Abstract
Certain immune-driven mutations in HIV-1, such as those arising in p24Gag, decrease viral replicative capacity. However, the intersubtype differences in the replicative consequences of such mutations have not been explored. In HIV-1 subtype B, the p24Gag M250I mutation is a rare variant (0.6%) that is enriched among elite controllers (7.2%) (P = 0.0005) and appears to be a rare escape variant selected by HLA-B58 supertype alleles (P < 0.01). In contrast, in subtype C, it is a relatively common minor polymorphic variant (10 to 15%) whose appearance is not associated with a particular HLA allele. Using site-directed mutant viruses, we demonstrate that M250I reduces in vitro viral replicative capacity in both subtype B and subtype C sequences. However, whereas in subtype C downstream compensatory mutations at p24Gag codons 252 and 260 reduce the adverse effects of M250I, fitness costs in subtype B appear difficult to restore. Indeed, patient-derived subtype B sequences harboring M250I exhibited in vitro replicative defects, while those from subtype C did not. The structural implications of M250I were predicted by protein modeling to be greater in subtype B versus C, providing a potential explanation for its lower frequency and enhanced replicative defects in subtype B. In addition to accounting for genetic differences between HIV-1 subtypes, the design of cytotoxic-T-lymphocyte-based vaccines may need to account for differential effects of host-driven viral evolution on viral fitness.
INTRODUCTION
Human leukocyte antigen (HLA) class I-restricted cytotoxic-T-lymphocyte (CTL) escape mutations that occur in conserved HIV-1 regions can compromise the structural or functional integrity of key viral proteins, affecting HIV-1 replication capacity. For example, mutations associated with escape from HLA-B*57/B*58:01-restricted responses to the TW10 epitope in p24Gag affect cyclophilin A binding ability (8), an important requirement for viral replication (24, 37, 41). Vaccines designed to focus immune responses against regions of HIV-1 susceptible to deleterious mutations may therefore represent a useful strategy to control viral replication in vivo (2, 3, 18).
To date, studies of immune-driven alterations in viral fitness have been undertaken predominantly on HIV-1 subtype B (7, 8, 11, 28, 32, 38, 39), the most common subtype in Western countries. Comparably fewer studies have examined subtype C (6, 21, 22, 48–50), despite the fact that it accounts for nearly 50% of infections globally (13, 44). Furthermore, no comparative studies of the replicative consequences of potential immune-driven mutations in subtypes B versus C have been reported, despite the relevance of this issue to the design of universal vaccines aimed at attenuating the disease course (18). Indeed, comparative studies of HIV-1 drug resistance and coreceptor tropism support critical differences between HIV-1 subtype B and C with respect to in vitro replicative capacity that have important implications for HIV treatment and clinical monitoring (1, 20, 40, 51), suggesting that similar intersubtype effects may be observed for immune-driven viral polymorphisms.
The M250I mutation, located one residue downstream of the TW10 epitope in p24Gag that is presented by numerous alleles of the B58 supertype including B*57 and B*58:01 (52), is one of a number of rare variants observed in HLA-B*57/B*58:01-expressing, subtype B-infected elite controllers, whose selection may contribute to viral attenuation and immune control in these individuals (33). In contrast, M250I appears to be a relatively common minor polymorphism in HIV-1 subtype C (35, 36, 48), whose relative fitness consequences were previously unknown. Moreover, M250I has been associated with lack of development of the typical T242N escape mutation in HLA-B*57/B*58:01-positive individuals in some (16, 28, 33) but not all (12, 14, 35) subtype B and C cohorts, supporting its potential role as a polymorphism that could serve to channel immune-driven HIV-1 evolution down unconventional pathways (2, 3, 18).
In the present study, we explore the relative consequences of M250I, alone and in combination with other immune-driven and putative compensatory mutations, in attenuating viral replication in HIV-1 subtypes B and C. We also investigate the structural basis for these defects. We confirm that M250I is very rare in HIV-1 subtype B, and that it is associated with substantial fitness costs that are not easily compensated due to profound alterations in the subtype B capsid structure. In contrast, M250I is more common in subtype C, and its fitness impact may be rescued by the selection of compensatory mutations. We also demonstrate additive fitness consequences of T242N in combination with M250I in HIV-1 subtypes B and C. These results have implications for the design of universal vaccines aimed at attenuating the disease course.
MATERIALS AND METHODS
Study subjects and data sources. (i) Subtype B chronic infection and elite controllers cohort.
The subtype B chronic infection cohort comprised 803 untreated individuals (median pVL of 5.1 log10 RNA copies/ml, with an interquartile range [IQR] of 4.7 to 5.5 log10 RNA copies/ml, and a median CD4 cell count of 273 cells/mm3 [IQR, 130 to 420 cells/μl]). Of these individuals, 762 (94.9%) represented the baseline (antiretroviral-naive) cross-section of the British Columbia HOMER cohort, while the remaining 41 (5.1%) represented chronically infected individuals recruited from Massachusetts General Hospital who were untreated at the time of sample collection (7). HLA class I typing and plasma HIV-1 RNA gag sequencing were performed as described previously (7, 19). Construction of Gag-protease recombinant NL4-3 viruses expressing participant-derived Gag-protease sequences, as well as evaluation of their replication capacity, was performed as described previously (7) and below.
HLA class I types and plasma HIV-1 RNA gag sequences from a cohort of 69 elite controllers (defined as chronically HIV-1-infected individuals maintaining pVL at <50 copies RNA/ml in the absence of therapy) recruited through Massachusetts General Hospital were also studied (described in references 31 and 34).
(ii) Subtype C chronic infection cohort.
The subtype C chronic infection cohort comprised 405 antiretroviral-naive chronically infected individuals from the Sinikithemba cohort in Durban, province of KwaZulu-Natal, South Africa. The median viral load was 4.77 (IQR, 4.15 to 5.27) log10 HIV RNA copies/ml, and the median CD4 count was 340 (IQR, 238 to 477) cells/μl. HLA class I typing was performed as described previously (26), and plasma HIV RNA gag sequencing was performed as described by Wright et al. (48). Construction of Gag-protease recombinant NL4-3 viruses expressing participant-derived Gag-protease sequences, as well as evaluation of their replication capacity, was performed as described in reference 48 and below.
(iii) Subtype C longitudinal seroconverter cohort.
The subtype C longitudinal seroconverter cohort comprised 53 women enrolled in the CAPRISA 002 observational cohort study, which was established to investigate the role of viral and immunological factors during acute/early subtype C infection on HIV-1 disease progression (47). The cohort includes high-risk HIV-negative women monitored monthly for recent HIV-1 infection using two rapid antibody tests and PCR (Roche Amplicor v1.5). HIV-1 infection was confirmed using an enzyme immunoassay test. Women were enrolled in the present study within 3 months of infection from both the HIV negative cohort and other seroincident cohorts in Durban, South Africa. The date of infection was estimated as the midpoint between the last HIV-1-negative test and the first antibody-positive test, or 14 days prior to the PCR-positive but antibody-negative date. Samples were collected at enrollment, weekly for 3 weeks, fortnightly for 3 months, monthly for a year, and quarterly thereafter. CD4+ T cell counts were assessed using a FACSCalibur flow cytometer, and viral loads were measured using a COBAS Amplicor HIV-1 Monitor Test v1.5 (Roche Diagnostics). The 53 individuals included in the present study represented seroincident individuals monitored for 12 months postinfection. The median CD4+ count and log viral load at 12 months postinfection (calculated as the average of the three time points closest to 12 months) were 416 cells/μl (IQR, 341 to 563) and 4.43 log10 HIV RNA copies/ml (IQR, 3.35 to 4.93), respectively. The present study was approved by the relevant institutional review boards overseeing the above cohorts.
RNA isolation, RT-PCR, and viral sequencing.
For the CAPRISA 002 cohort, viral sequencing was carried out as described previously (17). RNA isolated from plasma samples by using a Magna-Pure compact nucleic acid extractor (Roche, Mannheim, Germany) was reverse transcribed using a Thermoscript reverse transcription (RT) kit (Invitrogen). Limiting-dilution nested PCR was carried out by serial endpoint dilution of the cDNA (34a). PCR products were either directly sequenced or cloned using the pGEM-T Easy vector system (Promega). Sequencing was carried out using an ABI Prism dye terminator cycle sequencing kit (Applied Biosystems). Chromatograms were assembled using ChromasPro (www.technelysium.com.au/chromas.html) and aligned using CLUSTAL W (46). PCR and sequencing primers used are listed in Table S1 in the supplemental material.
HLA typing.
For the CAPRISA 002 cohort, high-resolution (four-digit) HLA typing was performed on all participants. DNA was extracted from either peripheral blood mononuclear cells (PBMC) or granulocytes using a Pel-Freez DNA isolation kit. HLA-A, -B, and -C typing was performed by sequencing exons 2, 3, and 4 using Atria HLA-AlleleSeqr kits (Abbott) or HLA-SBTexcellerator kits (Qiagen) and Assign-SBT v3.5 software (Conexio Genomics). Ambiguities resulting from polymorphisms outside the sequenced exons or identical heterozygote combinations were resolved using sequence-specific primers or resolved statistically using a published computational HLA class I completion algorithm trained on a data set of complete high-resolution HLA-A, -B, and -C types from >13,000 individuals of known ethnicity (27; http://research.microsoft.com/en-us/projects/bio/mbt.aspx#HLA-completion).
Viral replication assays of site-directed mutants and participant-derived subtype B and C sequences (NL4-3 recombinant system). (i) Generation of site-directed mutant viruses.
HIV-1 NL4-3 (subtype B) and consensus C (subtype C) p24 gag fragments were amplified using 100-bp-long primers CA_Chimeric Recomb F and CA_Chimeric Recomb R, matching the NL4-3 sequence upstream and downstream of p24, respectively (see Table S1 in the supplemental material). Amplicons were cloned into the pGEM-T Easy vector. Site-directed mutagenesis was used to introduce various mutations and combinations of mutations (T242N, M250I, G248A/M250I, and T242N/M250I for subtype B and T242N and M250I for subtype C) into the relevant p24 recombinant vectors using a QuikChange II site-directed mutagenesis kit (Stratagene). To generate recombinant NL4-3 viruses expressing the mutant p24 sequences, p24gag was reamplified from the mutated recombinant pGEM-T Easy plasmids using the primers described above.
The plasmid pNL4-3Δp24 was constructed by inserting unique BstEII restriction sites at the 5′ and 3′ ends of p24Gag using a QuikChange XL kit, followed by deletion of this region by BstEII digestion (New England BioLabs). This plasmid was maintained using Stbl3 Escherichia coli cells (Invitrogen). To generate recombinant viruses, 10 μg of BstEII-linearized pNL4-3Δp24 plus 50 μl of the above amplicon (∼5 μg) were mixed with 2.0 × 106 cells of a Tat-driven green fluorescent protein (GFP) reporter T-cell line of CEM origin (GXR 25 cells [9]) in 800 μl of R10+ medium (RPMI 1640 containing 10% fetal calf serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml) and transfected by electroporation using a Bio-Rad GenePulser II (exponential protocol: 300 V, 500 μF). After transfection, the cells were rested for 45 min at room temperature, transferred to 25-mm3 flasks in 5 ml of R10+ medium, and fed with 5 ml of R10+ medium on day 5. GFP expression was monitored by flow cytometry (FACSCalibur; BD Biosciences) and, once GFP+ expression reached >15% among viable cells, supernatants containing the recombinant viruses were harvested, and aliquots were stored at −80°C. The sequences of all recombinant virus stocks were verified via resequencing.
(ii) Generation of recombinant NL4-3 viruses expressing participant-derived sequences.
The same approach was previously used to generate recombinant viruses expressing participant-derived HIV RNA Gag-protease sequences from 803 chronically subtype B (7)- and 405 chronically subtype C (48)-infected individuals (data that were reanalyzed for the present study). Briefly, the Gag-protease region was RT-PCR amplified from plasma HIV-1 RNA using sequence-specific primers, followed by a second-round PCR using 100-bp-long polyacrylamide gel electrophoresis-purified recombination primers designed to match the NL4-3 sequence directly upstream of Gag and downstream of protease. Amplicons were cotransfected with linearized pNL4-3Δgag-protease into GXR T cells, and recombinant viruses were generated via homologous recombination as described above and previously (7, 48).
(iii) Recombinant virus titers and replication capacity assays.
Determination of virus titers and assessment of viral replication capacity of site-directed mutant and participant-derived recombinant viruses was performed as described previously (7, 48). Briefly, virus titers were determined by infecting GXR cells with chimeric virus stocks and measuring GFP expression by flow cytometry. The titer data were used to adjust input viral volumes in the subsequent replication capacity assay to obtain a multiplicity of infection (MOI) of 0.003 on day 2. GFP expression was monitored daily for a week. Replication capacity was expressed as the slope of the natural log of the percentage of GFP-expressing cells between days 3 and 6. This value was then divided by that of the appropriate control strain (wild-type NL4-3 or consensus C), yielding a normalized replication capacity value where 1.0 represented growth identical to that of the control strain, and values of <1.0 and >1.0 represented growth that was lower or higher than that of the control strain, respectively. Replication capacity assays of site-directed mutant viruses were performed in triplicate in a single experiment, while recombinant viruses were assayed in duplicate, and the results are reported as the average (mean) value.
Viral replication assays of subtype C site-directed mutants in a subtype C backbone (primary cell assay system). (i) Generation of molecular clones harboring Gag mutations in subtype C backbone.
Specific gag mutations were also engineered into a consensus gag sequence cloned into an HIV-1 subtype C infectious molecular clone pBR246-F10 backbone (provided by B. Hahn). The pBR246-F10 backbone contains HLA-B57/B*58:01 escape variants that have been previously reported to reduce replicative fitness (8, 28); therefore, we modified the backbone by replacing the gag gene with a synthetic consensus subtype C gag gene, as follows. The XhoI restriction site in the vector component of the original plasmid was deleted by digestion with XhoI, followed by blunting with Klenow and blunt-end ligation. A BssHII site at the 5′ end of gag and a XhoI site 60 nucleotides downstream of the gag stop codon were engineered using a QuikChange II site-directed mutagenesis kit. Quantitative PCR (qPCR) primer sites were similarly introduced into nef (6). The consensus subtype C full-length gag gene (www.hiv.lanl.gov) was synthesized (Integrated DNA Technologies) and ligated into the pGEM-T Easy vector. The mutations T242N, M250I, M250I/S252G/D260E, and T242N/M250I/S252G/D260E were introduced through site-directed mutagenesis and confirmed via DNA sequencing. The mutated genes were amplified using forward and reverse primers harboring BssHII and XhoI restriction sites, respectively, after which amplicons were digested and ligated into the modified pBR246-F10 backbone. E. coli STBL3 cells (Invitrogen) were used to propagate full-length proviral plasmids. In the subsequent experiments, the recombinant pBR246-F10 molecular clone harboring consensus subtype C Gag served as the control strain.
(ii) Generation of viral stocks of subtype C site-directed mutants.
Viral stocks were generated by transfection of HEK293T cells with 7.5 μg of plasmid DNA in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Atlantic Biologicals) using Lipofectamine 2000 (Invitrogen). Supernatants were harvested 72 h after transfection, and aliquots were stored at −80°C. HIV-1 p24 Capsid concentrations were quantified by a p24 enzyme-linked immunosorbent assay using the Vironostika HIV-1 Antigen MicroELISA system (bioMérieux, Netherlands).
(iii) Viral replication assays in primary cells.
PBMC isolated from five healthy HIV-negative donors were CD8 depleted using Dyna-Beads (Invitrogen). The PBMC were stimulated in RPMI 1640 supplemented with 20% FBS, 5 mg of phytohemagglutinin (Sigma, St. Louis, MO)/ml, and 20 U of interleukin-2 (Becton Dickinson Labware)/ml for 72 h prior to infection. After 72 h, the PBMC were washed with 1× phosphate-buffered saline supplemented with 1% (vol/vol) FBS (wash medium). A total of 5.0 × 105 PBMC were inoculated with viral stocks (MOI = 0.002) in 1 ml of RPMI 1640 supplemented with 20% FBS and 20 U of interleukin-2 (growth medium). After 24 h, the growth medium was removed, PBMC were washed, and fresh growth medium was added. Then, 250 μl of supernatant was removed for qPCR analysis and replaced with an equal volume of growth medium on days 2, 4, 6, 8, 10, and 12. Fresh PBMC were added on day 6.
(iv) qPCR.
RNA was extracted from 200 μl of supernatant as described above. cDNA was synthesized from 4 μl of RNA using the Superscript III RT-PCR kit (Invitrogen) in a total volume of 10 μl. qPCR was carried out as described earlier (6) on a LightCycler using the FastStart SYBR green kit (Roche) in a 20-μl reaction volume. Briefly, each reaction mixture consisted of 10.4 μl of distilled water, 1.6 μl of MgCl2 (25 mM), 2 μl each of 5 μM stocks of WT-F and WT-R primers, 2 μl of LightCycler SYBR green mix, and 2 μl of cDNA. The cycling conditions consisted of a 15-min initial activation and denaturation step at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C, and a melting curve involving 1 min at 60°C, followed by ramping to 95°C with continuous sampling. The quantification standards consisted of the modified infectious molecular clone pBR246-F10 linearized by BssHII digestion, followed by gel purification and spectrophotometric copy number quantification.
Identification of amino acids covarying with M250I in participant-derived subtype B and C sequences.
Published phylogenetically informed methods (14, 15) were used to identify HLA class I alleles associated with, and HIV-1 Gag amino acids significantly covarying with, M250I in chronic subtype B (7) and subtype C (48) cohorts. Multiple comparisons were addressed using q-values, the P value analogue of the false discovery rate (FDR) (42). The FDR is the expected proportion of false positives among results deemed significant at a given P value threshold; for example, at a q ≤ 0.2, we expect 20% of the identified associations to be false positives.
Conformational modeling of p24 helix 6 peptides.
Conformational modeling of peptide sequences was carried out using the program PEP-FOLD (29, 45) via the PEP-FOLD standalone server (http://bioserv.rpbs.univ-paris-diderot.fr/cgi-bin/PEP-FOLD). The HIV-1 consensus B p24 N-terminal domain nuclear magnetic resonance structure 1L6N (retrieved from the RCSB Protein Data Bank [www.pdb.org]) (43) was edited to include only residues from positions 242 to 252 and was used as a structural template for modeling helix 6. Helix 6 stretches from positions 242 to 252 inclusive in the structure 1L6N. Helix 6 peptides from positions 242 to 252 were extracted from HIV-1 clade B and HIV-1 clade C consensus sequences (www.hiv.lanl.gov) and submitted to PEP-FOLD for conformational modeling. PEP-FOLD reports the structure and energy of the 50 lowest energy conformations (i.e., the most stable) for each peptide sequence that it models. We regarded this collection of 50 structures reported by PEP-FOLD as a “cluster,” and we associated the pair of energy terms—[a,b]—with each cluster. In the pair [a,b], “a” is the lowest value in the cluster, and “b” is the highest energy value. The energy terms calculated by PEP-FOLD are in kcal/mol. Predictions using PEP-FOLD algorithms were qualitatively and semiquantitatively checked against predictions using the protein structure prediction server PSI-PRED (30; http://bioinf.cs.ucl.ac.uk/psipred).
Statistical analyses.
Wilcoxon rank-sum tests were used to compare viral loads and CD4+ T-cell counts between pairs of group. The Fisher exact test was used to compare differences in frequencies of the M250I mutation between study cohorts and other dichotomous groups. Statistical tests were implemented in GraphPad Prism 5.0 (GraphPad Software, Inc.).
RESULTS
M250I is rare in subtype B and is associated with reduced fitness in site-directed mutant and participant sequences.
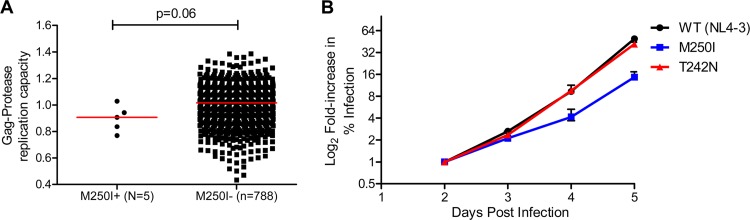
The Gag M250I mutation was observed in only 5 of 803 (0.6%) chronically subtype B-infected individuals (Fig. 1A). Of these, three harbored an M/I mixture, while the remaining two harbored I as the sole amino acid at this position (not shown). The median in vitro replicative capacity of recombinant viruses expressing participant-derived Gag/protease in a subtype B NL4-3 backbone was lower among M250I-containing viruses (n = 5) compared to those lacking M250I (n = 788), although this did not reach statistical significance (median of 0.91 versus 1.02, P = 0.06; Fig. 1A). When engineered alone into a NL4-3 backbone, M250I conferred a 22% reduction in replication capacity compared to wild-type NL4-3 (Fig. 1B). To provide context, a site-directed NL4-3 mutant virus expressing the well-characterized T242N mutation exhibited a 2% reduction in replication capacity compared to wild-type NL4-3 (Fig. 1B).
Fig 1.
Subtype B replication capacity. (A and B) Normalized replication capacities of recombinant viruses expressing Gag-protease from subtype B chronically infected patients carrying 250I (n = 5) and 250M (n = 788) (A) and NL4-3 site-directed mutants (T242N and M250I) (B). The error bars represent the standard errors for triplicate measurements.
Distribution and clinical correlates of M250I in subtype B are also consistent with a fitness cost.
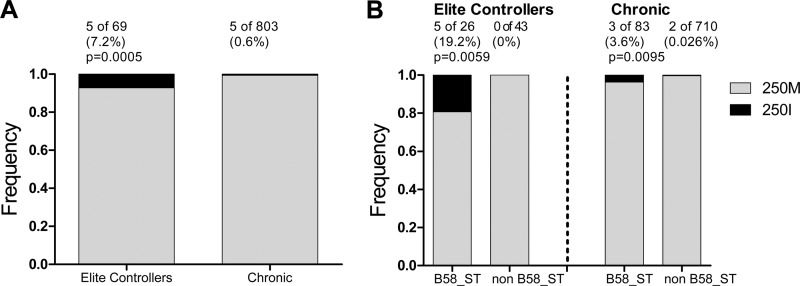
M250I was previously described as a rare mutation occurring in HIV-1 subtype B-infected elite controllers (33), the same cohort analyzed in the present study. Indeed, the frequency of this mutation was 12-fold higher among elite controllers (5/69 [7.2%]) compared to chronically subtype B-infected persons (5/803 [0.6%]) (Fisher exact test, P = 0.0005; Fig. 2A). Moreover, in both subtype B-infected elite controllers and chronically infected individuals, M250I was significantly enriched in individuals expressing alleles of the HLA-B58 supertype (Fig. 2B). Whereas 5/26 (19.2%) of the elite controllers expressing a B58-supertype allele were infected with viruses harboring M250I, the mutation was not observed in any of the 43 elite controllers who did not express B58-supertype alleles (estimated odds ratio [OR] of 22.3; Fisher exact test, P = 0.0059; phylogenetically corrected P = 0.0017). Similarly, while the mutation was observed in 3/83 (3.6%) chronically infected individuals expressing a B58-supertype allele, it was only observed in 2/710 (0.025%) individuals lacking such alleles (OR 13.3; Fisher exact test, P = 0.0095; phylogenetically corrected P = 0.004). A phylogenetically corrected comparison of the estimated ORs of B58-supertype mediated selection of M250I in controllers (OR = 22.3) versus chronically infected individuals (OR = 13.3) revealed that this nearly 2-fold difference was statistically significant (P = 0.017). Of note, the five B58-supertype expressing controllers harboring M250I were B*15:16/B*57:03, B*49:01/B*57:01, B*13:02/B*57:01, B*15:17/B*57:01, and B*52:01/B*57:03, while the three B58-supertype expressing individuals from the chronic cohort harboring M250I were B*15:01/B*57:01, B*08:01/B*58:01, and B*08:01/B*58:01. Taken together, the results support M250I as a rare B58 supertype-associated polymorphism in subtype B, whose odds of selection are nearly 2-fold higher among controllers compared to chronically infected individuals harboring the relevant HLA allele(s).
Fig 2.
Distribution of M250I by cohort and HLA in subtype B. (A and B) Distribution of the M250I mutation between elite controllers and chronic subtype B patients (A) and distribution between subjects with or without HLA-B58 supertype in the elite controller and chronic cohorts (B).
Furthermore, in our chronic subtype B cohort, a trend toward lower plasma viral loads was observed in individuals infected with viruses harboring the M250I mutation compared to individuals infected with viruses lacking this mutation, although this was not statistically significant (median of 0.58 versus 5.08 log10 HIV RNA copies/ml, P = 0.089). CD4 counts were comparable between the two groups (median of 300 versus 280; P = 0.4) (see Fig. S1 in the supplemental material).
In subtype C, M250I is associated with reduced fitness in site-directed mutant, but not participant sequences.
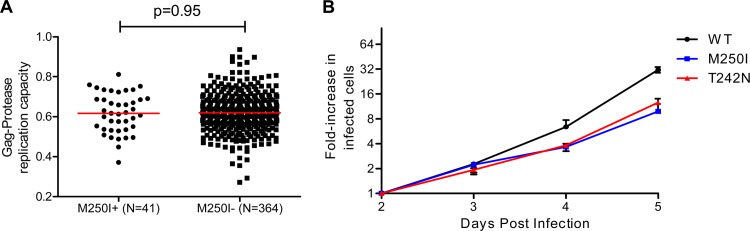
In subtype C, the M250I frequency in both recently transmitted and chronic viral sequences was considerably higher than in subtype B: 8/53 (15%) (median, 42 days postinfection) for recently transmitted viruses and 41/405 (10%) for viruses from chronic infections. The in vitro replicative capacities of NL4-3 derived recombinant viruses expressing subtype C Gag-protease sequences sampled during chronic infections were comparable between M250I-containing (n = 41) and non-M250I-containing (n = 364) sequences (median of 0.62 in both groups, P = 0.95; Fig. 3A). However, when introduced alone into a consensus subtype C p24gag (also in an NL4-3 backbone), M250I conferred a 25% reduction in replication capacity compared to consensus C p24gag sequence (Fig. 3B). To provide context, a mutant NL4-3 virus expressing T242N introduced into consensus C p24gag exhibited a 13% reduction in replication capacity compared to consensus C p24gag sequence (Fig. 3B).
Fig 3.
Subtype C replication capacity. (A and B) Normalized replication capacities of recombinant viruses expressing Gag-protease from subtype C chronically infected patients harboring M250I (n = 41) and M250 (n = 364) (A) and consensus subtype C p24 site-directed mutants (T242N and M250I) (B). The error bars represent the standard errors for triplicate measurements.
Distribution and clinical correlates of M250I in subtype C support a modest fitness cost.
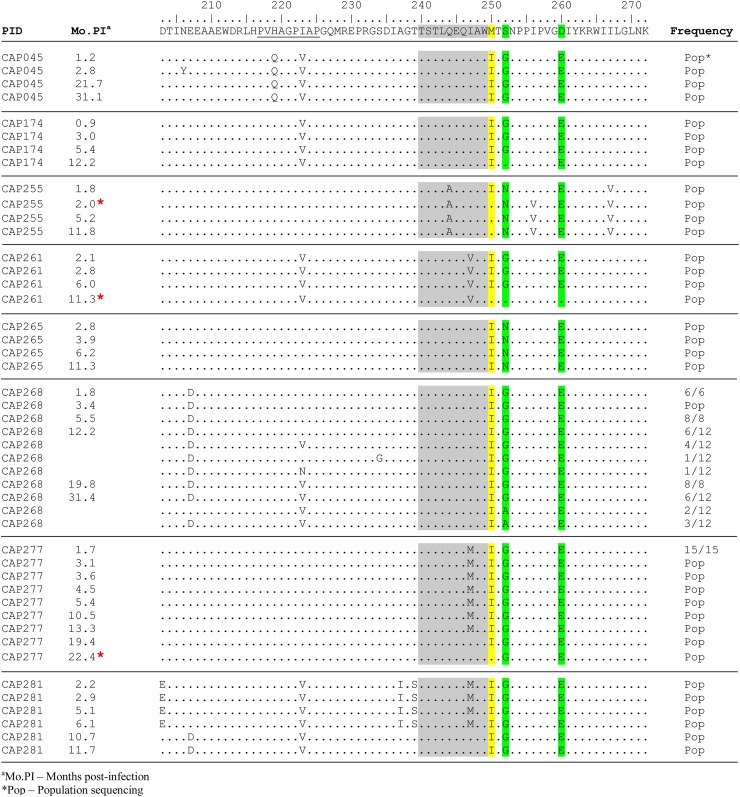
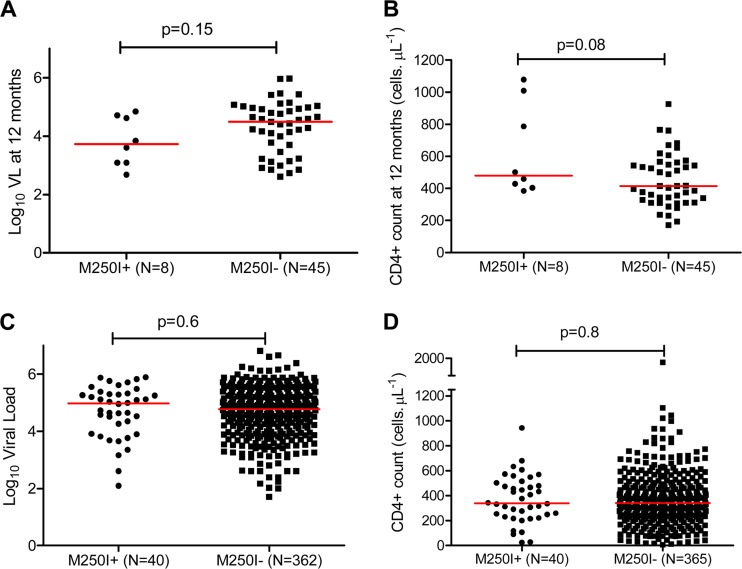
Analysis of longitudinal clonal Gag sequences from our subtype C seroconverter cohort (median [IQR] follow-up of 12.2 [11.7 to 31.1] months) revealed reversion of M250I mutations to wild type in three of eight participants harboring this polymorphism at their earliest time point (Fig. 4). These three reversions occurred at 2.0 (subject CAP255), 11.3 (CAP261), and 22.4 (CAP277) months postinfection. In our subtype C seroconverter cohort at 12 months postinfection, we observed modestly higher CD4 T-cell counts, although not statistically significant, among individuals harboring M250I (median of 480 versus 414 cells/μl, P = 0.08), whereas plasma viral loads were comparable between the two groups (median of 3.73 versus 4.49 log10 HIV RNA copies/ml, P = 0.15) (Fig. 5A and B). In our subtype C chronic infection cohort, no differences in pVL and CD4 counts were observed in individuals infected with M250I-expressing viruses compared to individuals infected with viruses lacking this mutation (median log10 HIV RNA copies/ml of 4.98 versus 4.78 [P = 0.6] and median CD4+ counts of 339 versus 340 cells/μl [P = 0.8], respectively) (Fig. 5C and D). Among chronically subtype C-infected individuals the M250I mutation was not associated with the expression of any specific HLA class I alleles (data not shown).
Fig 4.
Gag sequences of M250I mutants from the acute subtype C cohort. An alignment of p24 sequences of viruses carrying M250I mutation at transmission shows the reversion over time in three participants (red asterisk [CAP255, CAP261, and CAP277]). The B*57/B*58:01-restricted TW10 epitope is highlighted in gray. The CypA binding loop is underlined. Site 250 is highlighted in yellow, whereas sites 252 and 260 are highlighted in green.
Fig 5.
M250I mutation and disease progression in subtype C infections. Comparisons of the plasma viral loads and CD4+ T-cell counts between individuals infected with M250I mutants and those without in the acute infection cohort (A and B) and in the chronic cohort (C and D) are shown. The viral loads and CD4+ T-cell counts for the acute cohort were calculated as the means of three time points closest to 12 months postinfection.
Covariation between M250I and other sites in p24 Gag.
Fitness costs of certain mutations can be fully or partially restored through the selection of compensatory mutations at secondary sites (22, 39, 49). To identify potential compensatory mutations for M250I in subtypes B and C, we applied phylogenetically informed methods (14, 15) to identify Gag amino acids covarying with M250I in sequences from our chronic subtype B (7) and C (48) cohorts. No significant M250I-associated amino acids were identified in subtype B sequences at q < 0.2 (data not shown), which is likely due to the extreme rarity of this mutation in this cohort. In contrast, in our chronic subtype C cohort, significant covariation was detected between codon 250 and seven other Gag sites, all of them in p24 (Table 1). Of these, codons 252 and 260 were the most statistically significant. Specifically, strong positive associations were observed between M250I and both S252G and D260E, suggesting that these may function as compensatory mutations in a subtype C context. Independent analysis of our subtype C seroconverter cohort confirmed these covarying pathways, although all q-values were >0.2 in this analysis due to the small sample size (data not shown). We therefore hypothesized that S252G and D260E represent compensatory mutations for M250I in subtype C.
TABLE 1.
Covariation between codon 250 and other Gag sites in chronic subtype C sequencesa
| Codon 250 | Covarying codon | Association | TT | TF | FT | FF | Total | P | q-value |
|---|---|---|---|---|---|---|---|---|---|
| 250M | 252G | Negative | 55 | 317 | 34 | 7 | 413 | 7.4E–09 | 8.0E–06 |
| 260D | Positive | 264 | 108 | 4 | 37 | 413 | 1.9E–08 | 8.0E–06 | |
| 252S | Positive | 220 | 153 | 2 | 38 | 413 | 2.7E–07 | 7.0E–05 | |
| 247M | Negative | 0 | 372 | 4 | 36 | 412 | 2.5E–05 | 5.0E–03 | |
| 256V | Positive | 192 | 180 | 3 | 38 | 413 | 3.3E–05 | 5.0E–03 | |
| 207D | Positive | 38 | 334 | 0 | 41 | 413 | 2.2E–04 | 3.0E–02 | |
| 260E | Negative | 107 | 265 | 37 | 4 | 413 | 7.5E–04 | 8.0E–02 | |
| 250I | 252G | Positive | 33 | 6 | 55 | 318 | 412 | 1.4E–08 | 8.0E–06 |
| 260D | Negative | 4 | 36 | 264 | 110 | 414 | 1.4E–07 | 4.0E–05 | |
| 252S | Negative | 2 | 37 | 220 | 154 | 413 | 5.4E–07 | 1.0E–04 | |
| 256V | Negative | 3 | 37 | 192 | 181 | 413 | 5.0E–05 | 7.0E–03 | |
| 247 M | Positive | 4 | 35 | 0 | 374 | 413 | 5.9E–05 | 8.0E–03 | |
| 207D | Negative | 0 | 39 | 38 | 335 | 412 | 3.1E–04 | 3.0E–02 | |
| 260E | Positive | 36 | 4 | 109 | 265 | 414 | 8.2E–04 | 8.0E–02 |
TT, number of sequences with both codon 250 residueand covarying residue; TF, number of sequences with codon 250 residue butwithout covarying residue; FT, number of sequences without codon 250 residuebut with covarying residue; FF, number of sequences with neither codon 250residue nor covarying residue.
In addition, other amino acids potentially interacting with M250I have been described, including a negative association between M250I and T242N in both subtypes B and C (16, 28, 33) and a positive association between M250I and G248A in subtype B (16, 28, 33) (although other population-level studies have not reported these [10, 14, 35]). Given the proximity of M250I to the B58 supertype-restricted TW10 epitope and the association between this supertype and M250I in subtype B, we therefore also assessed the replicative consequences of M250I in combination with these mutations.
S252G and D260E mutations partially restore the fitness of subtype C M250I mutants.
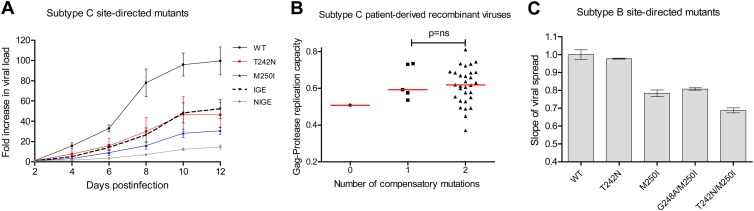
Our site-directed mutant experiments described thus far featured mutants engineered into a consensus C p24 sequence in an NL4-3 backbone. To confirm the effects of the M250I mutation and to assess replicative costs associated with putative compensatory and negatively associated mutations in a full subtype C backbone, we engineered the T242N, M250I, M250I/S252G/D260E (“IGE”), and T242N/M250I/S252G/D260E (“NIGE”) mutations into consensus C p24Gag and inserted this into the subtype C molecular clone backbone pBR246-F10 (see Materials and Methods). Furthermore, to explore the replicative costs of these mutations in a more biologically relevant context, we assayed the resulting recombinant viruses in primary PBMC over 12 days in culture (see Materials and Methods). The replication curves of the mutants in the subtype C backbone over 12 days in culture are shown in Fig. 6A. When the exponential slopes of viral spread in Fig. 6A were calculated and normalized to those of consensus C p24Gag, T242N- and M250I-containing viruses exhibited 15 and 18% reductions, respectively, in replication compared to the consensus C p24Gag control strain, values that were consistent with the replication of these mutants when they were engineered into the consensus C p24Gag/NL4-3 recombinant virus backbone and cultured in a reporter cell line (Fig. 3B). The subtype C triple mutant virus M250I/S252G/D260E (IGE) exhibited only an 8% reduction in replication compared to the control strain, supporting the combination of S252G and D260E as compensatory mutations for M250I (Fig. 6A and data not shown). Notably, the quadruple mutant carrying the T242N/M250I/S252G/D260E (“NIGE”) mutations exhibited a 31% reduction in replication compared to the control strain (Fig. 6A and data not shown), supporting a negative interaction between T242N and M250I mutations in subtype C.
Fig 6.
M250I compensation in subtype C and B viruses. (A) Replication capacities of site-directed mutants (T242N, M250I, M250I/S252G/D260E [IGE] and T242N/M250I/S252G/D260E [NIGE]) in a subtype C backbone showing the compensatory effect of the S252G and D260E mutations on the M250I mutant (IGE). Pooled CD8-depleted PBMC were inoculated in duplicate at an MOI of 0.002, and viral spread was measured by qPCR. The error bars represent standard errors for duplicate measurements. (B) Stratification of the subtype C Gag-protease replication capacities according to the number of the compensatory mutations (S252G and D260E). (C) Replication capacities of subtype B NL4-3 site mutants showing the minimal compensatory effect of the G248A mutation. The error bars represent the standard errors for triplicate measurements.
Examination of Gag sequences from our chronic subtype C cohort with a “full” M250I substitution (i.e., no mixtures) revealed that 30/36 (83%) harbored the full M250I/S252G/D260E (IGE) combination, 5/36 (14%) harbored M250I with either S252G or D260E, and only one sequence (3%) harbored M250I in the absence of either of these substitutions. Classification of participant-derived sequences according to the number of compensatory mutations they possessed at codons 252 and 260 (0, 1, or 2) revealed a dose-dependent increase in replication capacity over this range. However, due to the presence of only one virus harboring M250I alone, it was not possible to perform a statistical test on this overall trend (Fig. 6B). Moreover, of the viruses sampled from the 30 individuals chronically infected with subtype C that harbored the IGE combination with no amino acid mixtures, only one harbored the T242N mutation (data not shown), a finding consistent with the quadruple T242N/M250I/S252G/D260E (NIGE) combination being particularly detrimental (and thus rare) in subtype C viruses.
Compensation for M250I appears difficult in subtype B viruses.
As described above, no amino acids were identified to covary significantly with M250I in our chronic subtype B cohort. Furthermore, it was not relevant to introduce the putative subtype C-specific compensatory mutations S252G and D260E into subtype B, since a G at position 252 is never observed in naturally occurring subtype B sequences, and an E at position 260 is the subtype B consensus. Previous reports, however, have suggested that M250I may interact with certain HLA-B58-supertype associated polymorphisms in the upstream TW10 epitope such as T242N (for which a negative interaction had previously been suggested with M250I [28]) and G248A (observed frequently in sequences harboring M250I [33]). We therefore investigated the impact of these additional mutations on viral fitness when present with M250I in a subtype B context. When engineered into an NL4-3 backbone, the G248A/M250I double mutant exhibited 3% higher replicative capacity compared to M250I alone and yet still exhibited 19% lower replicative capacity compared to NL4-3 (Fig. 6C). These data suggest that G248A may only alleviate M250I-associated fitness costs to a very minor extent, if any. The T242N/M250I double-mutant virus had a replicative capacity 31% lower than that of NL4-3, suggesting that previously described negative associations between the two mutations are due to the severe fitness cost of the T242N/M250I double mutation.
The M250I mutation probably destabilizes helix 6 of p24.
The p24Gag N-terminal hairpin (Gag codons 133 to 145), cyclophilin A binding loop (Gag codons 217 to 225), and helix 6 (Gag codons 242 to 251), which includes the HLA-B58-supertype-restricted TW10 epitope) are conformationally coupled (28). It is believed that helix 6 stability is important in the binding of cyclophilin A by p24Gag (28) and that the modest replicative costs of T242N are due in part to disruption of the interactions between these p24Gag domains, thereby compromising the binding of helix 6 to cyclophilin A. Given that M250I lies only one amino acid downstream of TW10, we hypothesized that this mutation may also affect the stability of p24Gag helix 6 and its associated interactions. We therefore undertook conformational analysis of published consensus subtype B and C p24Gag helix 6 peptide sequences (www.hiv.lanl.gov) using PEP-FOLD (see Materials and Methods).
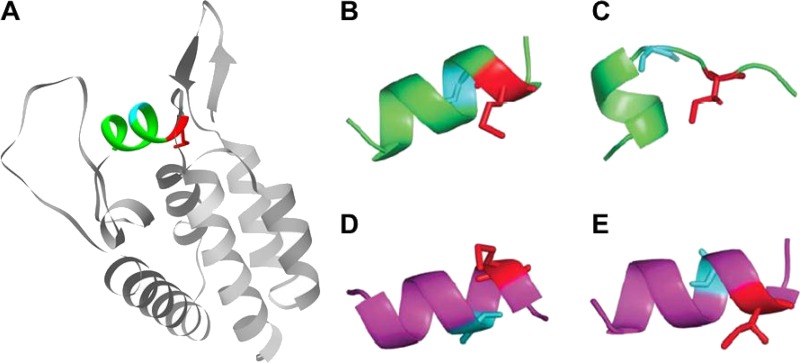
The predicted conformations of naturally occurring subtype B and C helix 6 peptides exhibited similar free energies (Table 2). When M250I was introduced into these sequences in silico, the predicted helical conformation of helix 6 was destabilized compared to the cognate “wild-type” conformation for both subtypes, but this effect was far more pronounced for subtype B compared to subtype C (Table 2 and Fig. 7). The subtype B and C consensus sequences differ at positions 248 and 252 (G248 versus A248 and N252 versus S252 in subtype B versus subtype C), suggesting that, in the context of M250I, either or both of these positions confer the greater stability of subtype C helix 6 peptide compared to that of subtype B. Further PEP-FOLD simulations on HIV-1 subtype B helix peptides harboring the subtype C consensus residues A248 and S252 revealed that A248 had a major stabilizing influence on helix 6 peptide and was predominantly responsible for minimizing the conformational differences between the wild-type and M250I-containing subtype C helix 6 (see Table S2 in the supplemental material). This finding may in part explain the frequent occurrence of G248A substitutions among subtype B M250I-containing sequences, since the former may act to restabilize helix 6 in a subtype B context.
TABLE 2.
Summary of helix 6 predictions by PEP-FOLD
| Description of helix 6 peptide | Sequence of helix 6 peptidea | Energy value (kcal/mol) in predicted cluster |
|
|---|---|---|---|
| Minimum | Maximum | ||
| HIV-1 subtype B | |||
| Wild type | TLQEQIGWMTN | –11.878 | –11.873 |
| M250I variant | TLQEQIGWITN | –10.195 | –10.116 |
| HIV-1 subtype C | |||
| Wild type | TLQEQIAWMTS | –15.614 | –15.610 |
| M250I variant | TLQEQIAWITS | –14.863 | –14.857 |
The peptide spans amino acids 242 to 252. Amino acids 248, 250 and 252 are underscored.
Fig 7.
Effect of M250I mutation on p24 helix 6. (A) N-terminal domain of p24 (PDB ID 1AK4 [23]) showing helix 6 (in color) and positions 248 (cyan) and 250 (red). (B) Conformation predicted for the wild-type HIV-1 subtype B helix 6 peptide. (C) Conformation predicted for the M250I variant. (D) Conformation predicted for the wild-type HIV-1 subtype C helix 6 peptide. (E) Conformation predicted for the M250I variant. In all panels, the residue at position 248 is shown in cyan and that at position 250 is shown in red. Stick representations are also shown at residue positions 248 and 250. In panels B and C, the helix 6 peptide has a glycine at position 248, and in panels D and E it has an alanine. The images were rendered using Pymol (www.pymol.org).
DISCUSSION
The extremely low frequency (0.6%) of M250I in chronic subtype B sequences renders it challenging to draw firm conclusions regarding its provenance and fitness impact in vivo. Despite this, multiple lines of evidence support a fitness cost in this subtype. First, even when it is present, M250I tends to occur as a mixture alongside M250 (i.e., it rarely achieves fixation in vivo), suggesting an inherent incompatibility between it and the subtype B genetic background. Second, patient-derived sequences harboring this mutation exhibit modestly (albeit not significantly) lower replication capacities and lower viral loads than those harboring M250. In both chronic (7) and controller (33) subtype B cohorts, M250I was enriched among individuals expressing an HLA allele belonging to the B58 supertype. Moreover, odds of B58 supertype-mediated selection of M250I was nearly 2-fold higher among controllers compared to chronically infected individuals, supporting M250I as a rare B58-supertype associated escape variant preferentially selected in elite controllers. The mechanism(s) behind this differential selection remain unclear; in addition, our observations are in contrast to those of a previous report of M250I occurring in a minority of hypermutated proviral (but not plasma HIV RNA or intact proviral) sequences from seven HLA-B*57-expressing elite controllers (5). Elite controller sequences from the present study are indeed derived from HIV RNA (a DNase step was used during RNA extraction to eliminate proviral DNA contamination [31]); as such, the enrichment and differential selection of M250I among elite controllers merits follow-up in independent cohorts.
The rarity of M250I (along with the fact that it is most often observed as an M/I mixture at position 250) may also explain its lack of identification in population-level studies of HLA-associated polymorphisms in subtype B (10, 25) and the difficulty of identifying (10, 25) potential compensatory mutations using computational approaches. Nevertheless, based on the observation that the upstream G248A mutation (a well-characterized B58-supertype-associated polymorphism [10, 25]) is often present, along with M250I in elite controller sequences (33), we attempted to evaluate G248A as a putative compensatory mutation for M250I. Introduction of G248A alleviated M250I-associated replicative defects only to a very modest extent (3%), if at all. Although the lack of statistical power due to the rarity of M250I represents a limitation of our analysis, results nevertheless suggest that M250I-associated fitness costs are not easily rescued via compensatory mutations in subtype B.
In contrast, in subtype C, M250I appears to represent a minor (≥10%) polymorphic variant where fitness costs can be alleviated through selection of compensatory mutations in vivo. First, individuals harboring M250I-containing viruses tended to have modestly (albeit not statistically significantly) higher CD4 counts during early (but not chronic) subtype C infection. Second, reversion of M250I mutations were observed in 40% of cases following transmission. Most notably, despite the M250I mutation conferring an 18% reduction in replicative capacity when introduced alone into a subtype C backbone, no differences in replication were observed between M250I-containing and non-M250I-containing subtype C viruses sampled during chronic infection. Furthermore, computational analyses identified two p24Gag polymorphisms —S252G and D260E—strongly covarying with M250I that restored the replicative capacity of M250I mutants from 82 to 92% of control strain levels when engineered into a subtype C backbone. The observation that all but one M250I-containing patient sequences sampled during chronic infection harbored one or both of these compensatory mutations and that the replication capacities of these were higher than the sequence harboring M250I alone was also suggestive of M250I-associated fitness costs being recouped by compensatory mutations.
We also investigated previous reports of a potential negative interaction between the T242N and M250I mutations reported in some (16) but not all (7, 10, 35, 48) cohorts. There were differences that may be attributable to variation in host/viral or other characteristics between cohorts and/or geographic regions. Of interest, the replicative capacity of the T242N/M250I double mutant was 13% lower than that of the M250I single mutant, thus providing a mechanism to explain the proposed negative interaction, if indeed one exists.
In silico analysis of the structural consequences of M250I revealed that it likely has a severe destabilizing effect on the subtype B p24Gag helix 6 structure that may be partially alleviated by the B58-supertype-associated mutation G248A. Notably, although G248A may partially stabilize helix 6 in subtype B, its ability to compensate for the replicative fitness costs of M250I appears to be marginal at best. In subtype C, the modest effect of M250I on helix 6 structure can be attributed to the stabilizing effects of the alanine residue at position 248, which is the consensus amino acid at this position in subtype C. This provides a possible explanation for the substantially higher frequency of the M250I mutation in subtype C sequences compared to subtype B sequences. The host factor(s), if any, driving the higher frequencies of M250I in subtype C sequences remain unknown but may be revealed through more statistically powerful analyses of larger subtype C cohorts or comparative analyses of subtype C cohorts from different geographical regions (similar to those undertaken for subtype B) (4).
In conclusion, M250I in HIV-1 subtype B appears to be a rare immune-driven mutation with fitness consequences, but in HIV-1 subtype C it is a relatively innocuous circulating polymorphism that is not selected by any known HLA alleles. Analysis of the structural basis for these observations suggests that M250I has a greater destabilizing effect on the subtype B p24gag helix 6 structure compared to subtype C, yielding greater replicative fitness costs. It is possible that M250I in subtype B reduces the ability of p24 to bind host factor cyclophilin A (21, 26); however, further studies will be required to explore this potential mechanism. Our data suggest CTL-based vaccines targeting functionally constrained HIV-1 regions that aim to drive viral evolution down fitness-attenuating pathways may need to account for intersubtype differences in the specific mutations selected under immune pressure and their relative impact on viral fitness.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by operating grants from the Canadian Institutes for Health Research (CIHR; MOP-93536 and HOP-115700 to Z.L.B./M.A.B.), a Jim Gray seed grant from Microsoft Research (to Z.L.B./M.A.B.), a National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), U.S. Department of Health and Human Services grant U19 A151794, and a NIAID International Research in Infectious Diseases (IRID) grant 1R01AI078936 (to C.W.). An India-South Africa collaborative grant was provided through the Technology Innovation Agency, South Africa (TIA), and the Centre for the AIDS Program of Research in South Africa (CAPRISA) (to C.W.). D.R.C. is a recipient of the Canada-HOPE fellowship from CIHR and Sanofi-Aventis and the Clinical Infectious Diseases Research Initiative (CIDRI) fellowship. A.Z. is funded by a Sydney Brenner fellowship. J.K.M. is funded by the National Research Foundation and the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University. T.N. holds the South African Department of Science and Technology/National Research Foundation Research Chair in Systems Biology of HIV/AIDS and an International Early Career Scientist Award from the Howard Hughes Medical Institute (HHMI). M.A.B. holds a Canada Research Chair, Tier 2, in Viral Pathogenesis and Immunity. Z.L.B. is the recipient of a CIHR New Investigator Award and a Michael Smith Foundation for Health Research (MSFHR) Scholar Award.
Footnotes
Published ahead of print 26 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abraha A, et al. 2009. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. J. Virol. 83:5592–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen TM, Altfeld M. 2008. Crippling HIV one mutation at a time. J. Exp. Med. 205:1003–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altfeld M, Allen TM. 2006. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 27:504–510 [DOI] [PubMed] [Google Scholar]
- 4. Avila-Rios S, et al. 2009. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology 6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey JR, Williams TM, Siliciano RF, Blankson JN. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boutwell CL, Rowley CF, Essex M. 2009. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol. 83:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brockman MA, et al. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J. Virol. 84:11937–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brockman MA, et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brockman MA, Tanzi GO, Walker BD, Allen TM. 2006. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J. Virol. Methods 131:134–142 [DOI] [PubMed] [Google Scholar]
- 10. Brumme ZL, et al. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol, and Nef proteins. PLoS One 4:e6687 doi:10.1371/journal.pone.0006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brumme ZL, et al. 2011. Reduced replication capacity of NL4-3 recombinant viruses encoding reverse transcriptase-integrase sequences from HIV-1 elite controllers. J. Acquir. Immune Defic. Syndr. 56:100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brumme ZL, Walker BD. 2009. Tracking the culprit: HIV-1 evolution and immune selection revealed by single-genome amplification. J. Exp. Med. 206:1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buonaguro L, Tornesello ML, Buonaguro FM. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 81:10209–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carlson JM, et al. 2008. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput. Biol. 4:e1000225 doi:10.1371/journal.pcbi.1000225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carlson JM, et al. 2012. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86:5230–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chopera DR, et al. 2011. Virological and immunological factors associated with HIV-1 differential disease progression in HLA-B 58:01-positive individuals. J. Virol. 85:7070–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chopera DR, et al. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033 doi:10.1371/journal.ppat.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chopera DR, Wright JK, Brockman MA, Brumme ZL. 2011. Immune-mediated attenuation of HIV-1. Future Virol. 6:917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cotton LA, et al. 2012. HLA class I sequence-based typing using DNA recovered from frozen plasma. J. Immunol. Methods 382:40–47 [DOI] [PubMed] [Google Scholar]
- 20. Coutsinos D, et al. 2009. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J. Virol. 83:2029–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crawford H, et al. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crawford H, et al. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gamble TR, et al. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294 [DOI] [PubMed] [Google Scholar]
- 24. Gatanaga H, et al. 2006. Altered HIV-1 Gag protein interactions with cyclophilin A (CypA) on the acquisition of H219Q and H219P substitutions in the CypA binding loop. J. Biol. Chem. 281:1241–1250 [DOI] [PubMed] [Google Scholar]
- 25. John M, et al. 2010. Adaptive interactions between HLA and HIV-1: highly divergent selection imposed by HLA class I molecules with common supertype motifs. J. Immunol. 184:4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiepiela P, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775 [DOI] [PubMed] [Google Scholar]
- 27. Listgarten J, et al. 2008. Statistical resolution of ambiguous HLA typing data. PLoS Comput. Biol. 4:e1000016 doi:10.1371/journal.pcbi.1000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Picado J, et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maupetit J, Derreumaux P, Tuffery P. 2009. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 37:W498–W503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405 [DOI] [PubMed] [Google Scholar]
- 31. Miura T, et al. 2008. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J. Virol. 82:8422–8430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miura T, et al. 2009. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J. Virol. 83:140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miura T, et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pereyra F, et al. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563–571 [DOI] [PubMed] [Google Scholar]
- 34a. Rodrigo AG, et al. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retroviruses 13:737–742 [DOI] [PubMed] [Google Scholar]
- 35. Rolland M, et al. 2010. Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One 5:e12463 doi:10.1371/journal.pone.0012463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rousseau CM, et al. 2008. HLA class-I driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 82:6434–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schaller T, et al. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7:e1002439 doi:10.1371/journal.ppat.1002439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schneidewind A, et al. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneidewind A, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soares EA, et al. 2009. Mutation T74S in HIV-1 subtype B and C proteases resensitizes them to ritonavir and indinavir and confers fitness advantage. J. Antimicrob. Chemother. 64:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sokolskaja E, Sayah DM, Luban J. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang C, Ndassa Y, Summers MF. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537–543 [DOI] [PubMed] [Google Scholar]
- 44. Taylor BS, Hammer SM. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 359:1965–1966 [DOI] [PubMed] [Google Scholar]
- 45. Thevenet P, et al. 2012. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 40:W288–W293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Loggerenberg F, et al. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954 doi:10.1371/journal.pone.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright J, et al. 2010. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J. Virol. 84:10820–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wright JK, et al. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J. Virol. 86:3193–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright JK, et al. 2011. Influence of gag-protease-mediated replication capacity on disease progression in individuals recently, infected with HIV-1 subtype C. J. Virol. 85:3996–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu HT, et al. 2009. Effects of the K65R and K65R/M184V reverse transcriptase mutations in subtype C HIV on enzyme function and drug resistance. Retrovirology 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yusim K, et al. 2009. HIV molecular immunology. Publication LA-UR 09-05941. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.