Abstract
Bacteriophages are central components in the development of molecular tools for microbial genetics. Mycobacteriophages have proven a rich resource for tuberculosis genetics, and the recent development of a mycobacterial recombineering system based on phage Che9c-encoded proteins offers new approaches to mycobacterial mutagenesis. Expression of the phage exonuclease and recombinase substantially enhances recombination frequencies in both fast-and slow-growing mycobacteria, facilitating construction of both gene knockout and point mutants; it also provides a simple and efficient method for constructing mycobacteriophage mutants. Exploitation of host-specific phages thus provides a general strategy for recombineering and mutagenesis in genetically naive systems.
Introduction
Genetic manipulation of Mycobacterium tuberculosis is complicated by its pathogenesis, slow growth rate, inefficient DNA uptake, and relatively high level of illegitimate recombination. Construction of gene knockout mutants can be achieved using plasmid-based systems and by phage-mediated specialized transduction, but the simple introduction of a linear double-stranded DNA (dsDNA) substrate by electroporation leads to a high proportion of ectopic integration events regardless of homology between the targeting substrate and the bacterial chromosome1, 2. Other types of mutagenesis such as the construction of defined isogenic strains with single point mutations are further complicated by the lack of generalized transducing phages that infect M. tuberculosis3.
A variety of techniques for constructing gene knockout or replacement mutants have been described which are designed to overcome the relatively high level of illegitimate recombination compared to homologous recombination observed with linear dsDNA allelic exchange substrates (AESs). These include the use of long linear dsDNA substrates4, as well as various plasmid and cosmid-based strategies4–8, and these are often coupled with the use of counter-selectable genetic markers such as sacB9, 10. Specialized transducing phages based on conditionally-replicating shuttle phasmids represent an alternative and highly effective approach to mutagenesis11, including the construction of isogenic M. tuberculosis strains differing by a single known point mutation12. While most of these methods are effective, they are constrained by the requirements for complex genetic constructions and/or multiple steps of manipulation and screening. Given the extremely slow growth rate of M. tuberculosis (doubling time of 24 hours), alternative methods that simplify its genetic manipulation are highly desirable.
Recombineering – genetic engineering using recombination proteins13 – is a powerful system for mutagenesis in Escherichia coli (as well as Salmonella and Shigella14, 15) in which recombination systems encoded by the lambda Red system or the RecET genes of the Rac prophage massively enhance the frequencies of homologous recombination. The high recombination efficiencies can be exploited in a variety of ways including the construction of chromosomal gene knockouts, point mutations, deletions, small insertions, in vivo cloning, and mutagenesis of bacterial artificial chromosomes and genomic libraries13, 16–19. Only relatively short segments (50 bp) of DNA homology are required, and mutagenic substrates constructed by PCR can be readily introduced into E. coli strains expressing the phage recombination proteins by electroporation.
Recombineering in E. coli using the λ Red system involves three phage-encoded proteins, Exo, Beta, and Gam, whereas the Rac prophage encodes just RecE and RecT, which are functionally equivalent to λ Exo and Beta, respectively20, 21. Exo and RecE are 5′→3′ exonucleases that degrade a single strand of a linear dsDNA substrate22, 23, exposing a 3′ single-stranded DNA (ssDNA) tail to which Beta or RecT can bind24, 25. Recombination is then mediated by association of these complexes with resident chromosomal or plasmid targets through pairing of complementary sequences, strand exchange, or strand invasion26–29. λ Beta and RecT proteins are members of a large family of single-stranded DNA annealing proteins (SSAPs)30; two other families of SSAPs that function similarly are defined by the P22 Arf protein and the eukaryotic protein, Rad52. While the RecET and λ Red systems confer similar levels of recombination in E. coli, the Beta and RecT proteins have a strong preference for their cognate Exo and RecE proteins, and it is likely that they function as protein complexes31, 32. While E. coli dsDNA recombineering requires both an exonuclease and its associated SSAP, recombination using ssDNA substrates requires only the SSAP. The ability to recombineer with short oligonucleotide-derived ssDNA substrates is especially useful19, and frequencies can be sufficiently high to enable identification of mutants without the need for direct genetic selection, especially when using strains defective in mismatch repair34. The third protein in the λ system, Gam, increases recombineering frequencies by binding to the RecB subunit of RecBCD and inhibiting degradation of dsDNA substrates 35, 36,18, 19. While other Gam functional analogues have been characterized, such as Abc2 of bacteriophage P2237, these proteins are more rare compared to the large superfamilies of SSAPs and their associated exonucleases30.
Mycobacteriophages have played key roles in circumventing the challenges of genetics in M. tuberculosis, including the development of shuttle phasmids38 for transposon mutagenesis39, reporter gene delivery40, 41, and specialized transduction11, as well as integration-proficient vectors for stable introduction of foreign genes42–45. Exploitation of these phages is simplified by the availability of more than 50 completely sequenced mycobacteriophage genomes44–47, and the amazingly high degree of genetic diversity provides at least 1,500 phamilies of unique genes. Because the E. coli based systems appear to function less well in distantly related bacteria, we turned to this collection of mycobacteriophage genomes to identify mycobacteriophage-encoded proteins that could be developed for mycobacterial recombineering. Although RecET homologues are rare among mycobacteriophages, Che9c encodes both proteins, and we have exploited them to develop recombineering systems for both fast-and slow-growing mycobacteria48, 49. This approach also provides a powerful strategy for constructing mutant derivatives of lytically-replicating mycobacteriophage genomes, including in-frame deletions, point mutations and small insertions.
Identification of mycobacteriophage recombineering functions
Mycobacteriophages are genetically diverse, possess architecturally mosaic genomes44–47, and are replete in predicted open reading frames of unknown function that have no detectable similarity to known proteins. While constructing shuttle phasmid derivatives of phage TM4, Jacobs and colleagues38 noted the high prevalence of recombinants following electroporation of phage libraries into M. smegmatis, suggesting the presence of a phage-encoded recombination system. However, bioinformatic analysis of the TM4 genomic sequence50 provides no clues as to which genes might encode recombination functions.
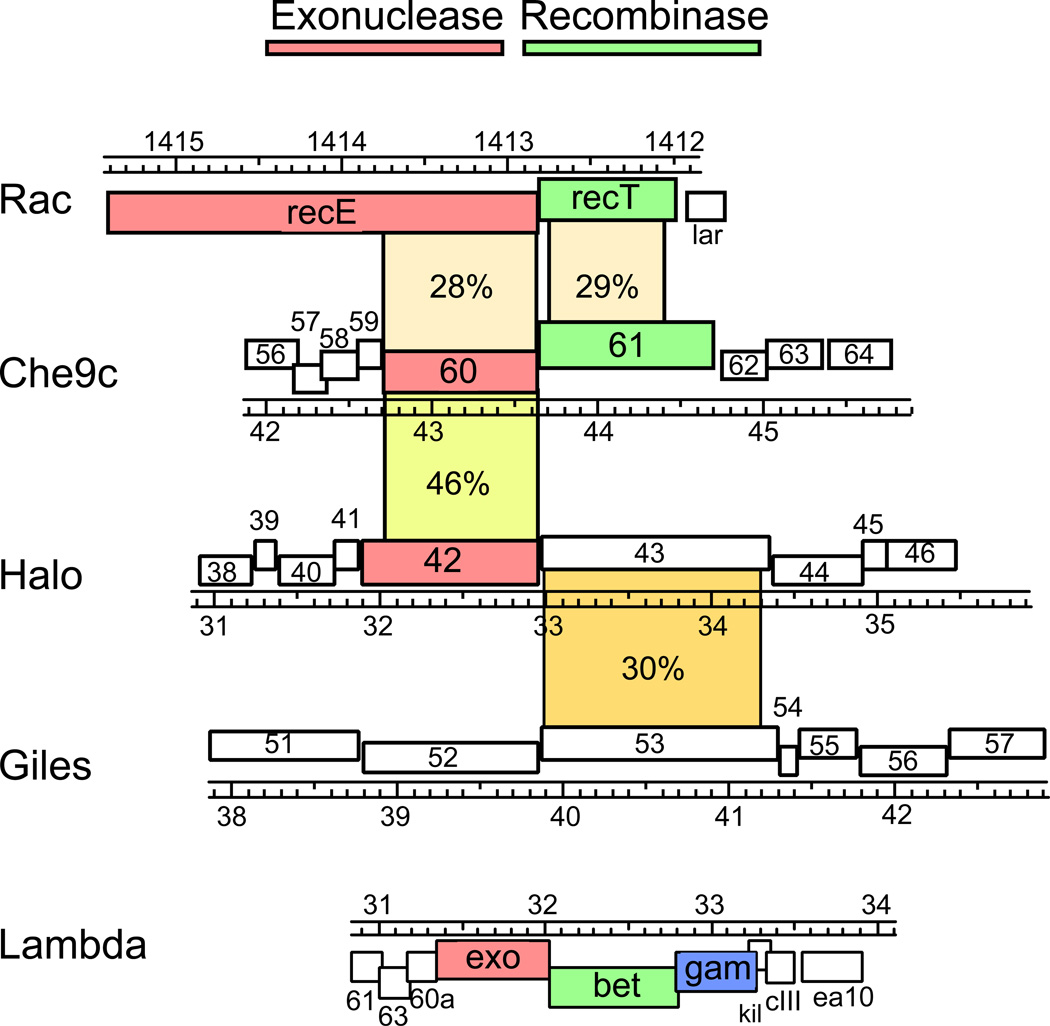
A search of the predicted open reading frames of all completely sequenced mycobacteriophage genomes reveals a small number of homologues of the E. coli RecET proteins but none related to the λ Red proteins. Che9c encodes for homologues of both RecE and RecT, the products of genes 60 and 61 respectively (Fig. 1a)47, 48; both are rather distant relatives and share less than 30% amino acid identity. Che9c gp60 (314 aa) is also much smaller than E. coli RecE (866 aa) and corresponds to just the C-terminal part encompassing the RecB-family nuclease domain (Fig. 1a); the N-terminal region of RecE that is absent in Che9c gp60 is not necessary for RecE exonuclease activity51. Che9c gp61 is clearly a member of the superfamily of SSAPs30 with 29% amino acid identity to E. coli RecT, but with a longer C-terminal extension (Fig. 1a). Although Che9c gp60 and gp61 are distant relatives of the RecET and λ Exo/Beta proteins, they possess similar biochemical functions48. Che9c gp60 has exonuclease activity in vitro that is dependent on the presence of dsDNA ends, and Che9c gp61 binds short (20 nucleotides) ssDNA as well as dsDNA substrates in the absence of Mg2+. It was also observed by electron microscopy to form toroidal multimers in the presence of ssDNA (Fig. 1b), a property exhibited by other SSAPs52, 53.
Figure 1. Mycobacteriophage-encoded recombination proteins.
A. Mycobacteriophages Che9c gp60 and gp61 are E. coli Rac prophage RecE and RecT homologues, respectively. Mycobacteriophage Halo encodes a RecE homologue that is 46% identical to Che9c gp60, while Giles gp52 belongs to the YqaJ family of exonucleases. Halo gp43 is similar to Giles gp53, but these are only distantly related to other phage RecT-like proteins. The λ Red recombination proteins are not closely related to the mycobacteriophage proteins or RecET.
B. Electron micrograph of Che9c gp61 protein multimers in the presence of ssDNA. Reactions containing gp61 protein (1.2 µM) incubated with ssDNA (100 nt; 2 µM) were absorbed to glow discharged 400 mesh formvar carbon coated copper grids, stained with 2% uranyl acetate, and examined by transmission electron microscopy. Images were collected at a magnification of 140,000×; four examples of toroid structures are shown alongside a size bar.
Two other mycobacteriophages, Halo and Giles, encode similar recombination systems. Halo gp42 is 46% identical to Che9c gp60 and 30% identical to RecE, while Giles gp52 contains a domain from the YqaJ family of phage-encoded exonucleases (Fig. 1a). However, while Halo gp43 and Giles gp53 are 30% identical, they are even more distantly related to other phage-encoded RecT homologues than Che9c gp61 (Fig. 1a). Interestingly, a prophage in the sequenced genome of M. avium strain 104 also encodes RecE/RecT-like proteins (MAV_0830 and MAV_0829, respectively) that are similar to Che9c gp60 and gp61 (41% and 29% identical) and also related to the Rac prophage proteins RecE and RecT (23% and 40% identity). Thus while the mycobacteriophage putative SSAP proteins are in each case associated with a gene encoding an exonuclease, these systems exhibit the same modularity seen broadly in other phage genomes54. None of these mycobacteriophage systems encode homologues of the λ Gam protein, and it is unknown if there are any mycobacteriophage functional analogues that are RecBCD-inhibitors similar to the Abc2 protein of phage P22.
Construction of recombineering strains of mycobacteria
Mycobacterial strains have been constructed for recombineering with both dsDNA and ssDNA substrates. These contain an extrachromosomal plasmid in which the phage recombination genes are under the control of an inducible acetamidase promoter55, and the most widely used configurations use the Che9c genes 60 and 61 for dsDNA recombineering48 or just Che9c 61 for ssDNA manipulations49. Optimal levels of recombination are obtained using plasmid derivatives expressing Che9c genes 60 and 61 from their endogenous translational signals (e.g. pJV53), though it is noteworthy that while some alternative constructions may give higher levels of expression, this does not necessarily result in higher levels of recombineering48. This has also been observed for the λ Red system where recombination activity correlates poorly with expression levels31.
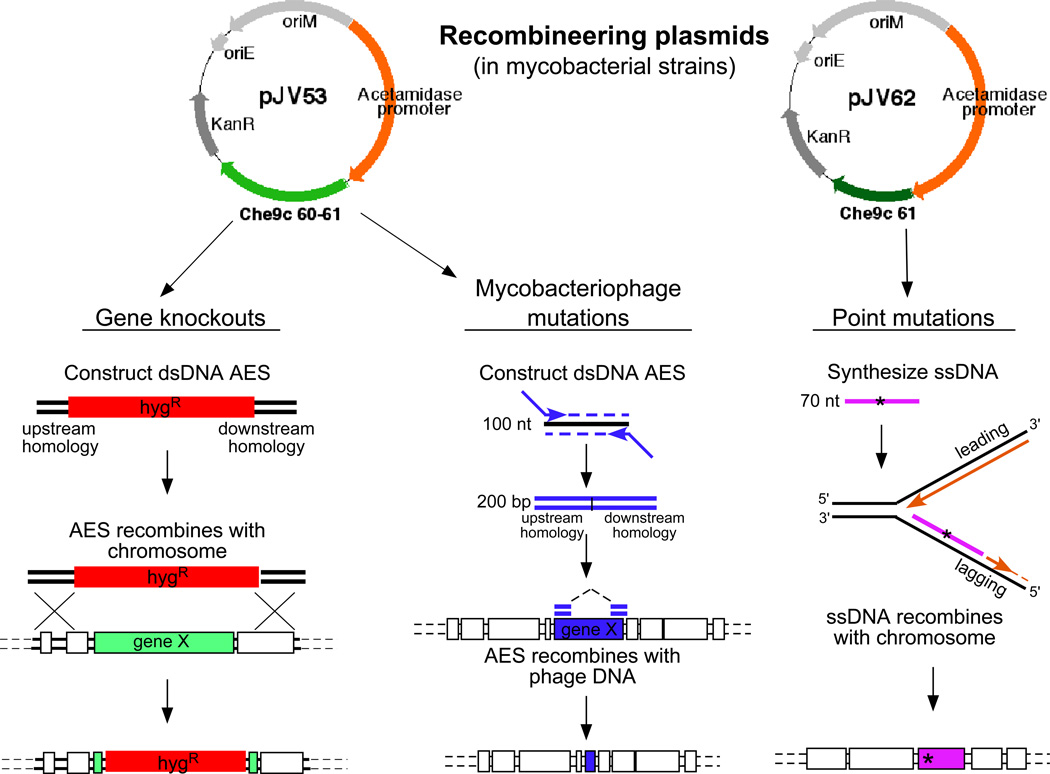
Plasmid pJV53 serves as a basic recombineering plasmid for regulated expression of gp60 and gp61 (Fig. 2), although other derivatives of this plasmid have been made, including those with different selectable markers or containing a sacB cassette to simplify removal of the plasmid by counter-selection following mutagenesis (plasmid pJV48). The acetamidase promoter is somewhat leaky, and there is a detectable level of Che9c gp61 expression in M. tuberculosis in some uninduced strains, although recombination frequencies are low without induction48. High expression levels of these proteins are toxic to M. smegmatis, and plasmids in which the constitutive Mycobacterium bovis BCG hsp60 promoter is linked to Che9c 60 and 61 do not transform mycobacteria. This toxicity appears to derive largely from the Che9c gp60 exonuclease since plasmids expressing only gp61 constitutively are tolerated, although they grow slowly. For ssDNA recombineering, plasmids such as pJV62 (Fig. 2) have been constructed that express gp61 under its own translation signals and the acetamidase promoter. In preparation for recombineering experiments, M. smegmatis or M. tuberculosis plasmid-containing strains in mid-logarithmic growth are induced with acetamide and harvested for electroporation48, 49.
Figure 2. Strategies for mycobacterial recombineering.
Plasmids pJV53 and pJV62 express Che9c gp60 and gp61 or just gp61 to facilitate dsDNA and ssDNA recombination respectively. Left: gene knockouts are made by targeted gene replacement with a linear dsDNA allelic exchange substrate (AES) that contains 500 bp of homology to the target locus flanking an antibiotic resistance gene (e.g. hygromycin; hygR). Middle: Mycobacteriophage mutants (either point mutants or deletion mutants) are made by synthesizing a 200 bp dsDNA AES by PCR using a 100 nt template and 75 nt extender primers. Right: point mutants constructed by synthesis of a 50–70 nt ssDNA substrate, centering the desired point mutation that will anneal to the lagging strand for DNA replication to undergo recombination.
Using dsDNA recombineering to construct mycobacterial gene knockout mutants
Targeted gene replacement mutants can be readily constructed by electroporation of linear dsDNA substrates into either M. smegmatis or M. tuberculosis recombineering strains (Fig. 2). The AESs typically contain regions of homology upstream and downstream of the target gene flanking a cassette for antibiotic resistance, such as hygromycin-resistance (HygR). Although SSAP recombinases such as gp61 can bind short regions of ssDNA, the frequency of recombineering is considerably lower with 50 bp than with 500 bp of homology, and with shorter lengths the DNA uptake frequency becomes limiting; we therefore recommend using a minimum of 500 bp of homology48. A simple protocol entails transformation of a linearized AES (100 ng) into electrocompetent mycobacterial cells containing pJV53 and induced for gp60/gp61 expression48,56. This typically yields 50–200 drug-resistant colonies of which >90% contain correctly targeted gene replacements; control transformations using 50 ng plasmid DNA with the same competent cells typically yields about 105 colonies. Similar frequencies are observed when targeting extrachromosomal plasmids, emphasizing the need to ensure that DNA with potential similarity to the pJV53 plasmid backbone (such as the E. coli origin of replication) is removed from the AES substrate. AESs can also be designed for subsequent unmarking of mutants, such as including γδ res sites flanking the antibiotic resistance cassette and a sacB cassette for counter selection11.
The dsDNA recombineering frequencies appear to be primarily limited by the relatively poor mycobacterial DNA uptake efficiencies rather than poor protein expression or degradation by host nuclease systems. With either M. tuberculosis57 or a high efficiency transformation M. smegmatis strain58 competencies as high as 1–5 × 106 transformants/µg plasmid DNA can be achieved48, 59, but only 1 in 1000 of all viable cells productively takes up DNA. Inactivation of the M. smegmatis recBCD system only modestly increases dsDNA recombineering frequencies, and expression of λ Gam does not influence the frequencies in any strain background that we have tested (our unpublished observations); mycobacterial recombineering is also RecA-independent as expected49. In practice, the recombination and DNA uptake frequencies with the Che9c system yield sufficient recombinants for constructing targeted gene replacement mutants in both M. smegmatis and M. tuberculosis.
Using ssDNA recombination to construct point mutations in mycobacteria
Recombineering using ssDNA substrates offers a simple method for constructing point mutations in mycobacterial genomes49. The overall efficiency of ssDNA recombineering is substantially higher than with dsDNA substrates for both chromosomal and plasmid targets. Moreover, the exonuclease function (Che9c gp60) is not required for ssDNA substrates, and strains containing plasmid such as pJV62 (Fig. 2) are suitable49.
In ssDNA recombineering, a choice needs to be made as to which DNA strand to target. In mycobacterial recombineering this is a critical issue, since ssDNA substrates targeting the same sequence but on different strands can differ in recombineering efficiencies up to 10,000-fold49; this far exceeds the 2–50 fold strand biases observed in the λ Red system19, 31. Recombination frequencies are always greater using a ssDNA substrate that anneals to the template for discontinuous DNA synthesis (lagging strand), presumably due to greater availability of ssDNA chromosomal DNA for pairing with gp61-ssDNA substrate complexes. It is not clear why the biases are so much larger in mycobacteria than in E. coli, but it may reflect fundamental differences in the DNA replication systems or in how gp61 interacts with the replication machinery. Substrates as short as 48 nt provide optimal recombination frequencies, and this recombination is also independent of host RecA functions49.
A simple experiment to test for ssDNA recombineering is using an oligonucleotide substrate to introduce a chromosomal point mutation conferring drug resistance. In a typical experiment using an oligonucleotide targeting the lagging strand, 100 ng ssDNA yields a similar number of drug resistant colonies as obtained using 100 ng extrachromosomal plasmid DNA (~105). Thus, in contrast to dsDNA recombineering, a high proportion of cells taking up ssDNA undergo recombination. The question then arises as to whether the overall frequency is high enough to construct point mutations that cannot be directly selected. The challenge with doing this arises not with the recombination frequencies per se, but because 99.9% of cell do not take up DNA.
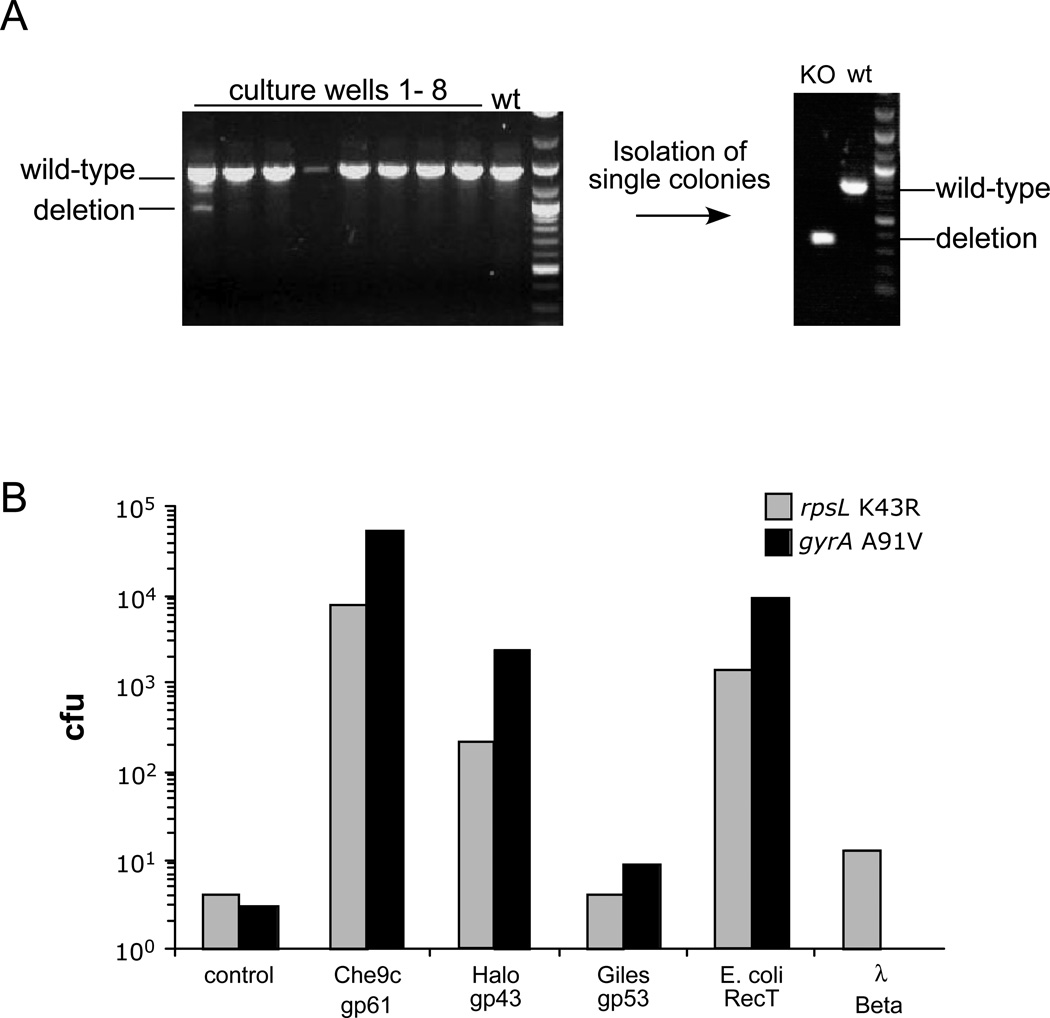
This problem can be conveniently circumvented using a co-selection strategy, in which a plasmid is co-transformed with the ssDNA and plasmid transformants selected; this effectively counter selects against non-competent cells in the population. The transformants can then be examined by PCR, and we find the mismatch amplification mutation assay PCR (MAMA-PCR)60, 61 that selectively amplifies the mutant allele to be especially effective. In our experience, 5–10% of the plasmid transformants also contain the newly introduced point mutation. A similar outcome can be achieved using a double-oligonucleotide configuration, in which two ssDNA substrates are used in the same electroporation; one introduces the desired point mutation in the chromosome, while the other repairs a mutant HygR gene present on the recombineering plasmid. The proportion of HygR colonies containing the point mutation is somewhat lower (~3%), but can still be readily identified by MAMA-PCR; the advantage is that only a single plasmid needs to be removed subsequently from strains for further analyses. A plasmid (pJV128) has been constructed containing Che9c gp61 fused to the acetamidase promoter, a KanR selectable marker, a mutant HygR gene and a sacB counter-selectable gene for this purpose49. This co-selection strategy can also be used with dsDNA substrates, and we have successfully constructed an unmarked deletion of the M. smegmatis leuD gene using a 200 bp dsDNA substrate with 100 bp leuD homology on each side of the deletion (Fig. 3a).
Figure 3. Mutagenesis of mycobacterial genomes.
A. Constructing an unmarked deletion of the M. smegmatis leuD gene using recombineering. Induced electrocompetent M. smegmatis/pJV76amber cells (pJV76amber is a derivative of pJV53 containing a mutant HygR gene) were co-electroporated with a 200 bp leuD deletion substrate along with an oligonucleotide that repairs the mutant plasmid HygR gene. After dilution to approximately 10 cells per sample, hygromycin selection and growth, at least one sample (#1) contained the mutant allele, and a leu− strain was readily recovered and verified by PCR.
B. Recombinases (SSAPs) were assayed for ssDNA recombineering activity in M. smegmatis by their ability to introduce streptomycin-resistant (rpsL K43R) and ofloxacin-resistant (gyrA A91V) mutations. SSAP genes are expressed from the acetamidase promoter and translationally-fused to the pLAM12 vector48 rather than using their native translation initiation signals49. The number of drug resistant colony forming units (cfu) obtained for each drug target are shown.
An intriguing use of the ssDNA recombineering technology is the study of mutations that confer antibiotic resistance. This is illustrated by introduction of several well characterized point mutations conferring resistance to isoniazid, rifampicin, streptomycin, and ofloxacin in M. smegmatis, as well as rifampicin and streptomycin in M. tuberculosis49. Recombineering with ssDNA thus offers a simple approach to determine whether any point mutation identified in a clinical strain of M. tuberculosis contributes to its drug susceptibility profile.
Identification of other phage recombinases
Although homologues of known recombinases are rare in mycobacteriophages, the Halo gp43 and Giles gp53 proteins are likely candidates for recombination activity (Fig. 1a). Introduction of drug-resistant point mutations by ssDNA recombineering provides a simple assay for testing these as well as SSAPs from non-mycobacteriophage sources including λ Beta and RecT (Fig. 3b). Halo gp43 is somewhat less active than Che9c gp61, and Giles gp53 even less so, further illustrating the utility of a large reservoir of phage genomes to optimize potential genetic tools49. Anecdotal reports that the λ Red system functions poorly in mycobacteria are confirmed by the low activity of λ Beta, although RecT works remarkably well in M. smegmatis (Fig. 3b), while still reduced in activity compared to Che9c gp61. Related studies report that Che9c gp61 performs relatively poorly in E. coli ssDNA recombineering assays62. The molecular basis for differing activities of SSAPs is not clear, although an intriguing possibility is that they act by interacting directly with the host DNA replication machinery.
Construction of mycobacteriophage mutants by recombineering
Constructing mutant derivatives of bacteriophages is often more difficult than manipulating the host chromosome, mostly because drug selection is not useful in lytically propagated viruses. The most powerful current method of manipulating mycobacteriophages is through the construction of shuttle phasmids63, and while these have many applications, they are of only limited use for determining phage gene functions. In one example, TM4 shuttle phasmids were used to make mutations in the TM4 tape measure gene, performing the mutagenesis in E. coli (by λ Red recombineering) and recovering the mutant phage in M. smegmatis64. However, most mycobacteriophage genomes are too large for shuttle phasmid construction, and this approach is not broadly applicable for functional genomic studies on mycobacteriophages. A homologous recombination approach was used to construct a firefly luciferase recombinant of mycobacteriophage L565, although the frequency of host-mediated recombination is sufficiently low that construction is inefficient and time-consuming. Mycobacterial recombineering can be employed for manipulating prophages, but since relatively few mycobacteriophages form stable lysogens, this is also not generally applicable.
The mycobacterial recombineering systems described above offer potentially powerful approaches to constructing mutants of any lytically growing mycobacteriophage. Recombineering of phage genomes has been described for E. coli phages such as lambda, using a strategy in which bacterial cells are infected with phage, competent cells are prepared, dsDNA or ssDNA substrate is introduced by electroporation and plaques analyzed for the presence of the mutation66. Since the frequency of DNA uptake in mycobacteria is substantially lower than the efficiency of phage infection, we have adopted an alternative approach based on the co-selection methods described above. While still under development, initial experiments suggest that this is a highly effective approach for mycobacteriophage functional genomics (LJM, JCV and GFH, manuscript in preparation).
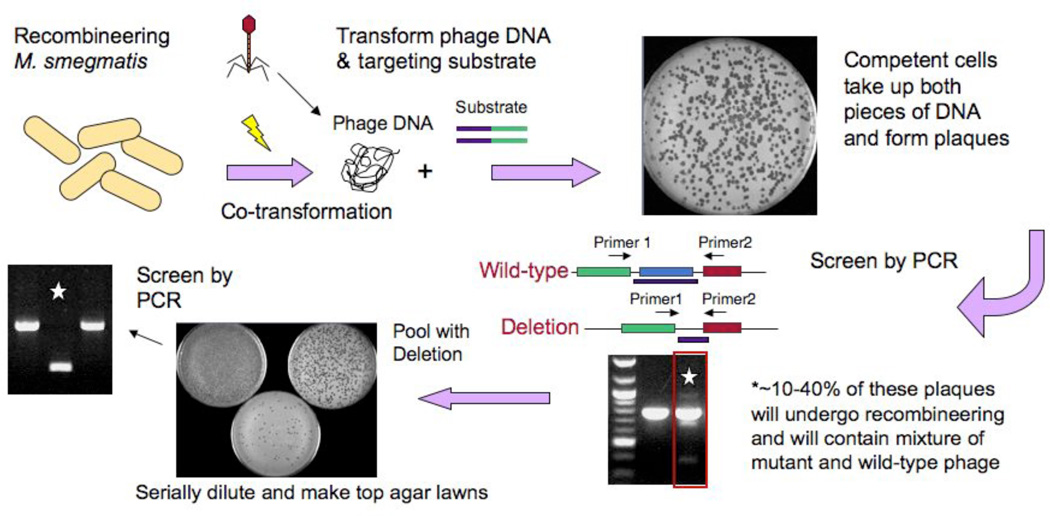
The scheme we have developed for mycobacterial mutagenesis is shown in Figure 4. An M. smegmatis strain carrying a recombineering plasmid such as pJV53 or pJV62 (Fig, 2) is induced with acetamide and electrocompetent cells prepared. Electroporation is performed with two DNAs; phage genomic DNA of the virus to be manipulated and a PCR-generated or synthetic DNA substrate. Plaques are recovered in an infectious center assay (i.e. prior to lysis) such that cells that have taken up phage DNA give rise to plaques on a lawn of M. smegmatis plating cells; ~100 plaques are obtained from 50–100 ng of phage DNA in such an experiment. All plaques contain wild-type phage DNA, but 10–40% also contain the mutant allele and can be easily identified by PCR screening of 12–18 individual plaques. Provided that the deleted region is non-essential for phage growth, then purified mutant phage derivatives can be isolated by plating serial dilutions of the mixed plaques and PCR analysis of the purified plaques (Fig. 4). Typically, analysis of 12–18 isolated plaques from this plaque-purification is sufficient to identify a homogenously mutant strain.
Figure 4. Mycobacteriophage recombineering.
Schematic illustration of the strategy utilized to adapt mycobacterial recombineering for use on mycobacteriophages. Induced electrocompetent M. smegmatis/pJV53 cells are co-electroporated with phage DNA (50–100 ng) and a 200bp dsDNA recombineering substrate (50–500 ng), mixed with M. smegmatis plating cells, and plated for individual plaques. Approximately 10–40% of plaques are mixed, containing both the mutant and wild-type alleles, and are distinguishable by PCR. A mixed plaque can be diluted and plated for analysis of individual plaques or for preparation of a lysate. If a non-essential gene is deleted then a mutant phage derivative can be readily identified by PCR, but if the gene is essential, then the mutant allele is no longer present in the lysate.
We have used phage recombineering to manipulate several different mycobacteriophage genomes and to generate gene knockouts, in-frame internal deletions, point mutations, and small insertions. Thus far, we find that when constructing deletion mutants a higher proportion of primary plaques contain the desired mutant allele when using dsDNA substrates than when using ssDNA substrates. Larger dsDNA substrates (200 bp) also appear to work better than shorter ones (100 bp) as with mycobacterial chromosomal recombineering48. We have also adapted this approach to determine whether mycobacteriophage genes are required for lytic growth. In one experiment, we used a 200 bp dsDNA substrate to generate a deletion in the lysin A gene of mycobacteriophage Giles that seemed likely to be an essential function. PCR analysis showed that approximately 30% of the initial plaques recovered contained mutants, but when lysates were prepared from re-plating of these mixed plaques, the mutant allele could not be identified even using a sensitive MAMA-PCR approach (see above). We conclude that the gene is essential for Giles growth, and that the mutation could be constructed and the mutant propagated in the presence of wild-type helper phage in the primary plaque, even though single particles do not give rise to plaques in the secondary plating. The full utility of these approaches for mycobacteriophage mutagenesis has perhaps yet to be realized, but the ability to modify mycobacteriophage genomes using mycobacterial recombineering will, for the first time, allow us to use broad functional genomic studies on mycobacteriophages.
Further development of mycobacterial recombineering systems
The ability to stimulate the levels of homologous recombination in both slow- and fast-growing mycobacteria by expression of mycobacteriophage-encoded recombination systems has the potential to be exploited in a variety of ways. First, it seems likely that the same approaches will work for manipulation of a variety of mycobacterial species other than those in M. smegmatis and M. tuberculosis described here, and possibly in other bacterial species that are closely related to the mycobacteria. Secondly, the relatively high efficiencies of chromosomal recombineering could facilitate construction of ordered gene knockout or replacement mutants, especially since identification of mutants by PCR is compatible with genome-wide robotic strategies. Lastly, the coupling of efficient point mutagenesis with mycobacterial nonsense suppressor strains67 offers the prospects of a relatively simple method for making conditional lethal mutants in mycobacterial genomes and their phages.
Acknowledgements
This work was supported by US National Institutes of Health Grant AI067649. We thank Dr. William Jacobs, Jr. and Dr. Donald Court for helpful discussions, and Matt Bochman for help with electron microscopy.
References
- 1.Aldovini A, Husson RN, Young RA. The uraA locus and homologous recombination in Mycobacterium bovis BCG. J Bacteriol. 1993;175:7282–7289. doi: 10.1128/jb.175.22.7282-7289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalpana GV, Bloom BR, Jacobs WR., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci U S A. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatfull GF. In: Tuberculosis. Eisenach K, Cole ST, Jacobs WR Jr, McMurray D, editors. Washington, DC: ASM Press; 2004. pp. 203–218. [Google Scholar]
- 4.Balasubramanian V, et al. Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 6.Hinds J, et al. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145(Pt 3):519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- 7.Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146(Pt 8):1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 8.Pashley CA, Parish T, McAdam RA, Duncan K, Stoker NG. Gene replacement in mycobacteria by using incompatible plasmids. Appl Environ Microbiol. 2003;69:517–523. doi: 10.1128/AEM.69.1.517-523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelicic V, Reyrat JM, Gicquel B. Positive selection of allelic exchange mutants in Mycobacterium bovis BCG. FEMS Microbiol Lett. 1996;144:161–166. doi: 10.1111/j.1574-6968.1996.tb08524.x. [DOI] [PubMed] [Google Scholar]
- 10.Sander P, Meier A, Bottger EC. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 11.Bardarov S, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 12.Vilcheze C, et al. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006 doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 13.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 14.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Ranallo RT, Barnoy S, Thakkar S, Urick T, Venkatesan MM. Developing live Shigella vaccines using lambda Red recombineering. FEMS Immunol Med Microbiol. 2006;47:462–469. doi: 10.1111/j.1574-695X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 16.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 17.Sarov M, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 18.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman MM, Gottesman ME, Gottesman S, Gellert M. Characterization of bacteriophage lambda reverse as an Escherichia coli phage carrying a unique set of host-derived recombination functions. J Mol Biol. 1974;88:471–487. doi: 10.1016/0022-2836(74)90496-3. [DOI] [PubMed] [Google Scholar]
- 22.Little JW. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 23.Joseph JW, Kolodner R. Exonuclease VIII of Escherichia coli. I. Purification and physical properties. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- 24.Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 26.Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda promotes strand exchange. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 27.Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 28.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci U S A. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rybalchenko N, Golub EI, Bi B, Radding CM. Strand invasion promoted by recombination protein beta of coliphage lambda. Proc Natl Acad Sci U S A. 2004;101:17056–17060. doi: 10.1073/pnas.0408046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 33.Muyrers JP, et al. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 2000;1:239–243. doi: 10.1093/embo-reports/kvd049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy KC. The lambda Gam protein inhibits RecBCD binding to dsDNA ends. J Mol Biol. 2007;371:19–24. doi: 10.1016/j.jmb.2007.05.085. [DOI] [PubMed] [Google Scholar]
- 36.Marsic N, Roje S, Stojiljkovic I, Salaj-Smic E, Trgovcevic Z. In vivo studies on the interaction of RecBCD enzyme and lambda Gam protein. J Bacteriol. 1993;175:4738–4743. doi: 10.1128/jb.175.15.4738-4743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy KC. Bacteriophage P22 Abc2 protein binds to RecC increases the 5' strand nicking activity of RecBCD and together with lambda bet, promotes Chi-independent recombination. J Mol Biol. 2000;296:385–401. doi: 10.1006/jmbi.1999.3486. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs WR, Jr, Tuckman M, Bloom BR. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature. 1987;327:532–535. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 39.Bardarov S, et al. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banaiee N, et al. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003;52:557–561. doi: 10.1099/jmm.0.05149-0. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs WR, Jr, et al. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 42.Kim AI, et al. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol. 2003;50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 43.Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration- proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J. Bacteriol. 2008;190:2172–2182. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–2723. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatfull GF, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedulla ML, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 48.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 49.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–1107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 50.Ford ME, Stenstrom C, Hendrix RW, Hatfull GF. Mycobacteriophage TM4: genome structure and gene expression. Tuber Lung Dis. 1998;79:63–73. doi: 10.1054/tuld.1998.0007. [DOI] [PubMed] [Google Scholar]
- 51.Chang HW, Julin DA. Structure and function of the Escherichia coli RecE protein, a member of the RecB nuclease domain family. J Biol Chem. 2001;276:46004–46010. doi: 10.1074/jbc.M108627200. [DOI] [PubMed] [Google Scholar]
- 52.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci U S A. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD. Electron microscopic visualization of RecT protein and its complexes with DNA. J Mol Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- 54.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 56.van Kessel JC, Hatfull GF. Mycobacterial recombineering. Methods Mol Biol. 2008;435:203–215. doi: 10.1007/978-1-59745-232-8_15. [DOI] [PubMed] [Google Scholar]
- 57.Wards BJ, Collins DM. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- 58.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 59.Bibb LA, Hatfull GF. Integration and excision of the Mycobacterium tuberculosis prophage-like element, phiRv1. Mol Microbiol. 2002;45:1515–1526. doi: 10.1046/j.1365-2958.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- 60.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 61.Swaminathan S, et al. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 62.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobs WR, Jr, et al. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 64.Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol. 2006;62:1569–1585. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarkis GJ, Jacobs WR, Jr, Hatfull GF. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol Microbiol. 1995;15:1055–1067. doi: 10.1111/j.1365-2958.1995.tb02281.x. [DOI] [PubMed] [Google Scholar]
- 66.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Ghosh P, Wasil LR, Hatfull GF. Control of Phage Bxb1 Excision by a Novel Recombination Directionality Factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]