteritis in many young animals and humans (14, 41). Evidence supporting the regular occurrence of human infection by animal rotaviruses is accumulating (27, 28), with recent data suggesting that livestock herds may serve as potential reservoirs for human infections (30). The infectious triple-layered rotavirus particle, but not the noninfectious double-layered particle, recognizes gangliosides on the host cell surface (7). The outer capsid viral proteins, VP4 and VP7, are essential for receptor binding and cell entry. Enhancement of rotavirus infectivity is induced by proteolytic cleavage of VP4 which yields functional subunits VP8* and VP5* that remain associated with the virion (5, 12, 36). The carbohydrate-recognizing subunit VP8* plays an important role in receptor binding during cell entry by rotaviruses (8, 9, 12).
As sialidase treatment of cells markedly reduces binding and infectivity of some (mostly animal) rotaviruses to N-acylneuraminic acid-containing receptors on the host cell surface, these viruses have been termed sialidase sensitive. In contrast, human rotavirus infection typically increases following cellular sialidase treatment and has been termed sialidase insensitive (17). This phenotypic definition derives from the use of bacterial sialidases that efficiently remove only terminal and unbranched N-acylneuraminic acids from host cells in culture (29, 42). The ability of human rotaviruses to recognize and interact via VP8* with N-acylneuraminic acid is debated. In NMR studies, the VP8* of prototypic human rotavirus Wa showed direct interaction with the N-acylneuraminic acid of GM1 (17). This provided evidence that recognition of this sialic acid moiety is potentially important in all human and animal rotavirus infections. In contrast, the human rotavirus HAL1166 did not interact with sialylated glycans in glycan array experiments (18). The HAL1166 VP8* amino acid sequence is atypical among human rotaviruses and interestingly has some features usually associated with animal strains. These findings indicate the complexity of carbohydrate recognition by rotaviruses and have significant implications for cross-species transmission.
N-Acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) are the most common neuraminic acids in nature, being ubiquitously expressed on the surface of most mammalian cells. N-Acylneuraminic acids are usually located on the termini of glycoconjugates, where they mediate and regulate a wide range of cell-cell and macromolecule-cell interactions. Neu5Ac and Neu5Gc are utilized as receptors for host cell infection by a variety of pathogens, including rotaviruses (22, 34). Neu5Gc is distinguished from Neu5Ac by the addition of a hydroxyl group to the N-acetyl moiety. Although abundant in the animal kingdom, Neu5Gc is not synthesized in normal human tissues due to mutation of CMP–N-acetylneuraminic acid hydroxylase. However, dietary Neu5Gc uptake occurs in human intestinal epithelium, the target of rotavirus infection (2, 39), possibly facilitating human infection by Neu5Gc-utilizing rotaviruses. As shown in Table 1, sialidase-sensitive rotavirus strains from cattle (Nebraska calf diarrhea virus [NCDV]), monkeys (rhesus rotavirus [RRV]), and pigs (porcine rotavirus strain OSU) show distinct binding preferences for Neu5Ac or Neu5Gc, whether these are presented as monosaccharides or embedded within ganglioside glycans (7, 8, 35). The VP8* region from position 64 to 224 (VP8*64-224) is responsible for sialoside binding. The preference of NCDV and OSU for the Neu5Gc-containing glycan of ganglioside GM3 (aceramido-GM3Gc [a-GM3Gc]) over the Neu5Ac-containing GM3 (a-GM3Ac) glycan contrasts with the higher affinity of the RRV VP8*64-224 for Neu5Ac than for Neu5Gc (Table 1). Recently, we showed that the N-glycolyl-containing a-GM3Gc glycan is a more effective inhibitor of infection by porcine rotavirus CRW-8 than RRV and that a-GM3Ac exhibits the converse trend (43). It is anticipated that such specificity differences result from particular amino acid characteristics in the sialoside binding sites among those strains. The influence of amino acid type at two key positions (157 and 187) of the VP8* carbohydrate binding site is of interest, since their close proximity to the N-acyl portion of the bound ligand gives them the potential to impact N-acylneuraminic acid preference and/or ligand binding affinity. Rotavirus strain comparisons, notably for the amino acid at 187, indicate a possible association and likely involvement with N-acylneuraminic acid preference (Table 1). However, this has not been examined experimentally.
TABLE 1.
Diversity of amino acids at positions 157 and 187 in VP8* sequences of animal rotaviruses in relation to sialidase sensitivity and N-acylneuraminic acid preference
| Rotavirus strain(s)a | Origin | Sialidase sensitivityb | N-Acylneuraminic acid preference | Amino acid residue at position: |
|
|---|---|---|---|---|---|
| 157 | 187 | ||||
| CRW-8, OSU, YM | Porcine | + | Neu5Gc > Neu5Ac c | Pro | Gly |
| SA11,d NCDV | Simian, bovine | + | Neu5Gc > Neu5Ac | Ser | Gly |
| RRV | Simian | + | Neu5Ac > Neu5Gc | Pro | Lys |
| UK | Bovine | −e | Neu5Ac | Thr | Lys |
Other sialidase-sensitive rotaviruses from dogs (CU-1, K9), cats (CAT97 and FRV64), cattle (C486) and horses (H1) also show Pro157 and Gly187 (accession numbers: K9, D13400; CU-1, D13401; CAT97, D13402; FRV64, D14723; C486, Y00127; H1, D16341). Other sialidase-sensitive rotaviruses from cattle show Thr157 and Gly187 (BRV033) or Ser157 and Asp187 (RF), and equine rotavirus strain L338 has Pro157 and Ala187 (BRV033, U62155; RF, U65924; L338, D13399).
From published data that define sialidase insensitivity as preserved infectious virus titers on cultured cells treated with sialidase (3).
The preference of YM has not been reported.
SA11 isolates vary in sequence. For example, SA11-H96 has the listed sequence, whereas SA11-Both has Pro157 and Ala187 (38).
Although UK is classified as sialidase insensitive, it atypically has Phe190 and binds gangliosides with a specificity dependent on the neuraminic acid moiety (7).
Studies of rotavirus-sialoside interactions are almost invariably conducted with cell culture-adapted viruses. Limited data are available on amino acid sequence changes selected in animal rotavirus proteins during culture adaption. Studies analyzing VP8* sequence indicate that Pro157 in the original Adamson NCDV isolate NCDV-V mutated to serine during the cell culture passage that produced the NCDV (Cody) strain and the RIT 4237 (Lincoln) strain of NCDV. This was the sole amino acid change identified between these three NCDV isolates in the VP8*64–224 region (33). Sequences of 25 pig and cattle primary isolates obtained directly from stools exhibited proline or threonine at position 157 (30). Additionally, we previously observed a change from Pro157 to Ser157 in CRW-8 VP8* after additional cell culture passage (37). This amino acid forms part of the sialoside binding site in CRW-8 VP8* and lies adjacent to the N-acetyl or N-glycolyl group. These studies show that mutation at residue 157 in some animal rotaviruses is associated with cell culture adaption, and we propose that such mutation could alter sialoside recognition.
Methyl-α-d-N-acetylneuraminide (Neu5Acα2Me) is representative of a minimal sialoside that binds VP8* from animal rotaviruses (1, 8). Here we report the X-ray crystallographic structures of the VP8*64-224 of CRW-8 and RRV bound to the N-glycolylneuraminic acid derivative, methyl-α-d-N-glycolylneuraminide (Neu5Gcα2Me). These structures have led to our proposed hypothesis for the molecular basis of VP8* preference for particular N-acylneuraminic acids. The effect of the Pro157-to-Ser157 mutation on the carbohydrate specificity and binding affinity of CRW-8 VP8*64-224 was analyzed, and these proteins demonstrated distinct binding affinities. These studies provide insight into the flexibility of the sialoside binding cleft as well as the conformation of constituent amino acids and their resultant effects on ligand binding and carbohydrate specificity. In combination with results from saturation transfer difference nuclear magnetic resonance spectroscopy (STD NMR), isothermal titration calorimetry (ITC), and rotavirus infectivity blockade by Neu5Acα2Me and Neu5Gcα2Me, these data show the correlation between virus carbohydrate recognition phenotypes, VP8* sequences and structural characteristics. These studies provide further evidence that VP8* determines the ability of rotaviruses to recognize host cell carbohydrates and reveal that the specificities shown by various animal rotavirus strains at the level of VP8* are translated to those exhibited by the infectious virion.
MATERIALS AND METHODS
Rotaviruses and cells.
The origins, cultivation in MA104 cells, and infectivity assays of porcine rotavirus strain CRW-8 (G3P9[7]), monkey rotavirus RRV (G3P5B[3]), and bovine rotavirus NCDV-Lincoln (G6P6[1]) have been described previously (4, 19, 20, 32). The original CRW-8 isolate contained Pro157 in VP8* (19). We utilized CRW-8 passage 31 (p31) virus with Ile, Gly, and Ser at VP8* positions 99, 150, and 157 in VP8*, respectively, which is identical in sequence to our VP8* expression clone obtained from CRW-8 passage 30 (p30) (37). We also used CRW-8 passage 12 (p12) virus, which was shown by DNA sequencing to contain VP8* forms that differ at amino acid positions 99 (Ile or Thr), 150 (Gly or Glu), and 157 (Pro or Ser), with Pro157 strongly dominating over Ser157. Otherwise, the CRW-8 p12 and p31 VP8* amino acid sequences are identical. CRW-8 isolates obtained from later plaque purification either were identical in sequence to p31 virus or contained the same sequence mixtures at positions 99, 150, and 157 as the p12 virus but had Ser157 predominating over Pro157. As CRW-8 with Pro157 alone could not be isolated, CRW-8 p12 and p31 were used to analyze the effect of the Pro157Ser mutation on glycan receptor recognition.
Generation of VP8* proteins and site-directed mutagenesis of CRW-8 VP8*.
The protein segment comprising amino acids 64 to 224 for RRV VP8*, CRW-8 (passage 30 with serine at position 157) (CRW-8 VP8*_S157) and the NCDV VP8* were expressed and purified as we reported previously (15, 37, 44). To generate the CRW-8 VP8*_P157 protein, the construct encoded Leu64-Leu224 of the CRW-8 VP8* fused to an N-terminal glutathione S-transferase (GST) tag. Following expression and initial purification, the protein was treated with 10 μg/ml trypsin to cleave off the GST tag, generating a form of VP8* that contained a two amino acid N-terminal extension (Gly62 and Ser63) derived from the GST linker. The CRW-8 VP8* Ser157Pro construct was made using a forward primer comprising ACG GGA AAT TAT ACG CAG CAT GGG CCC TTG TTT TCT ACA CCG AAA TTG TAC (Integrated Sciences) and a complementary reverse primer (mutated bases are in bold, and the underlining indicates an extra restriction enzyme digest region for screening). To confirm the success of this point mutation, the mutants possessed an extra digest region for restriction enzyme Bme1580I. Gene integrity was confirmed by standard DNA sequencing. The construct was fused into the pGEX-4T-1 vector in frame downstream of GST. VP8* protein was purified as before (37), and purity was assessed by SDS-PAGE.
Crystallization, X-ray diffraction data collection, and crystallographic structure determination.
Crystals of CRW-8 VP8*_S157 in complex with Neu5Gcα2Me were grown via hanging drop vapor diffusion using protein at a concentration of 20 mg/ml that was premixed with Neu5Gcα2Me (synthesized in house) in a protein-ligand ratio of 1:50, and an equal volume of reservoir solution comprising 0.1 M HEPES (pH 7.5) and 70% 2-methyl-2,4-pentanediol. Crystals grew within 1 to 2 days at 293 K and reached typical dimensions of 0.25 by 0.025 by 0.025 mm. Prior to data collection, the crystal was frozen by immersion in liquid nitrogen (the 70% MPD acting as a cryoprotectant). X-ray diffraction data of the CRW-8 VP8*_S157 Neu5Gcα2Me complex were collected to a resolution of 1.85 Å at the Advanced Light Source (100 K; λ = 1.1159 Å; beam line 8.3.1) with an ADSC charge-coupled device (CCD) detector (distance, 150 mm; 1° oscillation angle; 12-s exposure time).
To generate the structure of RRV VP8* in complex with Neu5Gcα2Me, crystals of the protein were first grown in complex with the N-acetyl-containing analogue, and then these were soaked in a solution that contained Neu5Gcα2Me. The RRV VP8* Neu5Acα2Me complex crystals were produced via hanging-drop vapor diffusion using equal volumes of protein at a concentration of 40 mg/ml premixed with ligand (at a 1:50 ratio) and reservoir solution comprising 1.7 M (NH4)2SO4, 2.4% (vol/vol) polyethylene glycol (PEG) 400 and 0.1 M PIPES, pH 6.5. Crystals were then soaked in the reservoir solution containing 50 mM Neu5Gcα2Me for 18 h. Prior to data collection, the crystals were briefly soaked in reservoir solution supplemented with 20% glycerol (as a cryoprotectant) and 10 mM Neu5Gcα2Me. X-ray diffraction data of the RRV VP8* Neu5Gcα2Me complex were collected (at 100 K and a 0.3° oscillation angle, with an exposure time of 2 min) to a resolution of 2.0 Å on an in-house (Eskitis Institute, Griffith University) Rigaku MicroMax007-HF diffractometer (CuKα λ = 1.5418 Å) equipped with an R-Axis IV++ detector (distance set at 150 mm) and X-Stream cryoequipment.
The X-ray diffraction data sets were integrated using MOSFLM (26) and scaled with SCALA (11) as implemented in the CCP4 suite of crystallographic software (2a). Atomic models were refined using REFMAC (31), and visualization of electron density and model building were done using Coot (10). Electron density is very well defined for ligands and the protein structures, with an exception in one localized region of the RRV Neu5Gcα2Me structure. The carbonyl group of Tyr155 competes with a water molecule for site occupancy, facilitated by rotation about the ψ angle. The electron density is ambiguous for exact positioning of this carbonyl group. However, this does not interfere with the ligand and does not affect our structure-based conclusions reported here. Structure validation was done using SFCHECK and PROCHECK (25). Secondary structure analysis was performed using DSSP (21). LIGPLOT (40) was used to assess hydrogen bonds and hydrophobic properties. Table 2 reports data collection and refinement statistics. Figures were created using PyMOL (6).
TABLE 2.
Crystallographic data and refinement statistics of VP8*64–224 of CRW-8 and RRV in complex with Neu5Gcα2Me
| Parameter | Result for proteinin complex with Neu5Gcα2Me |
|
|---|---|---|
| CRW-8 VP8*_S157 | RRV VP8* | |
| Crystal system, space group | Orthorhombic, P212121 | Tetragonal, P41212 |
| Unit cell parameters (Å) | a = 53.98, b = 59.65, c = 111.96 | a = 48.42, b = 48.42, c = 130.91 |
| Resolution (Å) | 59.65–1.85 (1.95–1.85) | 45.41–2.0 (2.11–2.0) |
| Total no. of observations | 125,888 (16,780) | 70,515 (11,231) |
| Total no. of unique observations | 31,649 (4,531) | 8,759 (1,572) |
| Redundancy | 3.98 (3.70) | 8.05 (7.14) |
| Completeness (%) | 99.9 (99.6) | 99.8 (98.8) |
| I/σ (I) | 5.5 (1.7) | 4.0 (3.4) |
| Rmerge (%)a | 9.7 (35.4) | 10.0 (17.1) |
| Refinement | ||
| Resolution (Å) | 40.03–1.85 (1.90–1.85) | 38.93–2.0 (2.052–2.0) |
| Total no. of unique observations | 29,988 (2152) | 10,639 (641) |
| Rcryst (%) | 18.7 | 18.3 |
| Rfree (%) | 22.5 | 24.9 |
| No. of atoms | ||
| Protein | 2,628 | 1,304 |
| Sialoside ligand | 46 | 23 |
| Water molecules | 363 | 216 |
| Sulfate | 10 | |
| MPD | 32 | |
| HEPES | 15 | |
| Benzamidine | 9 | |
| Na+ | 2 | |
| Average B factors (Å2) | ||
| Protein | 12.2 | 10.6 |
| Sialoside ligand | 12.8 | 10.3 |
| Water molecules | 29.5 | 23.0 |
| Na+ | 11.7 | |
| RMS deviations | ||
| Bond length (Å) | 0.007 | 0.008 |
| Bond angle (°) | 1.05 | 1.25 |
| Ramachandran plot statistics (%) | ||
| Favored | 95.7 | 95.0 |
| Allowed | 4.3 | 4.4 |
| Outliers | 0 | 0.6 |
Isothermal titration calorimetry.
CRW-8 VP8* and ligands were prepared in TNE buffer (20 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA [pH 8.0]). Samples were degassed (ThermoVac; MicroCal Inc.) prior to use. Experiments were conducted with 0.5 mM CRW-8 VP8*_S157 and 30 mM ligand or 0.2 mM CRW-8 VP8*_P157 and 18 mM ligand. Titrations were performed in a VT-ITC microcalorimeter (MicroCal, Inc.) using 22 to 25 injections applied 240 s apart. Replicated titration data and blank data were collected at 4°C, and binding isotherms were fitted by nonlinear regression using a single-site model (Origin software; MicroCal, Inc.). The stoichiometry of the interaction (N), equilibrium constant (Ka) and change in enthalpy (ΔH) were floated during the fitting of the data until the χ2 factor remained constant with values for the blank subtracted. The, equilibrium dissociation constant (Kd) was calculated as a reciprocal of Ka. The Kd values were obtained when n was fixed at 1, since this stoichiometry correlates with the binding of one ligand molecule per VP8* molecule (43). Based on crystallographic structures of VP8* in complex with mono- and also trisaccharides, the assumption of this stoichiometry is appropriate, since one ligand bound per VP8* at a site consistent for both CRW-8 and RRV (9, 24, 43).
Saturation transfer difference nuclear magnetic resonance (STD NMR).
All STD NMR spectra were acquired by using our previously reported protocols (15) on a Bruker 600 MHz Avance spectrometer with a conventional 1H/13C/15N gradient cryoprobe system at 288 K. Deuterium oxide (99.9% deuterium) was purchased from Novachem Pty. Ltd. (Collingwood, Australia). Neu5Acα2Me was synthesized as previously described (23), and Neu5Gcα2Me was prepared in-house. NMR samples were prepared by dissolving VP8*64–224 proteins with ligand, giving a molar ratio of 1:100 in 600 μl, in NMR buffer (20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl). One-dimensional STD NMR was performed on Neu5Acα2Me and Neu5Gcα2Me with CRW-8 VP8*_P157 and NCDV VP8* as described previously for RRV (15).
Specifically, the VP8* was saturated at 7.13 ppm, and off-resonance was set at 33 ppm with a cascade of 40 selective Gaussian-shaped pulses of 50 ms duration. A 100-μs delay between each pulse was applied, giving a total of 2 s saturation time. Data were obtained with an interspersed acquisition of pseudo-two-dimensional on-resonance and off-resonance spectra. On- and off-resonance spectra were processed separately to obtain the final STD NMR spectrum by subtracting individual on- and off-resonance spectra.
Flow cytometry.
Cells from confluent MA104 cell monolayers were placed in suspension by treatment with phosphate-buffered saline (PBS) containing 0.75 mM EDTA (PBS-EDTA) for 10 min, washed in PBS-EDTA containing 0.5% (vol/vol) blocking agent (Sialix), resuspended in PBS containing 0.5% blocking agent, passed through a 40-μm filter, and quantitated in a hemocytometer by trypan blue exclusion. Cells (8 × 105) were stained at 4°C with polyclonal chicken Neu5Gc-specific IgY antibody or matched isotype control antibody (Sialix) at 30 μg/ml, followed by Alexa Fluor 647-conjugated donkey anti-chicken IgY (Jackson Laboratories), and fluorescence was detected in a FACSCalibur analyzer. The level of Neu5Gc antibody bound to cells relative to the negative control was expressed as the relative linear median fluorescence intensity (RLMFI), with an RLMFI value of ≥1.20 being considered positive, as determined previously (13).
Assays for inhibition of rotavirus infectivity.
For all assays, rotavirus infectivity was activated with porcine pancreatic trypsin (Sigma) at 10 μg/ml for 20 min at 37°C. For glycan blockade, Neu5Acα2Me or Neu5Gcα2Me, at a range of concentrations, or Dulbecco's modified Eagle's medium (DMEM) containing 2 mM l-glutamine (Sigma-Aldrich), 20 mM HEPES (Roche), and antibiotics (diluent) was incubated with trypsin-activated rotavirus for 1 h at 37°C. Confluent MA104 cell monolayers containing 4 × 104 cells were inoculated with the virus mixtures at a multiplicity of infection of 0.02. After incubation for 1 h at 37°C and inoculum removal, cells were incubated for a further 15 h prior to acetone fixation and indirect immunofluorescent staining of infected cells, as previously described (17). For antibody blockade, MA104 cells were incubated with antibody or DMEM for 1 h at 37°C prior to virus inoculation. For sialidase experiments, sialidase from Vibrio cholerae (Sigma) was prepared in DMEM, which was brought to pH 5.5 with HCl. MA104 cells were treated with sialidase at 0.52 U/ml or maintained in DMEM (pH 5.5) for 1 h at 37°C before being cooled to 4°C and inoculated with virus at 4°C.
Statistical analysis.
Student's t test was used to assess virus infectivity blockade data. Significance was set at the 95% level. Data given for cell-based assays represent the means for triplicate samples from at least two independent experiments.
Protein structure accession numbers.
The X-ray crystallographic structures of VP8*64–224 of CRW-8 VP8*_S157 and RRV in complex with Neu5Gcα2Me have been deposited in the Protein Data Bank (http://www.pdb.org) with accession codes 3TAY and 3TB0, respectively.
RESULTS AND DISCUSSION
The X-ray crystallographic structures of VP8*64–224 from CRW-8 and RRV in complex with Neu5Gcα2Me.
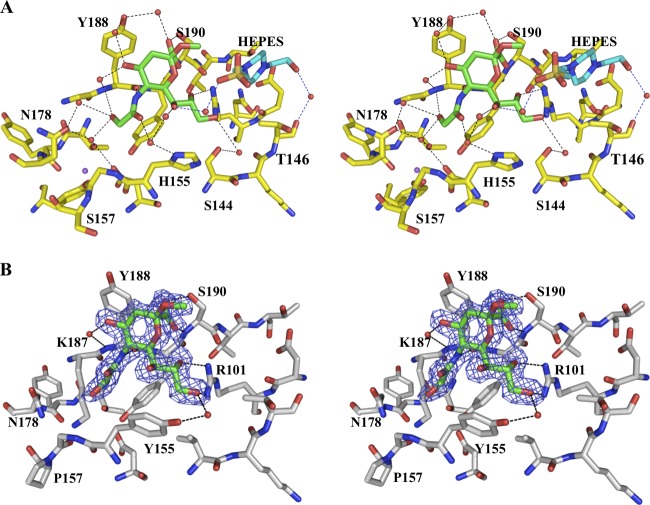
The atomic structures of VP8*64–224 of CRW-8 with S157 and RRV in complex with Neu5Gcα2Me were determined and revealed for each a well-defined ligand bound at the carbohydrate-binding site that is formed at one edge of the anti-parallel β-sandwich (Table 2; Fig. 1). In the CRW-8 VP8*_S157 Neu5Gcα2Me complex, there are two VP8* protein molecules (molecules A and B) in the crystallographic asymmetric unit. Molecule A uniquely has an additional HEPES and benzamidine bound in the vicinity of the carbohydrate-binding region of the VP8* (Fig. 1A). These do not cause major disruptions, and the overall structures of the two VP8* molecules are comparable, with only a minor influence due to the HEPES sulfate on water-mediated hydrogen bonding networks involving the ligand glycerol side chain (in molecule A only) (Fig. 2A). The orientation of the bound Neu5Gcα2Me on each molecule is consistent with that for the N-acetyl-containing Neu5Acα2Me when in complex with CRW-8 VP8* (PDB code 2I2S) (1) and the anticipated direct protein-ligand hydrogen bonds, including engagement with Arg101 and Ser190, are apparent along with several water-mediated bonds that anchor the Neu5Gcα2Me to the protein (Fig. 2A). Importantly, in contrast to NeuAcα2Me, the Neu5Gcα2Me exhibits additional hydrogen bonds, notably a direct contact with the Tyr188 backbone amide and interactions within a water network that are mediated by the ligand's glycolyl hydroxyl group.
Fig 1.
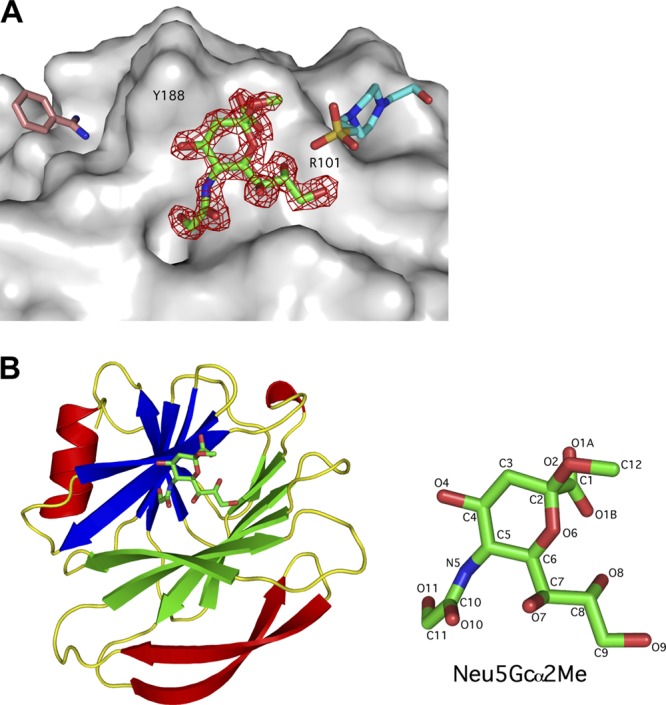
Structure of CRW-8 VP8*_S157 in complex with Neu5Gcα2Me. (A) Connolly surface (gray) representation of the sialoside binding region and adjacent areas of the CRW-8 VP8*_S157 complex (molecule A, which is unique in that it has small molecules bound as well as the N-acylneuraminic acid derivative, is depicted). The Neu5Gcα2Me, benzamidine, and HEPES [N-(2-hydroxyethyl) piperazine-N′-(2-ethane-sulfonic acid)] molecules are depicted as stick representations, with oxygen (red), nitrogen (blue), and sulfur (yellow) shown and the remaining atoms being carbons. Difference electron density (|Fo| − |Fc| αcalc; red, 3σ level), calculated prior to inclusion of the Neu5Gcα2Me in the model refinement, is depicted. The HEPES sulfate group binds proximal to the methyl aglycon group of Neu5Gcα2Me of the monosaccharide derivative, and the remainder extends along the cavity. (B) The compactly folded globular domain exhibited by VP8*64–224 comprises two β-sheets that form a 12-stranded anti-parallel β-sandwich (molecule A of the CRW-8 VP8*_S157 complex is depicted as representative). Secondary structure analysis assigned Tyr194-Asp195-Thr196 as a 310 helix. The C-terminal α-helix packs against one β-sheet, and two anti-parallel β-ribbons pack against the other. The molecular structure with relevant atom numbers of Neu5Gcα2Me is depicted on the right, with the distinguishing N-glycolyl hydroxyl oxygen being O11.
Fig 2.
(A) Stereo diagram of the sialoside binding site region of CRW-8 VP8*_S157 in complex with Neu5Gcα2Me. The stereo diagram shows the direct and water-mediated protein-ligand interactions of molecule A of the CRW-8 VP8*_S157 complex. In addition to the five direct hydrogen bond interactions that are identical to those in the Neu5Acα2Me-bound 2I2S structure (1), an additional hydrogen bond is formed between the glycolyl O11 and backbone amide of Tyr188. A HEPES molecule (present in molecule A only) is located adjacent to the methyl aglycon group of Neu5Gcα2Me that contacts the glycerol side chain O7 and Thr146 carbonyl via water bridges. The O10 and the carboxylate group are water-bridged to His155 Nδ1 and the hydroxyl group of Tyr 188, respectively, and the O7 is linked to His155 Nδ1 via a hydrogen bond network engaging two water molecules. The glycolyl group O11 is involved in a water network mediated by the apical water molecule. Hydrogen bonds are represented as dashed lines, water as a red sphere, and Na+ as a purple sphere. Direct hydrogen bond network (not illustrated) is identical between molecule A and B of the CRW-8 VP8*_S157 complex. (B) Stereo diagram of the sialoside binding site region of RRV VP8*64–224 in complex with Neu5Gcα2Me. The 2|Fo| − |Fc| αcalc map (blue mesh) contoured at 1.0 σ is shown for the ligand (the density for the protein is not depicted, in order to facilitate visibility of the ligand density). The N-glycolyl group of the bound Neu5Gcα2Me of the RRV VP8*64–224 complex does not partake in any direct interaction, with VP8* being instead directed outward from the binding pocket. Direct hydrogen bond interactions are identical to those in the Neu5Acα2Me-bound RRV VP8* structure (9, 23). Lys187 extends into the cavity that normally accommodates the Na+ and the apical water, displaces the water, and physically hinders the N-glycolyl hydroxyl group from being directed into the pocket.
In the CRW-8 VP8*_S157 complex, there is a water molecule that hydrogen bonds to the N-glycolyl group of the N-glycolylneuraminic acid derivative, and this “apical” water forms part of the octahedral coordination sphere of a Na+ ion (Fig. 2A). The biological role of this ion in rotavirus infection is unknown, but it is consistently present in our CRW-8 and also RRV VP8* structures. In molecule B, this water molecule is only partially present, because the side chain of Asn178 adopts two alternative conformations, and one projects toward the N-glycolyl group coordinating the Na+ (2.3 Å) and deposing the water molecule. Thus, when present, this apical water molecule can mediate hydrogen bonding, which facilitates the anchoring of the glycolyl group. In the alternative case, the Asn178 side chain is able to hydrogen bond to the glycolyl hydroxyl group, albeit very weakly (3.5 Å). Either way it is clear that there are beneficial interactions with the glycolyl group.
In stark contrast to the CRW-8 VP8* Neu5Gcα2Me complex, the N-glycolyl group of the bound Neu5Gcα2Me in the RRV VP8* complex crystal structure indicates a direction outward from the binding cleft and consequently is incapable of engaging in a direct interaction with the VP8* protein (Fig. 2B). It is anticipated that for CRW-8 VP8*, the combination of direct and water-mediated interactions made with the Neu5Gcα2Me glycolyl group dictate this porcine virus preference for N-glycolylneuraminic acid-containing glycans.
Characterization of the neuraminic acid binding preference of animal rotavirus VP8* and the role of the amino acid at position 187.
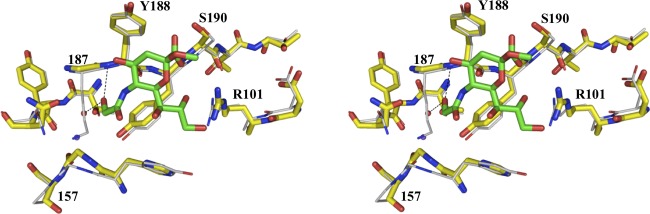
Although the VP8* subunits of CRW-8 and RRV demonstrate different preferences for Neu5Acα2Me and Neu5Gcα2Me, their binding affinities for N-acetylneuraminic acid are comparable. The CRW-8 VP8*_P157 exhibits a Kd value (determined by ITC) with Neu5Acα2Me of 0.27 ± 0.03 mM, and as we previously reported, RRV VP8* has a Kd of 0.30 mM (24). CRW-8 VP8*_P157 had a Kd for Neu5Gcα2Me of 0.068 ± 0.011 mM, and the values for CRW-8 VP8*_S157 were 1.35 ± 0.25 and 0.42 ± 0.095 mM for Neu5Acα2Me and Neu5Gcα2Me binding, respectively. It is clear that the N-glycolyl group enables interactions that would be impossible with the alternative N-acetyl group of Neu5Acα2Me. The crystal structure of RRV VP8* reveals a bound Neu5Gcα2Me molecule. However, since RRV VP8* interaction with this N-glycolyl ligand was not measurable by ITC (data not shown), RRV VP8* binding to this ligand is weak. The measurable Kd in ITC for RRV VP8* interaction with the N-acetyl ligand demonstrates the preference for, and stronger binding of, RRV VP8* to the N-acetylneuraminic acid. Based on our structural results, the preference for Neu5Gc-containing glycans by CRW-8 VP8* can be explained to a large extent by the detected interactions with the N-glycolyl hydroxyl group. The favorable interactions revealed in the CRW-8 VP8* Neu5Gcα2Me complex partially account for the distinct carbohydrate preferences of the two animal strains but do not explain why the same manner of engagement does not occur in RRV VP8*. Critical insight is given by our crystal structure of RRV VP8* in complex with Neu5Gcα2Me, which reveals that the direction of the N-glycolyl hydroxyl group could be influenced by potential for physical interference by the side chain of lysine 187, which additionally dislodges the apical water. Such steric interference would occur even if the Lys187 side chain had sufficient flexibility to move away from the ligand since it is the fixed Cβ atom of the Lys187 that is most proximal, 2.1 Å, to where the hydroxyl group of the N-glycolyl would lie if it emulated the positioning in the CRW-8 VP8* complex structure. Thus, in RRV the resulting outward projection of the hydroxyl group provides a rationale for the lower binding affinity of RRV VP8* for N-glycolylneuraminic acids (Fig. 3). The equivalent site in CRW-8 VP8* has a glycine which would not hinder the functional group from interaction with the protein. These results are of broad significance, since the rotaviruses OSU, CRW-8, and NCDV, which show a preference for N-glycolylneuraminic acid, have a glycine at the VP8* position 187, while RRV and the bovine UK virus, which favor the N-acetylneuraminic acid, have a lysine at this position (Table 1). Thus, there is a clear correlation among these animal rotaviruses, with the small amino acid glycine facilitating a preference for N-glycolylneuraminic acid, as the hydroxyl moiety can be accommodated within the cavity. Our data show the effect of Lys187 in conveying the RRV specificity for Neu5Acα2Me over Neu5Gcα2Me, which is anticipated to translate to RRV recognition of Neu5Ac- and Neu5Gc-containing glycans. It is likely that the majority of sialidase-sensitive animal rotaviruses studied to date will show a preference for N-glycolylneuraminic acid, based on the presence of glycine at position 187 in their VP8* (Table 1).
Fig 3.
The sialoside binding site of superimposed crystallographic structures of CRW-8 VP8*_S157 (carbons are in yellow) in complex with Neu5Gcα2Me (green) and rhesus RRV VP8* (carbons are in gray). The glycolyl group O11 of Neu5Gcα2Me forms two hydrogen bonds, one to the Tyr188 backbone amide and the other to the apical water (red sphere) that coordinates the sodium ion (purple). In RRV VP8*64–224, the Gly156-Pro157 region can exhibit either a cis or trans configuration. In contrast, the Gly156-Ser157 region of this Ser157 mutant of CRW-8 VP8*64–224 exhibits only the trans configuration.
Role of the VP8* amino acid at position 157 in carbohydrate binding affinity and specificity.
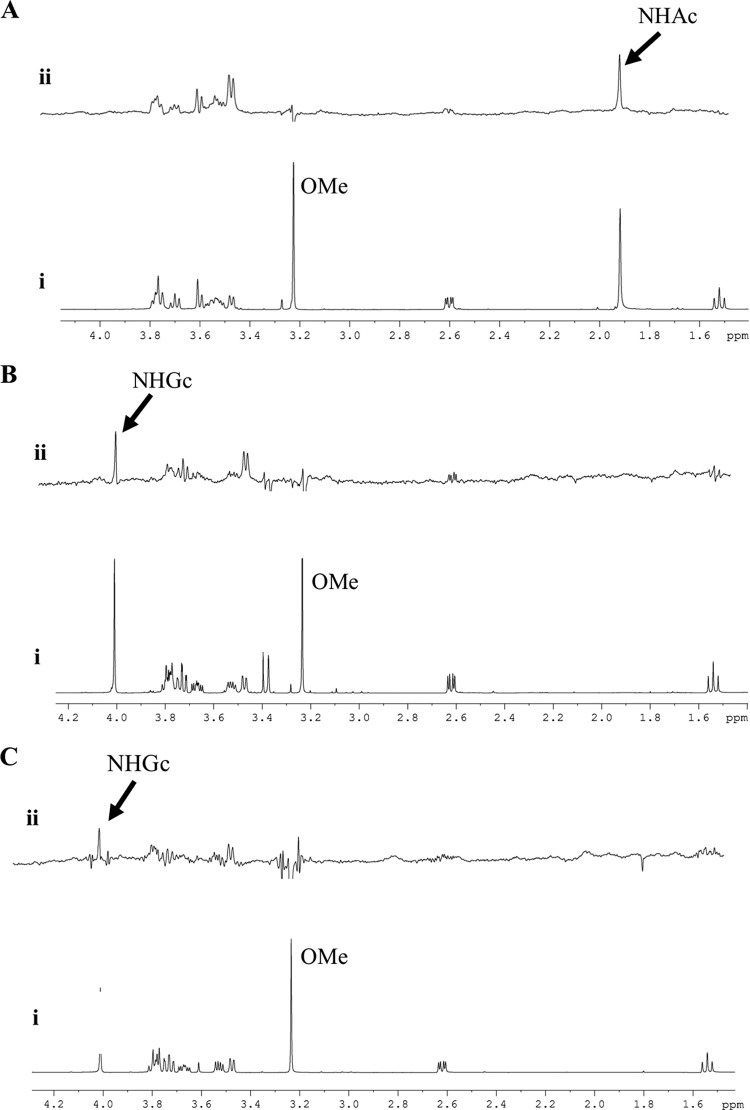
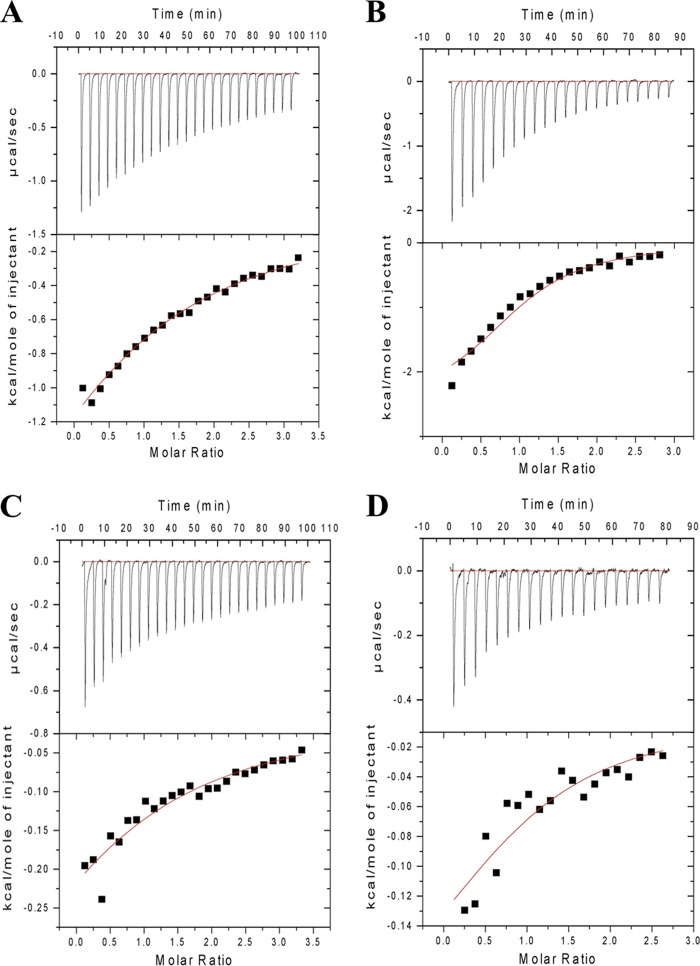
The nature of other amino acids in the region around the N-acyl portion of the ligand could also influence specificity and/or affinity toward carbohydrates. The proline in the VP8* binding site at amino acid position 157 of early-passage CRW-8 and RRV VP8* lies adjacent to the ligand N-acetyl or N-glycolyl group (Fig. 2). Proline is the predominant amino acid occurring at this site in sialidase-sensitive animal rotaviruses, with serine being the main alternative (Table 1). The crystal structure of CRW-8 VP8* (passage 30) that is presented with the bound monosaccharide has serine at position 157 (Fig. 2A). In this CRW-8 VP8*_S157 structure, Gly156-Ser157 is fixed in the trans conformation, with the carbonyl of Gly156 coordinating the Na+. A proline at position 157 in CRW-8 VP8* would permit flexibility, due to cis-trans isomerization, in the protein chain, which in turn would enable alteration of the direction of the Gly156 carbonyl group, making it then unable to coordinate the ion. In crystal structures of RRV VP8*, such flexibility is seen, the trans conformer being dominant but with evidence of the occurrence of the cis isomer (9, 24). Site-directed mutagenesis (proline replacing serine) was done to investigate the effect of such potential conformational change associated with the VP8* 156-157 segment on the local environment of the carbohydrate binding cleft and ascertain whether the sequence variation influences the carbohydrate specificity and/or binding affinity of Neu5Acα2Me and Neu5Gcα2Me. STD NMR and ITC were employed to assess the interaction of CRW-8 VP8*_P157 and CRW-8 VP8*_S157 with the N-acylneuraminic acid derivatives. It is clear that this mutation did not prevent the binding of Neu5Acα2Me or Neu5Gcα2Me to either CRW-8 VP8*64–224 sequence, and these ligands exhibited the same ligand binding features, with strong STD effects for the N-acetyl (Fig. 4A) and the N-glycolyl (Fig. 4B) forms. ITC conclusively showed that the CRW-8 VP8*_P157 has a greater affinity (∼5-fold higher) than the serine mutant for both Neu5Gcα2Me and Neu5Acα2Me and also that both VP8* subunits show at least a 3-fold-higher affinity for the N-glycolyl over the N-acetyl derivative (Fig. 5; also, see the Kd values above). It is clear that the amino acid sequence character and adaptability of the binding cleft have an inherent influence on interaction with carbohydrates. Our structure-based hypothesis that the specific amino acid at VP8* position 187 could influence specificity toward N-acylneuraminic acids is described above. In contrast, the character of either serine or proline at position 157 is not critical in determining the carbohydrate specificity of CRW-8 VP8*, as the preference for the N-glycolyl form is retained. However, the amino acid type does affect the ability of the VP8* to bind the ligand, since a proline in CRW-8 VP8* facilitates significantly stronger binding than a serine to both of the N-acylneuraminic acid containing glycans. Our results demonstrate unambiguously that the presence of serine compared to proline at position 157 affects VP8* binding affinities but not the carbohydrate specificity.
Fig 4.
STD NMR of VP8* with N-acetyl and N-glycolylneuraminic acids. (A) STD NMR analysis of CRW-8 VP8*_P157 with Neu5Acα2Me. (i) 1H NMR spectrum of Neu5Acα2Me; (ii) STD NMR spectrum of Neu5Acα2Me in the presence of VP8* at a protein-ligand molar ratio of 1:100. The methyl group of the NHAc moiety gives an STD signal at ∼1.9 ppm. (B) STD NMR analysis of CRW-8 VP8*_P157 with Neu5Gcα2Me. (i) 1H NMR spectrum of Neu5Gcα2Me; (ii) STD NMR spectrum of Neu5Gcα2Me in the presence of VP8* at a protein-ligand molar ratio of 1:100. The methylene group of the NHGc gives a signal at ∼4.0 ppm. (C) STD NMR analysis of NCDV VP8*64–224 with Neu5Gcα2Me. (i) 1H NMR spectrum of Neu5Gcα2Me; (ii) STD NMR spectrum of Neu5Gcα2Me in the presence of VP8* at a protein-ligand molar ratio of 1:100. The methylene group of the NHGc gives a signal at ∼4.0 ppm.
Fig 5.
ITC analysis of CRW-8 VP8*64–224 binding to Neu5Acα2Me or Neu5Gcα2Me. (A) ITC analysis of CRW-8 VP8*_P157 with Neu5Acα2Me. (B) CRW-8 VP8*_P157 with Neu5Gcα2Me. (C) CRW-8 VP8*_S157 with Neu5Acα2Me. (D) CRW-8 VP8*_S157 with Neu5Gcα2Me. The average Kd value for each combination was calculated with n set to 1 (see text for specific values).
Recognition of N-acetyl- and N-glycolylneuraminic acids by porcine and monkey rotaviruses.
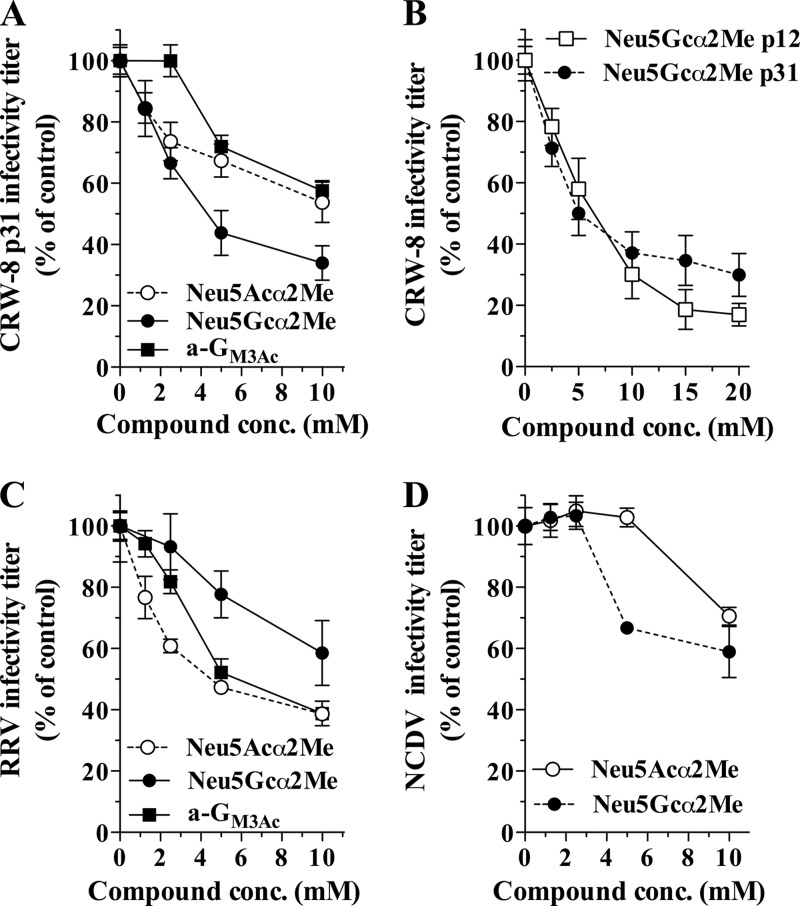
The N-glycolyl-containing the a-GM3Gc glycan is two to three times more effective than a-GM3Ac in the blockade of porcine rotavirus OSU binding to host cells (35). OSU and CRW-8 share VP8* serotype and genotype specificity. To examine the role of VP8* in receptor recognition by infectious rotaviruses and assess specificity differences for different carbohydrates, the ability of anti-Neu5Gc antibody, Neu5Gcα2Me, and Neu5Acα2Me to inhibit CRW-8 and RRV infection was determined. These experiments were conducted in permissive MA104 cells, which were shown by staining with anti-Neu5Gc antibody and flow cytometry to express high levels of Neu5Gc, with an RLMFI of 33.9. This antibody inhibited the infectivity of CRW-8 p31 and RRV by 22% ± 5% and 6.9% ± 3.1% (means ± standard errors of the means [SEM]), respectively, compared with the matched isotype control antibody or medium alone (P ≤ 0.003 and P = 0.045, respectively). This blockade by antibody was substantially less efficient than that seen with the corresponding glycan. CRW-8 p31 infectivity was reduced in a dose-dependent fashion, by 65% with 10 mM Neu5Gcα2Me (Fig. 6A and B) and 40% with 10 mM Neu5Acα2Me (Fig. 6A). In contrast, RRV infection was reduced by maxima of 40% with Neu5Gcα2Me and 60% with Neu5Acα2Me (Fig. 6C). At a 2.5 mM concentration, Neu5Gcα2Me inhibited CRW-8 p31 infection to a 1.3-fold-greater extent than Neu5Acα2Me, whereas RRV infection was reduced to a 5.8-fold-greater extent by Neu5Acα2Me than by Neu5Gcα2Me. These results demonstrate the importance for RRV infectivity of cellular N-acetylneuraminic acids and of N-glycolyl forms as CRW-8 p31 receptors. The limited ability of antibody to Neu5Gc to inhibit CRW-8 and RRV infection is likely to result from the difficulty in blocking virus access to cell surface Neu5Gc, and/or the variation in antigen specificity and affinity in this polyclonal antibody preparation. Antibody to GM3 glycan is similarly less effective than the GM3 glycan itself in inhibiting RRV infection (43).
Fig 6.
Inhibition of rotavirus infection by Neu5Acα2Me and Neu5Gcα2Me. (A) CRW8 p31 infectivity blockade by Neu5Acα2Me and Neu5Gcα2Me at 1.25 mM to 10.0 mM. (B) CRW-8 p12 and CRW-8 p31 blockade by Neu5Gcα2Me at 2.5 mM to 20.0 mM. (C and D) RRV (C) and NCDV (D) blockades with Neu5Acα2Me or Neu5Gcα2Me at 1.25 mM to 10.0 mM. Data obtained for a-GM3Ac blockade of CRW-8 and RRV infection that we reported recently (43) are included in panels A and C, as all these experiments were performed contemporaneously under the same conditions with the same rotavirus inocula. The infectivity titer of rotavirus treated with ligand is expressed as the mean percentage of the titer obtained in the absence of ligand (control). Error bars show standard deviations.
Infectivity blockade by Neu5Gcα2Me and Neu5Acα2Me was compared with the blockade by a-GM3Ac that we recently reported (43). Interestingly, infectivity blockade by the N-acetyl-containing monosaccharide is comparable to that of a-GM3Ac at 5 mM and 10 mM (Fig. 6A and C). This indicates that the N-acetylneuraminic acid moiety is the main interaction component of the trisaccharide, a result that is supported by our elucidation of the structure of CRW-8 p30 VP8* in complex with the N-acetyl-containing glycan of the aceramido-GM3 (a-GM3Ac) (43). This result also correlates with our finding that a-GM3Gc more effectively inhibits CRW-8 p31 than RRV infection and that a-GM3Ac exhibits the converse pattern (43).
Role of the amino acid residue at position 157 in recognition of N-glycolyl- and N-acetylneuraminic acids by CRW-8 rotavirus.
The effect of the Pro157Ser mutation in CRW-8 VP8* on virus recognition of glycan cell receptors was examined using CRW-8 p12 (predominantly P157) and CRW-8 p31 (S157). Sialidase treatment of MA104 cells reduced the infectivity of these viruses by 68 ± 4.7% and 81 ± 5.0% (means ± SEM), respectively. Although CRW-8 p12 was significantly less sialidase sensitive than CRW-8 p31 (P < 0.0001), both viruses showed a high degree of sialidase sensitivity. The ability of Neu5Gcα2Me and Neu5Acα2Me to inhibit the infectivity of the two viruses was analyzed in parallel within the same experiments. At concentrations of 1.25 mM to 10 mM, Neu5Gcα2Me inhibited CRW-8 p12 and p31 to a similar extent (P > 0.05) (Fig. 6B). At 15 mM and 20 mM Neu5Gcα2Me, CRW-8 p12 infectivity was reduced by 81% ± 7% and 83% ± 2% (means ± SEM), respectively, whereas CRW-8 p31 infectivity was reduced by 65% ± 8% and 70% ± 3%, respectively (Fig. 6B). The differences in Neu5Gcα2Me blockade between these viruses were significant at both 15 mM and 20 mM (P = 0.0037 and P = 0.0024, respectively). Thus, Neu5Gcα2Me inhibits the infectivity of CRW-8 p12 to a greater extent than CRW-8 p31, and CRW-8 with predominantly Pro157 shows greater Neu5Gcα2Me inhibition than CRW-8 with Ser157. This demonstrates the likely biological relevance of the enhanced affinity of CRW-8 VP8* Pro157 over CRW-8 Ser157 for Neu5Gcα2Me, as measured by ITC. Tighter Neu5Gcα2Me binding involving Pro157 in CRW-8 virus is the most likely explanation for the more effective Neu5Gcα2Me infectivity blockade. The sequence variation at positions 99 and 150 between CRW-8 p12 and p31 is unlikely to influence this property, as they are not in the immediate vicinity of the ligand when it is bound to VP8*. At concentrations of 1.25 mM to 20 mM, Neu5Acα2Me inhibited CRW-8 p12 and p31 to a similar extent (P > 0.05; data not shown). The lack of effect of the Pro versus Ser at VP8* position 157 on infectivity blockade by Neu5Acα2Me may relate to the inefficiency of the Neu5Acα2Me infectivity blockade relative to that of the Neu5Gcα2Me blockade (Fig. 6A). It is notable in this context that both the Pro157 and Ser157 CRW-8 VP8* proteins show a 3- to 4-fold-lower affinity for Neu5Acα2Me than Neu5Gcα2Me (as measured by ITC; see above).
This novel finding may have wider implications. Cell culture passaging which led to the proline-to-serine mutation in CRW-8 VP8* at site 157 (37), produced the same mutation in bovine NCDV (33). NCDV has been reported to bind the Neu5Gc-containing GM3 glycan but not the Neu5Ac-containing GM3 glycan (7). We have demonstrated here for CRW-8 that a mutation to serine at position 157 is associated with a weakened VP8* and viral ability to bind carbohydrates, particularly Neu5Gcα2Me. The predominance of proline at position 157 in low-passage-number, culture-adapted animal rotaviruses (Table 1) suggests that this property might provide a selective advantage. These other viruses with Pro157 also might exhibit stronger VP8* binding to Neu5Gcα2Me and other carbohydrates. It is possible that this enhanced receptor binding would facilitate efficient infection, particularly in the fluid intestinal environment. In contrast, higher binding affinity for N-acylneuraminic acids may not be advantageous for rotavirus replication in cell culture, perhaps due to a greater expression and/or accessibility of Neu5Gc on cultured cells, or more efficient cell-to-cell spread in cultures due to reduced retention of exiting virions on the cell membrane.
Bovine rotavirus NCDV preference for Neu5Gc receptors correlates with its VP8* preference.
Several other animal rotaviruses, such as NCDV, show preferential binding specificity toward Neu5Gc-containing glycoconjugates (Table 1). We examined the abilities of Neu5Gcα2Me and Neu5Acα2Me to inhibit NCDV infection (Fig. 6D). Neu5Gcα2Me produced 33% and 41% infectivity blockades at 5 mM and 10 mM concentrations, respectively. In contrast, a Neu5Acα2Me blockade was evident at 10 mM only. Our STD NMR analysis of bovine NCDV VP8* in the presence of Neu5Gcα2Me (Fig. 4C) provides the first demonstration of a direct interaction by NCDV VP8* with a N-acylneuraminic acid. Significantly, while NCDV VP8* bound Neu5Gcα2Me with close contact to the compound's methylene hydroxyl group, an interaction with the Neu5Acα2Me counterpart was not detected. NCDV VP8* either interacted only with the N-glycolyl derivative or bound both but with a much weaker interaction with the N-acetyl-containing ligand, such that it was below the limit of detection by this technique. Either way, this clear preference for the N-glycolyl-containing monosaccharide is in accord with our infectivity blockade data for NCDV (Fig. 6D).
Structure-function based hypothesis accounts for the N-acetyl- or N-glycolylneuraminic acid preference of CRW-8, RRV, and NCDV.
The preferences for either N-acetyl- or N-glycolylneuraminic acids shown by RRV, CRW-8, and NCDV can be accounted for by the structure-based hypothesis presented here. We propose that this explanation could apply also to the behavior of other sialidase-sensitive animal rotavirus strains. It is clear that VP8* has a critical role in rotavirus cell recognition, with our data showing the importance for RRV infectivity of cellular N-acetylneuraminic acids and of N-glycolyl forms as CRW-8 receptors (Fig. 6). These findings provide direct evidence that specificities exhibited by various animal rotavirus strains at the level of VP8* are translated to those exhibited by the whole virus. The striking correlation between the specificities of the VP8* protein and infectious rotavirus for these two different N-acylneuraminic acid derivatives has importance with respect to interaction with gangliosides, supporting the results of our recent studies that found that almost the entire interactions between VP8* from CRW-8 and RRV and the biologically important receptors Neu5Ac- and Neu5Gc-containing a-GM3 are contributed by the N-acylneuraminic acid (16, 43). Further, we have demonstrated that proline at position 157 confers a higher CRW-8 VP8* binding affinity for N-acylneuraminic acids, and this is of particular interest, as proline is commonly found at this position in animal rotaviruses. This leads to the intriguing concept that the presence of proline at this site confers a selective advantage in rotavirus interaction with host-cell carbohydrates.
ACKNOWLEDGMENTS
We thank Alex Szyczew and Paul Madge for providing Neu5Acα2Me and Neu5Gcα2Me and Istvan Toth and Daniel Coles (University of Queensland) for access and advice on the microcalorimeter. The CRW-8 isolates were a gift from the late Ian H. Holmes.
H.B. acknowledges financial support for access to synchrotron facilities under the Access to Major Research Facilities Program within the International Science Linkages Program of the Australian Government. We thank staff at beam line 8.3.1 at the Advanced Light Source (ALS) for assistance in data collection. Beam line 8.3.1 at the Lawrence Berkeley National Laboratory, ALS was funded by the NSF, the University of California and Henry Wheeler. This work was supported by the Australian Research Council (ARC; DP0774383) and the National Health and Medical Research Council of Australia (NHMRC; ID597439). B.S.C. is a Senior Research Fellow (ID 628319) of the NHMRC. M.V.I thanks the ARC for the award of an Australian Federation Fellowship.
Footnotes
Published ahead of print 3 October 2012
REFERENCES
- 1. Blanchard H, Yu X, Coulson BS, von Itzstein M. 2007. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*). J. Mol. Biol. 367:1215–1226 [DOI] [PubMed] [Google Scholar]
- 2. Byres E, et al. 2008. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456:648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a. CCP4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50:760–763 [DOI] [PubMed] [Google Scholar]
- 3. Ciarlet M, et al. 2002. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coulson BS. 1993. Typing of human rotavirus VP4 by an enzyme immunoassay using monoclonal antibodies. J. Clin. Microbiol. 31:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crawford SE, et al. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeLano WL. 2002. The PyMOL molecular graphics system. Schrödinger, LLC, Palo Alto, CA [Google Scholar]
- 7. Delorme C, et al. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dormitzer PR, et al. 2002. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J. Virol. 76:10512–10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126. [DOI] [PubMed] [Google Scholar]
- 11. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 [DOI] [PubMed] [Google Scholar]
- 12. Fiore L, Greenberg HB, Mackow ER. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553–563 [DOI] [PubMed] [Google Scholar]
- 13. Graham KL, Takada Y, Coulson BS. 2006. Rotavirus spike protein VP5* binds α2β1 integrin on the cell surface and competes with virus for cell binding and infectivity. J. Gen. Virol. 87:1275–1283 [DOI] [PubMed] [Google Scholar]
- 14. Greenberg HB, Estes MK. 2009. Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136:1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haselhorst T, et al. 2007. STD NMR spectroscopy and molecular modeling investigation of the binding of N-acetylneuraminic acid derivatives to rhesus rotavirus VP8* core. Glycobiology 17:68–81 [DOI] [PubMed] [Google Scholar]
- 16. Haselhorst T, et al. 2011. Recognition of the GM3 ganglioside glycan by Rhesus rotavirus particles. Angew. Chem. Int. ed Engl. 50:1055–1058 [DOI] [PubMed] [Google Scholar]
- 17. Haselhorst T, et al. 2009. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 5:91–93 [DOI] [PubMed] [Google Scholar]
- 18. Hu L, et al. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang JA, Nagesha HS, Holmes IH. 1993. Comparative sequence analysis of VP4s from five Australian porcine rotaviruses: implication of an apparent new P type. Virology 196:319–327 [DOI] [PubMed] [Google Scholar]
- 20. Jolly CL, Beisner BM, Ozser E, Holmes IH. 2001. Non-lytic extraction and characterisation of receptors for multiple strains of rotavirus. Arch. Virol. 146:1307–1323 [DOI] [PubMed] [Google Scholar]
- 21. Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 22. Kelm S, Schauer R. 1997. Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 175:137–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kononov LO, et al. 1998. Synthesis of oligosaccharides related to HNK-1 antigen. 2. Synthesis of 3′′′-O-(3-O-sulfo-beta-d-glucuronopyranosyl)-lacto-N-neotetraose beta-propylglycoside. Bioorg. Khim. 24:608–622 (In Russian.) [PubMed] [Google Scholar]
- 24. Kraschnefski MJ, et al. 2009. Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein. Glycobiology 19:194–200 [DOI] [PubMed] [Google Scholar]
- 25. Laskowski RA. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 26. Leslie AGW. 2006. The integration of macromolecular diffraction data. Acta Crystallogr. D62:48–57 [DOI] [PubMed] [Google Scholar]
- 27. Matthijnssens J, et al. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 80:3801–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matthijnssens J, et al. 2006. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 44:1801–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendez E, Arias CF, Lopez S. 1993. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 67:5253–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Midgley SE, et al. 2012. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet. Microbiol. 156:238–245 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G, Vagin A, Dodson E. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240–255 [DOI] [PubMed] [Google Scholar]
- 32. Nagesha HS, Holmes IH. 1988. New porcine rotavirus serotype antigenically related to human rotavirus serotype 3. J. Clin. Microbiol. 26:171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishikawa K, et al. 1988. Comparative analysis of the VP3 gene of divergent strains of the rotaviruses simian SA11 and bovine Nebraska calf diarrhea virus. J. Virol. 62:4022–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olofsson S, Bergstrom T. 2005. Glycoconjugate glycans as viral receptors. Ann. Med. 37:154–172 [DOI] [PubMed] [Google Scholar]
- 35. Rolsma MD, Kuhlenschmidt TB, Gelberg HB, Kuhlenschmidt MS. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruggeri FM, Greenberg HB. 1991. Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. J. Virol. 65:2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scott SA, et al. 2005. Crystallization and preliminary X-ray diffraction analysis of the sialic acid-binding domain (VP8*) of porcine rotavirus strain CRW-8. Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 61:617–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Small C, Barro M, Brown TL, Patton JT. 2007. Genome heterogeneity of SA11 rotavirus due to reassortment with “O” agent. Virology 359:415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tangvoranuntakul P, et al. 2003. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A. 100:12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wallace AC, Laskowski RA, Thornton JM. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134 [DOI] [PubMed] [Google Scholar]
- 41. WHO 2008. Rotavirus surveillance—worldwide, 2001–2008. MMWR Morb. Mortal. Wkly. Rep. 57:1255–1257 [PubMed] [Google Scholar]
- 42. Yolken RH, Willoughby R, Wee SB, Miskuff R, Vonderfecht S. 1987. Sialic acid glycoproteins inhibit in vitro and in vivo replication of rotaviruses. J. Clin. Invest. 79:148–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu X, et al. 2011. Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J. Mol. Biol. 413:929–939 [DOI] [PubMed] [Google Scholar]
- 44. Yu X, et al. 2008. Crystallization and preliminary X-ray diffraction analysis of the carbohydrate-recognizing domain (VP8*) of bovine rotavirus strain NCDV. Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 64:509–511 [DOI] [PMC free article] [PubMed] [Google Scholar]