Abstract
Super-resolution techniques like PALM and STORM require accurate localization of single fluorophores detected using a CCD. Popular localization algorithms inefficiently assume each photon registered by a pixel can only come from an area in the specimen corresponding to that pixel (not from neighboring areas), before iteratively (slowly) fitting a Gaussian to pixel intensity; they fail with noisy images. We present an alternative; a probability distribution extending over many pixels is assigned to each photon, and independent distributions are joined to describe emitter location. We compare algorithms, and recommend which serves best under different conditions. At low signal-to-noise ratios, ours is 2-fold more precise than others, and 2 orders of magnitude faster; at high ratios, it closely approximates the maximum likelihood estimate.
Introduction
Techniques for ‘super-resolution’ fluorescence microscopy like PALM (photo-activation localization microscopy) [1] and STORM (stochastic optical reconstruction microscopy) [2] depend upon precise localization of single fluorophores. Such localization represents a challenge, as photons emitted from a point source are detected by a CCD to yield a pixelated image; then, relevant information in the pixels must be used to deduce the true location of the point source. The various localization methods currently in use differ in precision and speed. For example, minimizing least-square distances (MLS) and maximum likelihood estimation (MLE) fit a Gaussian distribution to pixel intensities before estimating a fluor’s location; MLS is the most popular but less precise, while MLE is more involved but can achieve the theoretical minimum uncertainty [3-5]. Both are iterative and so computationally intensive; consequently, attempts have been made to maximize accuracy and minimize computation time [6-9]. More problematic, fitting implies an underlying model, which can introduce errors, especially at low signal-to-noise ratios (S:N). The straightforward center-of-mass (CM) estimate [10] has the advantages of simplicity and speed, but is considered less accurate than the iterative methods (mistakenly, as we shall see); as a result, it is not being used for PALM/STORM.
Borrowing principles from ‘pixel-less’ imaging – a technique that uses a photomultiplier as a detector [11] – we present a non-iterative (and so rapid) way of localizing fluors imaged with a CCD. Each photon registered in the image carries spatial information about the location of its source. As this information is blurred by the point-spread function (PSF) of the microscope, we use the PSF to define many independent probability distributions that describe the emitter’s possible locations – one for each photon in the population (Fig. 1(a)). We then assume that all photons came from the same emitter (the usual and fundamental basis of localization), and aggregate probability distributions; the result is a joint distribution (JD) of the probability of the emitter’s location (Figs. 1(b) and 1(c)). Localization by JD is similar to a weighted form of CM, offering advantages in simplicity and speed, and – for the curious practitioner – we detail the differences between the two. We also compare the performance of the various methods both quantitatively (using computer-generated images) and qualitatively (using ‘real’ images). Our results enable us to recommend which approach to use with images containing different degrees of noise, depending on whether precision or speed is the priority. We find that the most popular – MLS – is never the algorithm of choice. At high signal-to-noise ratios, MLE yields the highest precision, while JD offers a quick, closed-form alternative; with very noisy images (where both MLS and MLE fail) JD proves the most accurate.
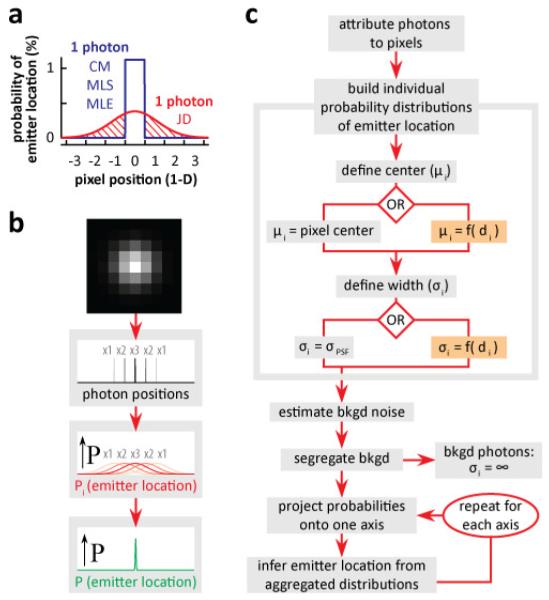
Fig. 1.
JD localization. (a) An individual photon carries information about the probability of an emitter’s location. CM/MLS/MLE (blue) assume equal probability throughout the pixel, with zero probability elsewhere; JD localization (red) uses a normal distribution to emulate microscope PSF (FWHM = 2.8x pixel width) which spreads beyond the pixel (shaded area The area under both curves is equal. (b) Principles behind JD localization. Each photon represented in a pixel is treated individually (1, 2, and 3, photons indicated by x1, x2, x3), individual probabilities of the location of the emitter, Pi (red curves) are aggregated to yield the joint probability, P, of emitter location (green). (c) Flow diagram for JD localization. Photons are attributed to each pixel dependent on intensity, and a probability distribution of the source of each photon is built using peak location (μi) and width (σi). The peak can be located at pixel centre (left) or anywhere within or outside the pixel (right); the width can be that of PSF (left) or any arbitrary value (e.g., as a function of distance, di, from the most intense pixel; right). Background (bkgd) noise is estimated and the influence of background photons nullified by setting σi = ∞. Then, probabilities of individual photons are projected onto a single axis, combined to infer the probability of emitter location for that axis, and the projection and inference repeated for each axis.
Results
Theory
In a typical single-molecule or PALM/STORM experiment, an image is acquired by collecting photons from temporally- and spatially-isolated emitters using a CCD. As many photons fall on one pixel, this is analogous to binning data into a histogram, with loss of sub-pixel (sub-bin) spatial information. We will think of individual photons as independent carriers of spatial information. Then, given a pixel that has registered one photon, conventional localization methods (such as MLE, MLS, and CM) would treat a photon as having a spatial distribution represented by the blue line in Fig. 1(a). The photon has a probability density function (i.e., the probability of the location of the source of that photon) that is uniformly flat over the whole area of the pixel, giving a 2-D rectangular or ‘top-hat’ distribution, with zero probability in neighboring pixels. [Note that this probability density function refers to one photon and not to many.] In other words, uncertainty is inaccurately recorded as a uniform distribution over just one pixel. In contrast, JD localization represents this uncertainty as a normal distribution that spreads over several pixels (see the one red curve in Fig. 1(a), and the many red curves in Fig. 1(b)).
The PSF serves as an initial estimate of the uncertainty imparted on the position of every photon by the microscope, and we initially use a normal distribution to approximate it [12] (as is common in the field). Such a distribution is uniquely described by center location (μi) and width (σi). Our default is to place μi at the center of a pixel and use σi equivalent to that of the PSF (Fig. 1(c), left); alternatively, μi and/or σi can be varied to suit the needs of a particular experiment (Fig. 1(c), right). After applying a distribution to each photon, distributions are aggregated to infer the probability of the location of the emitter (Figs. 1(b) and 1(c)). [Similar joining of independent probability distributions has been proposed for geolocation [13].] We have derived a simple equation to facilitate closed-form (non-iterative) – and so rapid – calculation.
| (1) |
Here, μo is the best estimate of the location of the emitter and N is the number of photons. [See Methods at the end of the manuscript for derivation.]
In contrast, methods such as MLS and MLE describe the probability of emitter location by fitting a curve to pixel intensities, which involves many sequential calculations, then deducing location information from that curve. We now benchmark test the different methods, first for precision and then for speed.
Quantitative comparison of precision
To assess precision, we use computer-generated images of a point source whose location is known. In each simulation, a ‘point source’ emits a known number of ‘photons’ that ‘pass’ through a ‘microscope’ (to be blurred by the PSF) to yield an image (initially 15×15 pixels) on a ‘CCD’ then, a specified number of ‘background photons’ are added. Using 10,000 such images for each condition analyzed, we go on to compute the 1-D root-mean-squared error (RMSE) between the true location of the emitter and the location estimated using each of the four methods.
In the first analyses (Figs. 2(a) and 2(b)), we apply JD using default settings (i.e., with μi set at the pixel center, and σi equivalent to that of the PSF); then, the JD equation simplifies to that used in CM (Methods). To aid comparison, we also plot the theoretical minimum uncertainty that is attainable under the particular conditions used – a lower bound (LB) computed using Eq. (6) of Thompson et al. [14]. This LB excludes effects of background noise, but includes those due to pixel size and PSF, and so differences from the LB reflect the influence of background noise on a method.
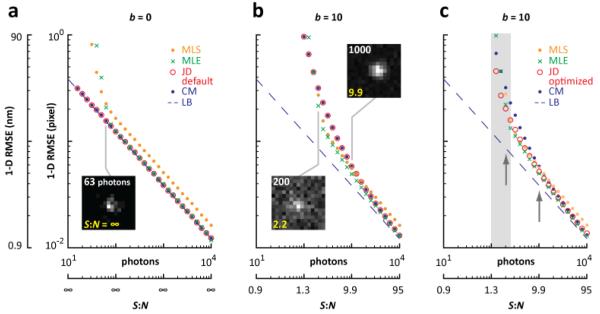
Fig. 2.
Qualitative comparison of methods. Computer-generated images (15×15 pixels) like those illustrated were generated using 10-104 emitter photons and different numbers of background photons (i.e., b = 0 or 10 photons/pixel) to give different signal-to-noise ratios (S:N); then, the root-mean-square error of localization in one dimension (1-D RMSE in nm or pixel units) was calculated (104 localizations per data point) using the methods indicated. Photon counts (top) and S:N (bottom) are indicated in some typical images. The lower bound, LB (blue dashed line), is computed using Eq. (6) of Thompson et al. [14] and plotted here and in subsequent Figures as a reference. (a) With no background (b = 0), the ‘default’ version of JD returns the same results as CM, and both track the LB; MLS and MLE fail at low photon counts. The failure of MLS without background is examined more in Supplemental Fig. 1(b). In the presence of background (b = 10), all methods fail at low photon counts; at moderate counts, MLE performs best, and at high counts MLS is the worst as the others converge to the minimum error. (c) An ‘optimized’ version of JD increases precision at low S:N while retaining precision at high S:N. The grey region is analyzed further in Fig. 3. Arrows: conditions used in Fig. 4.
We first consider the case where background is absent (b = 0; Fig. 2(a)). As expected, errors in localization given by all four methods decrease as the number of photons increases. Those given by CM and JD lie on the LB at all photon counts tested. Below ~30 photons, MLS and MLE ‘fail’ they either do not converge to a solution during the 200 iterations allowed, or yield a 1-D RMSE > 1 pixel (so values are not shown here) – and they sometimes even return a location outside the image (presumably because spot shape diverges significantly from a Gaussian; Supplemental Fig. 1, Media 1). As photon count increases, MLE is initially less accurate than MLS, but then errors fall progressively to reach the LB above ~100 photons. Errors given by MLS converge to a level 30% greater than the LB, as is well documented [4, 5, 15].
We now randomly add an average of 10 background photons per pixel (i.e., b = 10; Fig. 2(b)). At the very lowest signal-to-noise ratio, all methods fail (in the case of JD and CM, only because 1-D RMSE > 1 pixel). As the ratio progressively increases, JD and CM (when corrected for background; Methods) are the first to return a 1-D RMSE of less than 1 pixel, and then MLE and JD/CM (in that order) converge to the LB. Most PALM/STORM images are formed from data with S:N >5 (e.g., Löschberger et al. [16]), where MLE returns between 8 and 27% less RMSE than MLS.
As JD treats each photon separately, individual distributions can be tuned independently to optimize the precision and/or speed achieved at a given signal-to-noise ratio. As a first example, we eliminate the effects of outlying bright pixels that are likely to result from noise. As the PSF falls off precipitously from the central peak, few photons emitted by a point source will be detected in the image plane > 3σ distant from the true location. Then, we consider all signal detected > 3σ from the center of the brightest pixel to be noise (i.e., > 3.5 pixels away), and nullify its effects on the JD by ascribing σi = ∞ to each of its constituent distributions. This simple ‘optimized’ version of JD improves accuracy (compared with CM) over a wide range of S:N (Fig. 2(c)). It is also more precise than MLE at S:N < 2.7, than MLS at S:N < 3.0 and > 4.5, and it returns results within 5% of MLE at S:N > 7.
Tuning JD variables
When applying JD, we hitherto set μi = pixel center and σi = σPSF; we now tune each to maximize localization precision in noisy images (grey region in Fig. 2(c)) where MLS and MLE fail – first varying each one alone, and then both together. [Just as the default version of JD and CM produce the same results, we expect a tuned version of JD and an equivalent weighted CM variant (if developed) to do so too. However, we differentiate between JD and CM for several inter-related reasons: (i) By the strictest definition, CM weights each pixel position solely by a ‘mass’ equivalent to intensity; in contrast, in JD, μi and σi can be varied depending on distance from (and position relative to) the brightest pixel (with intensity determining the number of distributions to be joined together). (ii) Conceptually, CM applies statistics to a population of photons, whilst JD disaggregates the population into individual photons and then combines individual probabilities (with a consequential reduction in speed; below). (iii) In principle, it should be possible to derive a general form of CM that would allow tuning of the piecewise weightings of pixel positions to yield the same precision as the JD variants (below), but such a generalization would inevitably mean that the CM equation loses its characteristic simplicity.]
Consider Fig. 3(a), and the selected photon distributions (blue curves) in the cartoon on the left. By default, μi is placed at the center of the CCD pixel registering the photon (blue dots), even though that photon was probably emitted by a fluor in the central (brightest) pixel in the specimen plane. Therefore, the x- and y-coordinates of μi associated with all distributions – except those derived from the brightest pixel – are shifted between ⅕ − 1 pixel towards the brightest pixel (red dots mark new positions for a ½-pixel shift). Distributions from the brightest pixel are also shifted from the central default location by a distance proportional to the intensities of adjacent pixels (Methods, Eqs. (2) and (3)). [In all cases, σi remains constant and equal to σPSF.] A shift of ½-pixel width yields the least error (not shown), giving a ~5% reduction at S:N < 3 (Fig. 3(a), right).
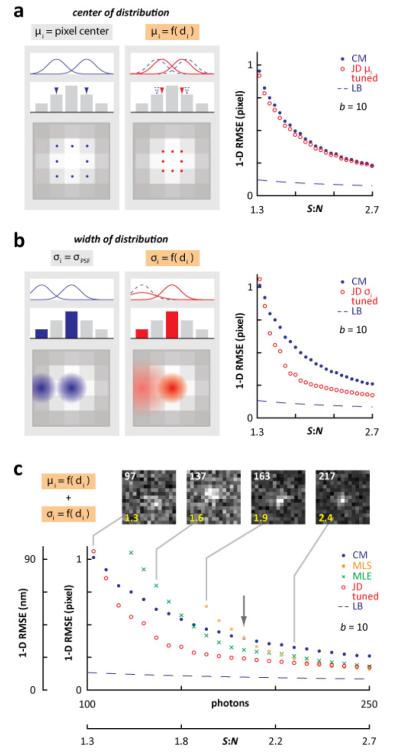
Fig. 3.
Tuning μi and σi to maximize localization precision near the detection limit. The 1-D RMSE in location of a point source was determined using 104 computer-generated images (15×15 pixels, 50-250 emitter photons, b = 10) per data point. JD localizations were determined by varying peak position (μi) and/or width (σi). Errors obtained using CM, MLS, and MLE, plus the lower bound (LB), are included for comparison (MLS/MLE plots smoothed by linear regression). (a) Varying μi depending on distance, di, from the most intense pixel (σi = σPSF for all distributions). Peaks of distributions are shifted towards the brightest pixel (in this example the central one) by ½ a pixel in both x- and y-dimensions. Left: cartoon illustrating how this shift applies to selected distributions (from blue curves/dots to red curves/dots). Right: JD localization yields 5% less error than CM for all S:N shown. (b) Varying σi as a function of distance, di, from the most intense pixel (as μi = pixel center for all distributions). Widths of distributions from the brightest pixel remain equal to σPSF, as those from surrounding ones expand. Left: cartoon illustrating these changes for selected distributions (from blue curves/halos to red curves/halos). Localization using JD yields up to 36% less error than CM. (c) Varying both peak position and width (as in (a) and (b), but using an x and y shift of ¼ pixel for μi). Representative images are shown (with photon counts given in white, and S:N in yellow). At this low S:N, JD localization yields less error than other methods. Arrow: condition used in Fig. 4. Error over the full range of S:N is shown in Supplemental Fig. 2. The effects of window size and offset of emitter from window center are shown in Supplemental Fig. 3.
Now consider Fig. 3(b). By default, σi is the width of the Gaussian that emulates the microscope’s PSF. As an emitter is most likely to lie in the brightest pixel, we expand distributions from other pixels (in the cartoon, the outer blue halo expands to give the outer dilated red one); distributions from pixels lying progressively further away from the brightest are expanded progressively more (Methods, Eqs. (4) and (5)). Distributions from the brightest pixel and its immediate neighbors remain unchanged (in the cartoon, the central blue halo gives an unchanged red halo). JD now yields up to 36% less error than CM (Fig. 3(b)); however, this comes at the price of higher error at higher signal-to-noise ratios (Supplemental Fig. 2, left).
We now combine both strategies. It turns out that an x-y shift in μi of ¼ pixel (not ½ pixel as in Fig. 3(a)) coupled with σi broadening (as in Fig. 3(b)) realizes up to 42% less error than CM – and 51% less than MLE – at S:N = 1.6 (Fig. 3(c)). Note that MLS begins to break down at S:N = 2.4 and fails completely below S:N = 1.9, while MLE never performs the best in this noisy region. In conclusion, this ‘tuned’ version of JD exhibits less error than (i) MLS at S:N < 2.4 and 5 < S:N < 38, (ii) MLE at S:N < 2.7, and (iii) CM at 1.3 < S:N < 24 (see also Supplemental Fig. 2, right (Media 1)).
Efficacy of localization algorithms is known to vary with image size [7] and the position of the spot within the image [10]; for example, at low S:N, CM favors the geometric center of the image. Therefore, we assessed the effects of reducing the size of the image window (from 15×15 to 7×7 pixels) and the position of the emitter relative to the center of the window (by up to 4 pixels), and found that the tuned version of JD still performs better and more robustly than the others under noisy conditions (Supplemental Fig. 3).
Computation speed
To assess computation speed, we compared (using windows with 15×15, 13×13, and 10×10 pixels) the number of 2-D localizations per second using images with two S:N ratios (indicated by arrows in Figs. 2(c) and 3(c)). As expected, higher S:N inevitably favors fast solution by the two iterative approaches (MLE and MLS), but both were slower than the ‘optimized’ and ‘tuned’ versions of JD, and much slower than CM (Fig. 4).
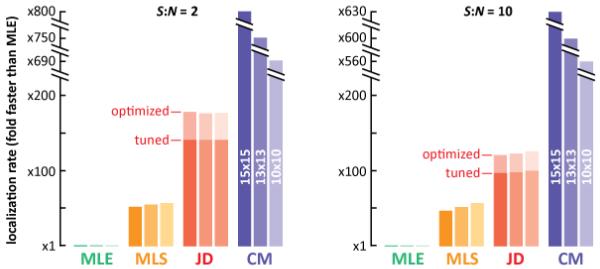
Fig. 4.
Computation speeds of the different methods (expressed relative to that of MLE). The times taken by the different methods to compute 2-D localizations were determined using 104 computer-generated images (15×15, 13×13, or 10×10 pixels) using conditions at the points indicated by the arrows in Figs. 2(c) and 3(c) (either 183 photons, b = 10, and S:N = 2; or 1,000 photons, b = 10, and S:N = 10). JD was tested using both ‘optimized’ and ‘tuned’ versions. Our MLS script gave 390 localizations per second with 15×15-pixel images, which is even faster than other reports on comparable computers [7]; it was also 50-times faster than an MLE script written by others [5] but implemented by us. JD applied using the ‘optimized’ conditions was 120- to 180-times faster than MLE, and the ‘tuned’ version was 100- to 140-times faster than MLE. As expected, CM (applied with background correction) proved the fastest, but both JD versions were faster than the two iterative techniques.
Localization using images of biological samples
We next compared performance of the four approaches using two kinds of images of biological samples; unfortunately, the true location of fluors in both samples cannot be known, so only qualitative comparisons can be made.
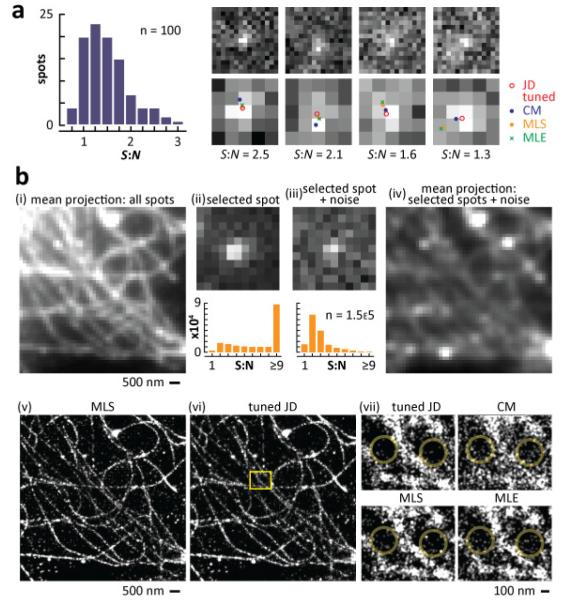
In the first example, RNA fluorescence in situ hybridization (RNA FISH) was used to tag, with Alexa 467, a nascent RNA molecule at a transcription site in a nucleus; then, images of the resulting foci were collected using a wide-field microscope. One-hundred images with a S:N < 3 were chosen manually, passed to the four algorithms, and the resulting localizations superimposed on each image; typical results are illustrated (Fig. 5(a); Media 2 gives results for all 100 spots). Visual inspection suggests that the tuned version of JD performs at least as well as, if not better than, the other methods.
Fig. 5.
Localization using ‘real’ images of transcription sites and microtubules in monkey cells (cos-7). (a) Nascent RNA at transcription sites. Cells expressing an EGFP gene containing an intron were fixed, and (nascent) EGFP transcripts detected using RNA FISH with probes targeting a short (sub-resolution) segment of the intron; images were collected using a wide-field microscope and CCD (90-nm pixels). One-hundred spots with S:N < 3 (histogram) were chosen manually, and four examples are shown at the top; the panels below illustrate the central 5x5 pixels in the upper panels, with 2-D localizations obtained by the different methods. As S:N decreases (left-to-right), localizations become more scattered (see Media 2 for results with all 100 spots). (b) Microtubules. Cells were fixed, microtubules indirectly immuno-labeled with Alexa 647, and a series of 30,000 images of temporally- and spatially-separated spots of one field collected using inclined illumination and an EM-CCD (155-nm pixels); 154,040 windows (11×11 pixels) containing 1 centrally-located spot were selected for analysis (using a Gaussian spot-finding algorithm). (i) Mean projection of all windows. (ii) One representative window (the histogram below illustrates the number of windows with different S:N). (iii) Individual windows were deliberately corrupted with noise (typical example and histogram shown). (iv) Mean projection of all resulting windows. Individual windows were now passed to each of the four methods, and localizations convolved with a 20-nm Gaussian intensity profile to aid visualization. (v, vi) Localizations obtained by MLS and the tuned version of JD yield roughly equivalent images. (vii) Magnified areas of the inset in (vi). Large circles in JD images contain fewer isolated results than the others, consistent with fewer mis-localizations (see also Supplemental Fig. 4(d)).
Microtubules imaged using direct STORM (dSTORM) [17] provide the second example. Tubulin in fixed cells was indirectly immuno-labeled with Alexa 647, 3×104 images of temporally- and spatially-separated single flours in the same field collected, and 1.5×105 windows (11×11 pixels) containing 1 centrally-located spot selected for analysis using a Gaussian spot-finding algorithm (Fig. 5(b)i illustrates a mean projection of all windows). A typical window contained one spot with S:N > 9 (Fig. 5(b)ii). Individual windows were then deliberately corrupted with a known level of noise (Fig. 5(b)iii and iv) – in this case to reduce S:N to less than 3 (after noise was added, spots had a mean S:N of 2.8 and a 71% had a S:N < 3). [See Supplemental Fig. 4 (Media 1) for a comparison of results obtained using uncorrupted and corrupted windows.] Despite the noisy images, all but 14% of spots are still detected by our simple spot-finding algorithm (not shown; spots found by the algorithm had a mean S:N of 2.8, and 71% had a S:N < 3, so the 14% were missed as a result of random chance and not low S:N). All windows were passed to the four algorithms, and localizations convolved with a 20-nm Gaussian intensity profile to aid visualization. MLS (chosen as an example because it is used most-often during the formation of STORM images) and the tuned version of JD yield roughly equivalent images (Fig. 5(b)v and vi), although analysis of nearest-neighbor distances indicates JD returns the most highly-structured images (Supplemental Fig. 4(g)). It also yields fewer isolated results than the others (yellow circles in Fig. 5(b)vi), which we assume are mislocalizations resulting from poor performance. We again conclude that JD performs better with noisy images than methods used traditionally.
Discussion
During the application of ‘super-resolution’ techniques like PALM and STORM, photons emitted from a point source pass through a microscope to yield an image on a CCD where they are registered by many pixels. Successful localization of the point source then depends on two critical steps. First, the pixelated ‘spot’ must be distinguished from others and the inevitable background; we have not studied this step (we apply it only in Fig. 5(b) where we rely on a cross-correlation-based ‘spot-finding’ algorithm to identify spots with S:N < 3). Second, the position of the point-source must be deduced using the relevant information in the isolated pixels. We introduce a method for performing this second step. Existing methods (e.g., MLE, MLS, and CM) inaccurately assume the probability of the location of each emitted photon is uniformly distributed over just one pixel; in contrast, our method represents this uncertainty as a normal distribution that spreads over several pixels (Fig. 1(a)). We then aggregate many probability distributions to yield a joint distribution (JD) of the probability of the location of the emitter (Figs. 1(b) and 1(c)).
Localization by JD has the advantage of flexibility; each individual probability distribution is defined solely by peak center (μi) and width (σi), and both can be tuned to improve precision to meet the needs of a particular experiment (Fig. 3). We anticipate that additional tuning of μi and σi (e.g., as functions of pixel intensity), and further optimization (e.g., of the rate at which σi increases as a function of distance) – will improve precision even further. In images where the PSF deviates from the ideal, different tuning parameters might maximize precision. Moreover, the use of smaller pixels should also increase precision, as μi could then be assigned more precisely. This can be accomplished, in spite of traditional knowledge that reducing pixel size decreases precision [18], by applying distributions that represent the PSF to each detected photon and summing overlapping regions to form complex images [11]. [Here, images have 90-nm pixels so as to meet the Nyquist criterion for a PSF with a 250-nm full width half maximum. Preliminary simulations indicate that a reduction in pixel width to 1 nm reduces the 1-D RMSE in localization by an additional 3%.]
All versions of JD provide computational simplicity and speed because emitter location is not calculated iteratively. Furthermore, all adeptly localize in windows with non-uniform background, as broadening individual distributions negates the influence of bright pixels distant from the brightest. They are also readily extended to both 3D localization (given a Gaussian-like PSF in the axial dimension, computation of a third dimension is straightforward because each axis is treated independently) and more than one color – and so to real-time imaging deep within living specimens. Nevertheless, they have several disadvantages. First, unlike the two fitting algorithms that ‘re-check’ spots selected by a spot-finding algorithm for an appropriate Gaussian intensity profile, JD (and CM) provide no such back-up. [Tests of various spot-finding algorithms suggest that local-maxima techniques are liable to return multiple spots in one window, but 2-D normalized cross-correlation with a Gaussian kernel robustly selected single spots from dSTORM data (not shown).] Second, the initial disaggregation of pixel intensity into individual photons followed by the aggregation of individual probabilities into a joint distribution inevitably makes JD slower than CM. Third, the greatest gains at low S:N (from the ‘tuned’ version) come at the cost of precision at high S:N. Fourth, the JD scheme fails completely when the brightest pixel in a window does not contain the emitter.
We compared accuracy and speed of localization achieved by various methods using images with a wide range of noise, and find that each has its own advantages and disadvantages (Figs. 2-4). Although widely used [19], we suggest MLS should rarely, if ever, be the algorithm of choice. At high signal-to-noise ratios, MLE – though the slowest – is the most accurate (as reported by others [3-5, 7, 15]). At the highest signal-to-noise ratios, CM is only marginally less accurate than MLE; at signal-to-noise ratios > 10, CM offers greater precision than MLS. [A variant of CM involving a limited number of iterative computations is even more accurate than the basic version [20].] If temporal resolution is of the greatest concern (e.g., during real-time computation), CM is by far the fastest (Fig. 4), and its simplicity makes it attractive to groups lacking sophisticated analysis software. [Other closed-form solutions also produce fast results, but at the cost of precision [6, 21]]. Most PALM/STORM images currently being analyzed have a S:N > 9 (as in the uncorrupted spot in Fig. 5(b)ii), where MLE yields the highest precision (13-27% and 16-0.5% less 1-D RMSE per pixel than MLS and CM, respectively). However, as the signal-to-noise ratio falls, both MLE and MLS fail to converge to a solution during the 200 iterations used, or yield an error > 1 pixel; then, the tuned version of JD becomes the most accurate. For example, when S:N = 1.6, the tuned version returns 42% less 1-D RMSE per pixel than CM, and offers a 2-fold improvement over MLE (Fig. 3(c)) – both significant increases in precision. Both versions of JD are also two orders of magnitude faster than MLE – again a significant increase (Fig. (4)).
In conclusion, we see no obstacles that might hinder the immediate adoption of JD for ‘super-resolution’ localization at low S:N; it allows use of spots in the noisier parts of the image that are now being discarded from data sets used to form PALM/STORM images, and will permit super-resolution imaging at the noisier depths of cells and tissues. We suggest that the signal-to-noise ratio be measured prior to localization to determine the best method to use. Then, if precision in location is the goal, MLE should be used at high ratios, and the tuned version of JD a low ratios. As the signal-to-noise ratio in any PALM/STORM image stack varies within one frame, and from frame to frame, the very highest precision can only be achieved by applying MLE and/or an appropriately-tuned version of JD to each spot depending on the immediate surroundings. Alternatively, if computation speed is paramount, we suggest CM be used because the gains realized by MLE over CM at high S:N are small, and the resulting STORM images are reasonably accurate (see Supplemental Fig. 4(b), (Media 1)). Finally, the ‘optimized’ version of JD provides a ‘one-size-fits-all’ compromise between simplicity, precision, and speed, which is more precise and faster than existing methods.
Methods
Computer and software specifications
Computations were conducted on a standard desktop PC (2.83 GHz ‘Core2 Quad’ CPU, Intel; 8 GB RAM; 64-bit Windows 7) using software written, compiled, and executed in MATLAB (Mathworks version 7.9.0.529; R2009b) without parallel computing. Software for implementing both ‘tuned’ and ‘optimized’ versions of JD is provided in Supplemental Material.
Image generation and analysis
To permit accurate measurements of precision, simulations were run on computer-generated images with known emitter locations. Except where specified otherwise, images contained 15×15 90-nm pixels (PSF FWHM = 250 nm, oversampled by 2.78, resulting pixel width = 90 nm). An ‘emitter’ was placed randomly (with sub-nanometer precision) anywhere in the central 1/9th of an image (i.e., in the 5×5 central pixels in a 15×15 image). Coordinates of ‘emitted photons’ were then randomly generated (again with sub-nanometer precision) using a 250-nm FWHM Gaussian distribution (the commonly-accepted representation of a PSF at the resolution limit of a microscope [1, 2, 4, 12, 14]), and photons binned into pixels to produce the final image. Where background noise was added, additional photon coordinates were randomly (uniformly) distributed over the entire image to obtain the average level indicated. Ten-thousand images were generated and analyzed for each data-point shown, except for those in Supplemental Fig. 3(b) where data from 104 images were sorted by distance into 0.1-pixel bins. Images were passed ‘blindly’ to localization algorithms, and the same image sets were analyzed by all methods. Images were also generated using an algorithm that first distributes photons normally in an image space, and then corrupts the image space with Poisson noise [7]. Both algorithms yield images that appear similar to the eye and result in identical localization error (not shown).
JD localization
JD begins by attributing different numbers of photon-events to each pixel using CCD intensity, and ascribing an individual (normal) probability distribution of emitter location to each photon (Fig. 1(c)). Such a distribution is uniquely described by center location (μi) and width (σi), and the default is to place μi at the pixel center and use σi equivalent to that of the PSF; alternatively, μi and/or σi can be varied. Here, we tune μi by shifting photons in all pixels (other than the brightest) towards the brightest one. Thus, in one dimension:
| (2) |
where μc is the location of the pixel center, di is the distance between the pixel and brightest pixel, So is pixel width, and C is an arbitrary scaling constant (in Fig. 3(a) C = 2, and in all other cases C = 4; 12 values of C between 1 and 5 were tested, and C = 4 yielded the highest precision under the conditions described in Fig. 3(c)). Distributions from the brightest pixel are also shifted from the central default location by a distance proportional to the intensities of adjacent pixels. In the x-dimension:
| (3) |
where Io is the intensity of the brightest pixel and IR and IL are the intensities of the adjacent pixels to its right and left, respectively (a similar shift is applied in the y-dimension relative to the pixel intensities above and below). We also tune σi as a function of distance from the brightest pixel. Thus, in one dimension, x:
| (4) |
Here, σPSF is the width of the PSF in terms of sigma, μmax is the center location of the brightest pixel, 2.5 is scaling factor chosen such that σi = σPSF for the maximum pixel, and g(x) is a piecewise Gaussian distribution function with a flat top:
| (5) |
Together, Eqs. (4) and (5) ensure that the distributions of photons from, and adjacent to, the brightest pixel are equal to the PSF, those from surrounding pixels become progressively wider the further away the pixels are, and those from distant pixels (i.e., > 3 σPSF) become infinitely wide (see Supplemental Fig. 5 for plots of these functions). To increase computation speed in the ‘optimized’ version of JD, the term 1/2.5g(x) in Eq. (4) is replaced with σPSF, and μi = μc.
Probabilistically speaking, individual probability distributions are random variables, independent and normally distributed. To infer the location of the emitter, individual probabilities are aggregated as a joint density, which is also normally distributed [22]. Given N variables, the joint density function in one dimension is:
| (6) |
Ignoring constant terms for simplicity (because they only affect the amplitude of the function, which is not of immediate interest here), we get:
| (7) |
Next we define:
| (8) |
so that
| (9) |
Factoring and, once again, ignoring constant terms:
| (10) |
After replacing k1 and k2, the joint distribution takes the form
| (11) |
which resembles a simple Gaussian distribution:
| (12) |
whose center is equal to the center of the joint distribution:
| (13) |
Projection and inference are repeated for each orthogonal axis. The width of the joint distribution does not provide a reliable estimate of localization precision, presumably because it does not account for effects of background noise.
To get to the CM equation from Eq. (13), first we must set σi to a constant for all photons, that is
| (14) |
The total number of photons in the image, N, can be rewritten for an m × n matrix as the sum of pixel intensities, I,
| (15) |
We then set μi equal to pixel center positions, xi:
| (16) |
Finally, the sum of all photons is equal to the sum of pixel intensities. This yields the estimated emitter location in one dimension, Cx:
| (17) |
which is equal to Eq. (3) in Cheezum et al. [10]. Another, more general, form of this equation would be required to incorporate weighting values equivalent to those implemented in the ‘tuned’ version of JD localization.
Background correction
MLE and MLS inherently correct for background, as background level is an intrinsic fitting parameter. In the presence of increasing background, emitter location estimated by CM and JD progressively diverge from the true location towards the geometric center of the image; therefore, high precision can only be achieved using these methods if background correction is included. For CM, a standard background correction [10] is used prior to localization: a noise threshold is defined (as the mean intensity plus two standard deviations in the two peripheral pixels around the circumference, which includes the 104 peripheral pixels in a 15×15 image) and subtracted from the intensity of every pixel in the image. Another background-correction algorithm tested (i.e., setting all pixels with intensity less than the threshold to zero and leaving the remaining pixels unaltered) did not perform as well (not shown). For the default version of JD, we first consider those pixels at or below the threshold (estimated as for CM); σi of their distributions is set to infinity, reducing amplitude to zero and negating any effect on localization. Then we consider pixels with intensity above the threshold; σi is set to infinity for the proportion of distributions corresponding to the fraction of intensity below the threshold. For the tuned and optimized versions of JD, background is removed similarly (note that distributions coming from ‘non-spot’ pixels with intensities between the noise ceiling and the brightest also have σi set to infinity in Eqs. (4) and (5).
Localization by CM, MLS, & MLE
CM was computed with an in-house program (described by Cheezum et al. [10]) as the mean of the locations of all pixel centers in the window weighted by their respective intensities. MLS fitting to a 2-D Gaussian intensity profile was also computed with an in-house program. Peak amplitude, background level, x- and y-width, plus x- and y-location were set as fitting parameters. Regression continued until changes fluctuated < 0.01% or until 200 iterations elapsed; when a solution was found, in most cases it was found within 10 iterations. MLE of a 2-D Gaussian intensity profile was implemented directly, as provided by others [5]. Neither fitting algorithm yielded a smooth line at low S:N in Fig. 3 even though 104 measurements were made for each data point; therefore, plots were smoothed by linear regression.
Precision measurements
Post-localization, estimates were compared with true locations of emitters and root-mean-squared errors (RMSE) computed. Signal-to-noise ratio is computed in different ways throughout the literature. Here,
| (18) |
where Io is the maximum pixel intensity, b is the background – the mean intensity of the two concentric sets of peripheral pixels in the image (i.e., the 104 peripheral pixels in a 15×15 image) – and Nb is the RMS intensity of the same peripheral pixels. This computation is as in Cheezum et al. [10], with two differences: (i) signal was measured as the maximum pixel intensity (instead of mean spot intensity) because images were generally so noisy, and (ii) noise was sampled from peripheral pixels (not across the whole image) to assess better the degree to which signal stands above fluctuations in background.
Speed test
All computations to assess speed were conducted serially on the same set of 1,000 images in this order: ‘optimized’ version of JD, ‘tuned’ version of JD, CM, MLS, and MLE. To test the possibility that residual computer memory loss retarded sequential computations, computations were repeated in reverse order and yielded identical results. The derivatives for MLS were computed by hand and implemented as linear equations to avoid built-in MATLAB functions known to be slow. The numbers of localizations/sec from our routine were compared with those reported by Smith et al. [7] (obtained using least-squares fitting on a single processor), and are similar (not shown). The mean computation rate of three independent trials is reported (in Fig. 4, standard deviations were < 1% in all cases).
RNA FISH images
Nuclear transcription sites containing nascent (intronic) RNA were detected using RNA FISH. Monkey kidney cells (cos-7) were transiently transfected with a plasmid encoding an EGFP gene (as in Xu and Cook [23]) with an intron containing sequences derived from intron 1 of human SAMD4A. One day after transfection, cells were seeded on to a coverslip etched with 0.1% hydrofluoric acid, and re-grown; 40 h post-transfection, cells were transferred to ‘CSK buffer’ for 10 min, and fixed (4% paraformaldehyde; 20 min; 20°C) [24]. Nascent (intronic) SAMD4A RNA was then detected by RNA-FISH using 50-nucleotide probes each tagged with ~5 Alexa 647 fluors (as in Papantonis et al. [25]). After hybridization, cells were mounted in Vectashield (Vector Laboratories) containing 1 μg/ml DAPI (4,6-diamidino-2-phenylindole; Sigma), and imaged using a Zeiss Axiovert microscope (63×/1.43 numerical aperture objective) equipped with a CCD camera (CoolSNAPHQ, Photometrics). Sub-diffraction spots marking nuclear transcription sites with a S:N < 3 were selected manually for analysis.
dSTORM Images
Direct STORM (dSTORM) images were kindly provided by S. Van De Linde [8]. Microtubules in fixed cos-7 cells were indirectly immuno-labeled with Alexa 647, and 30,000 images (excitation at 641 nm under inclined illumination, emission recorded between 665 and 735 nm) of spatially-separated sub-diffraction sized spots in one field collected (image acquisition rate 885 s−1) using an EM-CCD camera (Andor; EM-gain = 200; pre-amp-gain = 1). Spots were identified by 2-D cross-correlation with a randomly-generated 2-D Gaussian intensity pattern, and candidates for fitting selected by a minimum cross-correlation value. 154,040 windows (11×11 pixels) containing 1 spot were selected, and independently corrupted with noise until S:N measured < 3; then each window was passed to each of the four localization algorithms (Supplemental Fig. 4(a), (Media 1)). Localization results were rounded to the nearest nanometer, and used to reconstruct an image of the whole field using 1-nm pixels. To aid visualization, each of the resulting images was convolved with a 2-D Gaussian intensity profile with a 20 nm FWHM. Contrast and brightness of all images displayed are equal between methods.
Supplementary Material
Acknowledgments
We thank N. Publicover, J. Sutko, A. Papantonis, S. Baboo, B. Deng, J. Bartlett, S. Wolter, and M. Heilemann for discussions, and S. Van De Linde for dSTORM data. This work was supported by the Wellcome Trust (grant 086017).
Footnotes
OCIS codes: (180.2520) Fluorescence microscopy; (100.6640) Superresolution; (100.4999) Pattern recognition, target tracking.
References and links
- 1.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3(10):793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ober RJ, Ram S, Ward ES. Localization accuracy in single-molecule microscopy. Biophys. J. 2004;86(2):1185–1200. doi: 10.1016/S0006-3495(04)74193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham AV, Ram S, Chao J, Ward ES, Ober RJ. Quantitative study of single molecule location estimation techniques. Opt. Express. 2009;17(26):23352–23373. doi: 10.1364/OE.17.023352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortensen KI, Churchman LS, Spudich JA, Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods. 2010;7(5):377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedde PN, Fuchs J, Oswald F, Wiedenmann J, Nienhaus GU. Online image analysis software for photoactivation localization microscopy. Nat. Methods. 2009;6(10):689–690. doi: 10.1038/nmeth1009-689. [DOI] [PubMed] [Google Scholar]
- 7.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods. 2010;7(5):373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolter S, SchãoeTtpelz M, Tscherepanow M, Van De Linde S, Heilemann M, Sauer M. Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 2010;237(1):12–22. doi: 10.1111/j.1365-2818.2009.03287.x. [DOI] [PubMed] [Google Scholar]
- 9.Holden SJ, Uphoff S, Kapanidis AN. DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat. Methods. 2011;8(4):279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- 10.Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 2001;81(4):2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin JD, Publicover NG, Sutko JL. Photon event distribution sampling: an image formation technique for scanning microscopes that permits tracking of sub-diffraction particles with high spatial and temporal resolutions. J. Microsc. 2011;241(1):54–68. doi: 10.1111/j.1365-2818.2010.03406.x. [DOI] [PubMed] [Google Scholar]
- 12.Winick KA. Cramer-Rao lower bounds on the performance of charge-coupled-device optical position estimators. J. Opt. Soc. Am. A. 1986;3(11):1809–1815. [Google Scholar]
- 13.Elsaesser Derek. The discrete probability density method for emitter geolocation; Canadian Conference on Electrical and Computer Engineering; CCECE ’06 (IEEE, 2006). 2006.pp. 25–30. [Google Scholar]
- 14.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurence TA, Chromy BA. Efficient maximum likelihood estimator fitting of histograms. Nat. Methods. 2010;7(5):338–339. doi: 10.1038/nmeth0510-338. [DOI] [PubMed] [Google Scholar]
- 16.Löschberger A, van de Linde S, Dabauvalle M-C, Rieger B, Heilemann M, Krohne G, Sauer M. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 2012;125(3):570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- 17.Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 18.Pawley JB. Handbook of Biological Confocal Microscopy. 3rd ed. Springer; 2006. Points, pixels, and gray levels: digitizing image data. [Google Scholar]
- 19.Larson DR. The economy of photons. Nat. Methods. 2010;7(5):357–359. doi: 10.1038/nmeth0510-357. [DOI] [PubMed] [Google Scholar]
- 20.Berglund AJ, McMahon MD, McClelland JJ, Liddle JA. Fast, bias-free algorithm for tracking single particles with variable size and shape. Opt. Express. 2008;16(18):14064–14075. doi: 10.1364/oe.16.014064. [DOI] [PubMed] [Google Scholar]
- 21.Andersson SB. Precise localization of fluorescent probes without numerical fitting; 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2007; ISBI 2007. 2007; pp. 252–255. [DOI] [PubMed] [Google Scholar]
- 22.Hoel PG. Introduction to Mathematical Statistics. 5th ed Wiley; 1984. [Google Scholar]
- 23.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 2008;181(4):615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam R, Shopland LS, Johnson CV, McNeil JA, Lawrence JB. Fish. 1st ed Oxford University Press; USA: 2002. Applications of RNA FISH for visualizing gene expression and nuclear architecture; pp. 93–118. [Google Scholar]
- 25.Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol. 2010;8(7):e1000419. doi: 10.1371/journal.pbio.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.