Abstract
Classical cadherin cell-cell adhesion proteins play key morphogenetic roles during development and are essential for maintaining tissue integrity in multicellular organisms. Classical cadherins bind in two distinct conformations, X-dimer and strand-swap dimer; during cellular rearrangements, these adhesive states are exposed to mechanical stress. However, the molecular mechanisms by which cadherins resist tensile force and the pathway by which they convert between different conformations are unclear. Here, we use single molecule force measurements with an atomic force microscope (AFM) to show that E-cadherin, a prototypical classical cadherin, forms three types of adhesive bonds: catch bonds, which become longer lived in the presence of tensile force; slip bonds, which become shorter lived when pulled; and ideal bonds that are insensitive to mechanical stress. We show that X-dimers form catch bonds, whereas strand-swap dimers form slip bonds. Our data suggests that ideal bonds are formed as X-dimers convert to strand-swap binding. Catch, slip, and ideal bonds allow cadherins to withstand tensile force and tune the mechanical properties of adhesive junctions.
Keywords: single molecule biomechanics, force clamp, trans dimers, protein conformation, structure-function relationship
During tissue formation and wound healing, cells experience mechanical stress. They resist tensile force by adhering to neighboring cells using the classical cadherin family of Ca2+-dependent cell-cell adhesion proteins. Cadherins are essential for embryogenesis and for maintaining tissue integrity in multicellular organisms. Defects that alter adhesion lead to diverse diseases such as cardiovascular disease and metastatic cancers (1–4). Cell-cell adhesion is a dynamic process; cadherins tailor their adhesion in response to changes in the mechanical properties of their surrounding environment. However, the molecular mechanisms by which cadherins modulate adhesion and withstand mechanical stress are not fully understood.
Adhesion is mediated by the cadherin extracellular region; this region is comprised of five domains arranged in tandem (2, 5). Structural studies show that classical cadherins from opposing cells bind in two distinct trans conformations (6). In one conformation, known as a strand-swap dimer, opposing cadherins insert the side chain from a conserved Tryptophan at position 2 (W2) into a hydrophobic pocket on their adhesive partner (6–9). A second conformation, called an X-dimer, is formed by extensive surface interactions between the two outer domains of the cadherin extracellular region (6, 10–12). Although it has been proposed that X-dimers convert to a strand-swap conformation (6, 10), the mechanism by which this conversion occurs is unclear. Furthermore, although the structures of strand-swap and X-dimers have been resolved in atomic detail, the functional properties of these adhesive states are unknown.
Force measurements using mechanical probes show that single cadherins adhere strongly, indicating that cadherin trans dimers have long lifetimes in the presence of a pulling force (13, 14). In contrast, the binding affinities (5, 6) and lifetimes (15) of cadherin dimers in solution is remarkably low. This discrepancy between low binding affinities measured in solution under force-independent conditions and relatively strong adhesion is a major controversy in the cadherin field (16). However, the kinetics of cadherin interactions in the presence of pulling force has not yet been measured.
Theoretical studies predict that when a force is applied to interacting biomolecules, they respond in one of three distinct ways (17, 18): (i) they slip apart and their bonds become shorter lived when pulled (slip bonds), (ii) the interacting molecules catch and their bonds, counter intuitively, become longer lived in the presence of tensile force (catch bonds) or (iii) the bonds are insensitive to mechanical stress (ideal bonds). Slip bonds are the most commonly observed interactions in biology. Catch bonds, which provide a way for molecules to grip tightly and stabilize their attachments in the presence of mechanical stress, have been observed with motor proteins like myosin (19) and kinetochores (20) and with adhesive proteins like selectins (21–23), FimH (24, 25), and integrins (26), but not with cadherins. The third type of biomolecular interaction, ideal bonds, have also been proposed to play a role in permitting adhesive proteins to withstand mechanical stress (17). However, until now, ideal bonds were only a theoretical possibility (17, 18, 27); they had not been experimentally observed in any biological system.
Here, we use single-molecule force measurements with an atomic force microscope (AFM) to show that classical cadherins form bonds with catch, slip, and ideal mechanical properties. Cadherins switch between these bond types by changing their binding conformation; whereas X-dimers form catch bonds, strand-swap dimers form slip bonds. Our data suggests that ideal bonds are measured when X-dimers convert to strand-swap binding. The discovery of cadherin catch bonds reconciles the difference between cadherin binding properties measured by using solution and force measurements techniques. Catch, slip, and ideal bonds allow cadherins to withstand tensile force and tune the mechanical properties of intercellular junctions.
Results
Experimental Design.
We measured the force-dependent bond lifetimes of X-dimers, strand-swap dimers, and wild-type (WT) E-cadherin using single-molecule AFM force clamp spectroscopy. X-dimers were engineered by mutating the swapped amino acid W2 to alanine (W2A); X-ray crystallography has revealed that strand swapping is abolished in these mutants and they form an X-dimer structure (6). We trapped cadherins in a strand-swap dimer conformation by mutating Lys14 to glutamic acid (K14E); structural studies show that X-dimer formation is disrupted in these mutants (6). Finally, we also engineered the double mutant, W2A-K14E, where both strand-swapping and X-dimer formation is abolished (6).
Engineered cadherins, immobilized on glass coverslips and AFM cantilevers (Fig. 1A) (9), were allowed to interact for different periods of time by controlling the duration that the AFM tip and surface were in contact. Using a servo controlled feedback loop, the cadherin-cadherin bond was clamped at a constant force. The persistence time of the bond gave the lifetime at the clamping force (Fig. 1B). Because bond failure is a stochastic process, measurements were conducted approximately 1,000–2,000 times at five to nine different forces, and bond survival probabilities were calculated. We determined bond lifetimes at the different clamping forces from their mean lifetimes, the standard deviation of lifetimes, and exponential fits of the bond survival probability (23).
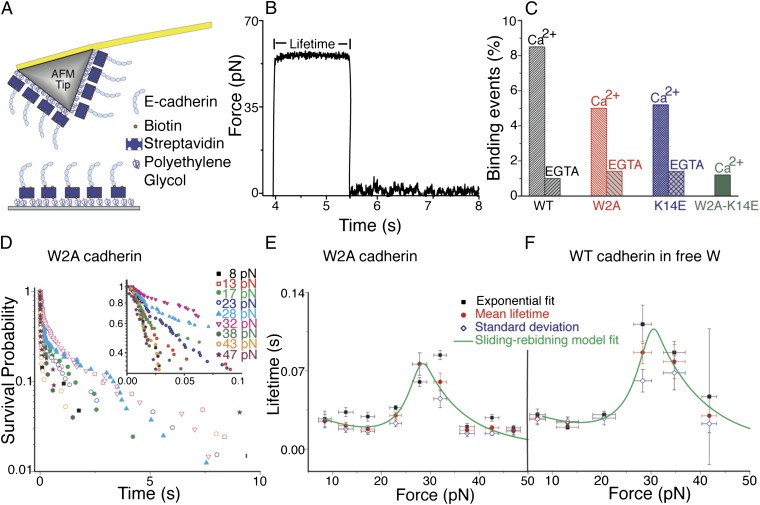
Fig. 1.
Single-molecule force clamp experiments show that cadherin X-dimers form catch-slip bonds. (A) Schematic of AFM tip and substrate functionalized with cadherins for single-molecule force clamp measurements. The tip and surface were functionalized with PEG linkers, some of which were decorated with streptavidin molecules. Biotinylated cadherin monomers were attached to the streptavidins. (B) Typical data obtained in an AFM force clamp experiment. A cadherin functionalized substrate and AFM tip were initially brought into contact to form a trans dimer. The AFM tip was then rapidly withdrawn from the surface and “clamped” at a predetermined force so that a constant pulling force was applied to the cadherin-cadherin bond. Bond lifetime at the clamping force was determined from the persistence time of the bond. Complete lifetime measurement cycle is shown in SI Appendix, Fig. S16. (C) Interaction of WT cadherins, W2A strand-swap mutants, and K14E X-dimer mutants are Ca2+ dependent. Less than 2% binding events were measured in the presence of EGTA, a Ca2+ chelator. Similarly, the frequency of nonspecific binding was less than 2% when the interaction of W2A-K14E double mutant was measured in the presence of 2.5 mM Ca2+; previous experiments have shown that both X-dimer and strand-swap dimer interactions are abolished in this mutant. (D) Bond survival probabilities for W2A cadherin were measured at nine different forces and approximately 1,000 to 2,000 measurements were conducted at each force. The probability P(t) that a pair of single molecules survive in the bound state at a constant force is given by 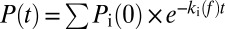 , where ki(f) is the off rate and 1/ki(f) is the bond lifetime. The measured bond survival probabilities were described by the sum of two exponentials. The high probability state occurred because of the unbinding of X-dimers or strand-swap dimers (Inset), whereas the low probability state occurred because of nonspecific adhesion. (E) Force-dependent bond lifetimes of the high probability state of W2A cadherin X-dimers exhibit a biphasic catch-slip behavior. Bond lifetimes initially increased with force indicating the presence of a catch bond. Beyond a critical force of ∼30 pN, lifetimes decreased with force, indicating slip bond behavior. Bond lifetimes at individual clamping forces were determined by using three methods: (i) by fitting the bond survival probabilities to a double exponential decay to obtain lifetime of the high probability, X-dimer state (black squares), (ii) by measuring the mean lifetime (red circles) of the high probability state, and (iii) from the standard deviation of lifetime of the high probability X-dimers (blue diamonds). The mean lifetime was fit to a sliding-rebinding model for two pairs of pseudoatoms (solid green line). (F) To confirm that the catch-slip behavior measured with W2A cadherins was not a mutagenesis artifact, WT cadherins were trapped in an X-dimer conformation by competitively inhibiting strand swapping using free W in solution. In the presence of 2 mM free W, the WT cadherins form catch-slip bonds. Error bars for bond lifetimes correspond to standard errors, whereas error bars for force correspond to standard deviation.
, where ki(f) is the off rate and 1/ki(f) is the bond lifetime. The measured bond survival probabilities were described by the sum of two exponentials. The high probability state occurred because of the unbinding of X-dimers or strand-swap dimers (Inset), whereas the low probability state occurred because of nonspecific adhesion. (E) Force-dependent bond lifetimes of the high probability state of W2A cadherin X-dimers exhibit a biphasic catch-slip behavior. Bond lifetimes initially increased with force indicating the presence of a catch bond. Beyond a critical force of ∼30 pN, lifetimes decreased with force, indicating slip bond behavior. Bond lifetimes at individual clamping forces were determined by using three methods: (i) by fitting the bond survival probabilities to a double exponential decay to obtain lifetime of the high probability, X-dimer state (black squares), (ii) by measuring the mean lifetime (red circles) of the high probability state, and (iii) from the standard deviation of lifetime of the high probability X-dimers (blue diamonds). The mean lifetime was fit to a sliding-rebinding model for two pairs of pseudoatoms (solid green line). (F) To confirm that the catch-slip behavior measured with W2A cadherins was not a mutagenesis artifact, WT cadherins were trapped in an X-dimer conformation by competitively inhibiting strand swapping using free W in solution. In the presence of 2 mM free W, the WT cadherins form catch-slip bonds. Error bars for bond lifetimes correspond to standard errors, whereas error bars for force correspond to standard deviation.
Cadherin binding frequency was adjusted to ∼7% by controlling the cadherin density on the tip and substrate. Under similar experimental conditions, cadherin surface density was determined to be 65 ± 18 cadherins per µm2, which corresponds to an average distance of 124 nm between neighboring cadherins (9). Because the separation between neighboring cadherins is an order of magnitude larger than the radius of curvature of the AFM tip, it is expected that the measured lifetimes correspond to the interaction of single cadherins immobilized on the surface and the AFM tip, respectively. Based on the measured binding frequency, Poisson statistics predicts that more than 96% of measured events occur because of the rupture of single bonds. Binding of WT cadherin and the W2A and K14E mutants were highly specific and Ca2+ dependent; less than 2% binding events were measured when cadherins were eliminated from either the tip or the substrate or in the presence of EGTA, a Ca2+ chelator (Fig. 1C).
As observed in previous cadherin force measurements (13, 14, 28, 29), the measured bond survival probabilities were described by the sum of two exponentials (Fig. 1D; SI Appendix, Figs. S2–S6); an F-test confirmed the double exponential fits with 95% confidence (SI Appendix, Tables S2–S6) (30). The coefficient of the second exponential did not systematically vary with force (SI Appendix, Figs. S13–S15), suggesting that the two exponentials correspond to the existence of two bound states rather than being an intrinsic part of the kinetics of single-state binding (31). The adhesive state with higher binding probability was weaker and had a shorter lifetime compared with the lower probability binding state.
We hypothesized that the high probability state occurred because of the unbinding of X-dimers or strand-swap dimers, whereas the low probability state was due to nonspecific adhesion. To test this hypothesis, we measured the force-dependent bond lifetimes of the W2A-K14E double mutant (SI Appendix, Fig. S7); previous analytical ultracentrifugation experiments have shown that both X-dimer and strand-swap dimer interactions are abolished in this mutant (6). We also measured the force-dependent lifetimes of nonspecific interactions between WT cadherins immobilized on an AFM tip (Fig. 1A) and a substrate that lacked cadherins (SI Appendix, Fig. S8). The measured survival probabilities of both nonspecific interactions and of the double mutant binding showed a single exponential decay indicating that they interact in only one state (SI Appendix, Fig. S7 and S8 and Tables S7 and S8). The force-dependent lifetimes of this state were identical to the long-lived state measured with WT, W2A, and K14E cadherin binding, confirming that this low-probability state arises because of nonspecific interactions (SI Appendix, Fig. S9 and Table S9). In further agreement with this data, the total binding probability of the W2A-K14E double mutant was <2%, which is comparable to the nonspecific binding probabilities measured with the WT, W2A, and K14E cadherins in the absence of Ca2+ (Fig. 1C). The remainder of this paper only focuses on the high-probability, specific binding state.
X-Dimers Form Catch-Slip Bonds.
W2A cadherin mutants immobilized on the AFM tip and substrate were allowed to interact for 0.3 s to form X-dimers, and their force-dependent bond lifetimes were measured (SI Appendix, Fig. S2 and Table S2). The X-dimers exhibited a biphasic relationship between lifetime and force (Fig. 1 D, Inset and E). The bond lifetimes initially increased with force, indicative of a catch bond. After reaching a maximum at a critical force of ∼30 pN, the lifetimes decreased with force, indicating slip bond behavior (Fig. 1E).
To confirm that the catch-slip behavior was not an artifact caused by introducing an Ala in place of Trp in the W2A mutants, we forced the WT cadherin into an X-dimer conformation by competitively inhibiting strand swapping using free W in solution. In the presence of 2 mM free W, the WT cadherins interacting for 0.3 s also formed catch bonds at low forces that transitioned to slip bonds beyond a critical force of ∼30 pN (Fig. 1F).
We hypothesized that X-dimers form catch bonds because the application of a tensile force changes the orientation of cadherins in the X-dimer: As they are pulled apart, the cadherins form transient bonds with an alternate binding site that slows unbinding and allows stochastic reformation of a dimer (SI Appendix, SI Materials and Methods, Fig. S1, and Table S1). The measured biphasic bond lifetimes were well described by such a “sliding-rebinding” model (32, 33) (Fig. 1 E and F).
Strand-Swap Dimers Form Slip Bonds.
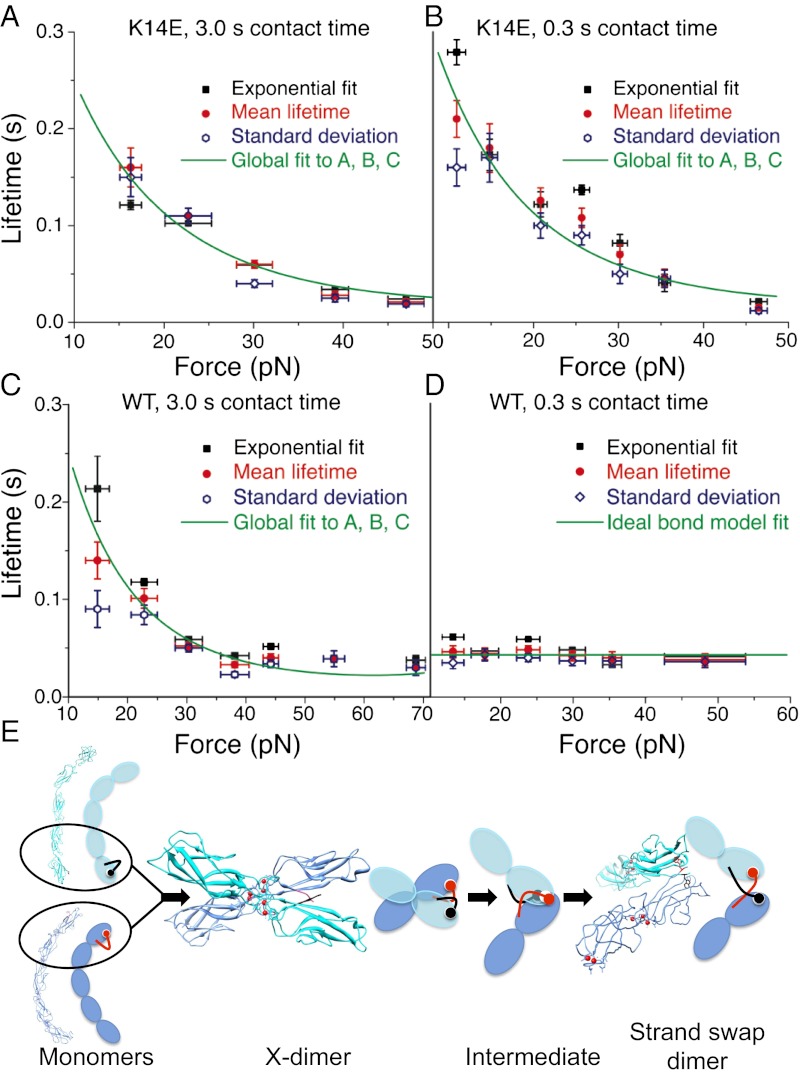
Unlike cadherin X-dimers, the K14E mutants, which only form strand-swap dimers, showed a traditional slip bond behavior; their bond lifetimes decreased with increasing tensile force (Fig. 2 A and B). Slip bonds were measured at both long (3.0 s; Fig. 2A and SI Appendix, Fig. S3 and Table S3) and short interaction times (0.3 s; Fig. 2B and SI Appendix, Fig. S4 and Table S4).
Fig. 2.
Strand-swap dimers form slip bonds. Ideal bonds are measured when X-dimers transition to a strand-swap conformation. (A) Force-dependent lifetimes of the high probability state of K14E mutants that are allowed to interact for 3.0 s show a slip bond behavior; their lifetimes decreases with force. Structural studies show that X-dimer formation is disrupted in these mutants, and they form strand-swap dimers. (B) The high probability state of K14E mutants that are allowed to interact for 0.3 s also form slip bonds. (C) The high probability state of WT cadherins that interact for 3.0 s show a similar slip bond behavior, indicating that they too form strand-swap dimers. Bond lifetimes at the different clamping forces were determined from the mean lifetime (red circle), from the standard deviation of lifetime (blue diamond), and from exponential fits of the bond survival probabilities (black squares). The mean lifetime data in A–C was globally fit to a microscopic, slip bond model (green solid line). (D) The high probability state of WT cadherins that interact for a shorter period (0.3 s), form ideal bonds; their lifetimes are independent of force. Bond lifetimes at the different clamping forces were determined from the mean lifetime (red circle), from the standard deviation of lifetime (blue diamond), and from exponential fits of the bond survival probabilities (black squares). The mean lifetime was fit to an ideal bond model (solid green line). Error bars for bond lifetimes correspond to standard errors, whereas error bars for force correspond to standard deviation. (E) Schematic of the proposed pathway for conversion of X-dimers to strand-swap dimers. Because the on-rate for X-dimer formation is greater than the rate of strand swapping, cadherins first form X-dimers that subsequently convert to strand-swap dimers via an intermediate that forms ideal bonds.
Next we used AFM force clamp experiments to measure the force-dependent bond lifetime of WT cadherins. Previous studies have shown that although strand-swapping occurs at a slower rate than X-dimer formation, the final strand-swapped dimer has a higher binding affinity (6). Consequently, when WT cadherins interact for a long period (3.0 s), they formed slip bonds characteristic of a strand-swap dimer (Fig. 2C and SI Appendix, Fig. S5 and Table S5). WT cadherin that interact for 1.0 s also formed identical slip bonds (SI Appendix, Fig. S12).
The WT cadherin slip bonds were indistinguishable from the slip bonds formed by K14E strand-swap dimers (Fig. 2 A–C). We globally fit the WT and the K14E cadherin data with a microscopic slip bond model (34) to obtain best-fit parameters across all datasets. The global fits yielded an intrinsic lifetime of 0.63 s (Fig. 2 A–C and SI Appendix, Table S10).
Ideal Bonds Are Measured When WT Cadherins Interact for a Short Time Period.
When their interaction time was decreased to 0.3 s, the WT cadherins formed ideal bonds; their lifetimes were independent of force (Fig. 2D and SI Appendix, Fig. S6 and Table S6). The intrinsic lifetime of the ideal bond was 0.04 s (Fig. 2D). We hypothesize that the ideal bonds correspond to an intermediate state as X-dimers transition to a strand-swap conformation (Fig. 2E); when strand-swapping is competitively inhibited, the WT cadherins form X-dimers (Fig. 1F). Ideal bonds are predicted to occur when the interaction energies of the bound state and the transition state of a receptor–ligand complex are harmonic with identical elastic constants and resting lengths (17). Ideal bond like behavior is also predicted in a multidimensional energy landscape when the extension of the receptor-ligand pair along the pulling coordinate is identical for the bound state and the transition state (27).
Next, we measured whether WT cadherins that were permitted to interact for a very short period (0.001 s) form ideal bonds (SI Appendix, Fig. S10); this interaction time corresponds to the fastest data acquisition rate of our force measurement apparatus. Even at this short interaction time, the WT cadherins formed ideal bonds, suggesting that the transition from X-dimer to the intermediate state occurs at a rapid rate.
Because the WT cadherins were loaded at a constant ramp rate to a constant hold force, we also tested the ramp-rate dependence of ideal bond formation; a recent study has shown that a mechanical bond that behaves as a catch-slip bond at low ramp rates may transform to slip-only bonds at high ramp rates (35). When we increased the ramp rate by an order of magnitude, the WT cadherins with 0.3 s interaction time continued to behave as ideal bonds, indicating the rate of force application did not affect ideal bond behavior (SI Appendix, Fig. S11).
Discussion
In summary, we used single-molecule force measurements to show that depending on their binding conformation, cadherins form catch bonds that adhere tightly in the presence of force, slip bonds that have a longer lifetime in the absence of force, and ideal bonds that are insensitive to force. Whereas X-dimers form catch bonds, strand-swap dimers form slip bonds. We hypothesize that ideal bonds are formed as X-dimers convert to strand-swap binding.
Cadherin catch bonds presumably allow cells to grip tightly and lock in place when pulled. We anticipate that cadherin catch bonds are widespread because nonclassical cadherins, which account for 81% of cadherins in the vertebrate genome, lack the sequence determinants for strand swapping and likely form X-dimers (11). In fact, recent crystallographic studies show that the nonclassical T-cadherin adheres in an X-dimer conformation that is virtually indistinguishable from the W2A X-dimer (11). Because catch bonds strengthen in the presence of force, our experiments reconcile the apparently contradictory finding that the lifetime of trans dimers measured in solution is low (15), whereas their lifetimes measured using mechanical probes is large (13, 14).
Our model for catch bond formation is that the application of a tensile force reorients cadherins in the X-dimer so that they form transient bonds with an alternate binding site and lock more tightly. Consequently, the biphasic catch-slip behavior of X-dimers can be fit with a sliding-rebinding model (32, 33) (Fig. 1 E and F). We were unsuccessful in fitting the data to other models for catch-slip bond formation, including two-pathway models, where force drives the system away from its native dissociation pathway into an alternative pathway involving a higher energy barrier (21, 36–38), and an allosteric model that suggest that force alters X-dimer conformation and induces changes in the cadherin binding site (31).
We propose that ideal bonds are formed by an intermediate state when X-dimers transition to a strand-swap conformation (Fig. 2E). This intermediate state adopts a structure where its extension along the pulling coordinate is identical for both the bound state and the transition state; consequently, its bond lifetime is independent of pulling force (27). In contrast, X-dimers and strand-swap dimers pulled along the same direction form catch-slip and slip-only bonds, respectively. The rate of force application does not affect ideal bond behavior. Furthermore, ideal bonds are measured even when WT cadherins interact for the short time of 1 ms, suggesting that the transition from X-dimer to the intermediate state occurs rapidly. However, the structure of this state and the molecular contacts responsible for ideal bond formation still need to be resolved.
It is important to note that the bond survival probability of WT, W2A, and K14E cadherins decay as the sum of two exponentials. Such a double exponential decay could either be an intrinsic part of the kinetics of a single bond (31, 38) or correspond to the existence of two distinct bound states (13, 14, 28). If a double exponential survival probability arises from a single bound state, the coefficient of the second exponential would increase monotonically with force (31, 38). However, in our data, the probability of the long-lifetime component does not systematically vary with force, suggesting that the WT, W2A, and K14E cadherins interact in two distinct bound states (SI Appendix, Figs. S13–S15). The lifetime vs. force curve for the long-lived state is identical to the force-dependent lifetimes of nonspecific interactions (SI Appendix, Fig. S9 and Table S9). These data demonstrate that the long-lived, low-probability state arises because of nonspecific adhesion.
It should be noted that the bond lifetimes measured in our experiments are not a result of failure of the noncovalent streptavidin–biotin interactions that are used to immobilize the cadherins on the AFM tip and surface. Previous studies have shown that streptavidin–biotin linkages form slip bonds, the lifetime of this bond, measured at a range of clamping forces up to 150 pN, is more than two orders of magnitude greater than the lifetimes reported here (39).
Our results suggest a physical mechanism that cadherins use to resist tensile forces as cells rearrange during tissue formation and wound healing. As cells migrate, cadherins bind rapidly to form X-dimers; the X-dimer catch bonds allow migrating cells to grip strongly under load. Over time, the X-dimers proceed to form more robust strand-swap dimers that have a high affinity in the absence of force; this conversion is facilitated by an intermediate conformation that is insensitive to tensile force (Fig. 2E). Ideal, catch, and slip bonds allow cadherins to resist mechanical stress and tune the mechanics of intercellular junctions.
Materials and Methods
Engineering and Purification of Cadherin Constructs.
Engineering, purification, and biotinylation of WT E-cadherin has been described (9). Briefly, the full extracellular region of E-cadherin with a C-terminal Avi-tag was cloned into pcDNA3.1(+) vectors by using primers containing a Tev sequence and His-tag. This cloning resulted in an ORF of the complete E-cadherin/Avi/Tev/His sequence. Cadherin mutations (W2A, K14E, and W2A-K14E) were introduced in the EC1 domain of E-cadherin/ATH by point mutation using QuikChange kit (Agilent). The engineered cadherin sequences were transfected into HEK 293 cells that were selected using 400 μg/mL Genecitin (G418; Invitrogen). Cells were grown to confluency in high glucose DMEM containing 10% (vol/vol) FBS and 200 g/mL G418 and then exchanged into serum-free DMEM. Conditioned media was collected 4 d after media exchange. Cell debris was removed by centrifugation and filtration.
His-tagged E-cadherin was purified from conditioned media by using a Nickel NTA resin (Invitrogen); the media was incubated with the resin for 2 h, washed with Hepes buffer containing 50 mM imidazole and eluted with 250 mM imidazole. The protein was further purified by running through a Superdex200 10/300 GL column (GE Healthcare) equilibrated with 25 mM Hepes buffer at pH 7.4 containing 100 mM NaCl, 10 mM KCl, and 1 mM CaCl2.
After protein purification, the Avi-tag sequence on the E-cadherin/ATH was biotinylated by using BirA enzyme (BirA500 kit; Avidity). After biotinylation for 1 h at 30 °C, free biotin was removed by using a Superdex 200 10/300 GL column. Before biotinylation, the protein sample was exchanged into a pH 7.4 low-salt buffer containing 25 mM Hepes, 5 mM NaCl, 1 mM CaCl2, and concentrated to approximately 40 μM.
Single-Molecule Force-Clamp Experiments.
Engineered cadherins were immobilized on glass coverslips and the silicon nitride (Si3N4) tip of AFM cantilevers (Olympus) using protocols described (9). Briefly, AFM cantilevers and glass coverslips were sequentially cleaned by using a 25% H2O2:75% H2SO4 solution, deionized water, 1 M potassium hydroxide solution, deionized water, and acetone. The AFM cantilevers and coverslips were functionalized with amine groups by using a 2% vol/vol solution of 3-aminopropyltriethoxysilane (Sigma) dissolved in acetone. The silanized cantilevers and coverslips were functionalized with polyethylene glycol spacers (PEG 5000; Laysan Bio) containing an amine-reactive N-hydroxysuccinimide ester at one end. Seven percent of the PEG spacers presented biotin molecules on their other end.
The biotins were incubated with 0.1 mg/mL streptavidin for 30 min. Biotinylated cadherins were bound to the streptavidin molecules on the AFM tip and surface. After cadherin immobilization, free biotin binding sites on streptavidin was blocked by using a 10 µM solution of biotin. Before beginning an experiment, the functionalized coverslips and AFM tips were incubated in 0.1 mg/mL BSA for 12 h to minimize nonspecific protein binding.
An Agilent 5500 AFM with a closed-loop piezoelectric scanner was used in the single molecule force-clamp experiments. Spring constants of the AFM cantilevers were measured with the thermal fluctuation method (40). Forces were measured in a pH 7.5 buffer (10 mM Tris, 100 mM NaCl, and 10 mM KCl) in either 2.5 mM CaCl2 or 2 mM EGTA.
Bond survival probabilities were calculated by using two alternate approaches. First, we distributed the measured bond lifetimes into force bins and calculated the clamping forces and bond survival probabilities for the events in each force bin; similar bond lifetimes were obtained by using different bin sizes. Second, we calculated bond survival probabilities at the exact clamping forces at which the experiments were done. Both approaches yielded similar results.
Supplementary Material
Acknowledgments
We thank Agilent Technologies for technical support. This work was supported in part by a Basil O’Connor Starter Scholar Award from the March of Dimes Foundation and an American Heart Association National Scientist Development Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208349109/-/DCSupplemental.
References
- 1.Harris TJC, Tepass U. Adherens junctions: From molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11(7):502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1(3):a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6(8):622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 4.Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91(2):691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol. 2012;22(6):299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison OJ, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17(3):348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vendome J, et al. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18(6):693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296(5571):1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA. 2009;106(1):109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17(8):1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciatto C, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17(3):339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380(6572):360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 13.Shi QM, Chien YH, Leckband D. Biophysical properties of cadherin bonds do not predict cell sorting. J Biol Chem. 2008;283(42):28454–28463. doi: 10.1074/jbc.M802563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi QM, Maruthamuthu V, Li F, Leckband D. Allosteric cross talk between cadherin extracellular domains. Biophys J. 2010;99(1):95–104. doi: 10.1016/j.bpj.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vunnam N, Flint J, Balbo A, Schuck P, Pedigo S. Dimeric states of neural- and epithelial-cadherins are distinguished by the rate of disassembly. Biochemistry. 2011;50(14):2951–2961. doi: 10.1021/bi2001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leckband D, Sivasankar S. Biophysics of cadherin adhesion. Subcell Biochem. 2012;60(2):63–88. doi: 10.1007/978-94-007-4186-7_4. [DOI] [PubMed] [Google Scholar]
- 17.Dembo M. On Peeling an Adherent Cell from a Surface. Lectures on Mathematics in the Life Sciences, Some Mathematical Problems in Biology. Vol 24. Providence, RI: Am Math Soc; 1994. pp. 51–77. [Google Scholar]
- 18.Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234(1274):55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 19.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci USA. 2006;103(26):9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468(7323):576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarangapani KK, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279(3):2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 22.Yago T, et al. Catch bonds govern adhesion through L-selectin at threshold shear. J Cell Biol. 2004;166(6):913–923. doi: 10.1083/jcb.200403144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 24.Le Trong I, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell. 2010;141(4):645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109(7):913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 26.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185(7):1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y, Dudko OK. Single-molecule rupture dynamics on multidimensional landscapes. Phys Rev Lett. 2010;104(4):048101. doi: 10.1103/PhysRevLett.104.048101. [DOI] [PubMed] [Google Scholar]
- 28.Chien YH, et al. Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region. J Biol Chem. 2008;283(4):1848–1856. doi: 10.1074/jbc.M708044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perret E, Leung A, Feracci H, Evans E. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc Natl Acad Sci USA. 2004;101(47):16472–16477. doi: 10.1073/pnas.0402085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck JV, Arnold KJ. 1st Ed. New York: Wiley; 1977. Parameter Estimation in Engineering and Science. [Google Scholar]
- 31.Thomas W, et al. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006;90(3):753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou JZ, et al. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174(7):1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou JZ, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92(5):1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96(10):108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 35.Sarangapani KK, et al. Regulation of catch bonds by rate of force application. J Biol Chem. 2011;286(37):32749–32761. doi: 10.1074/jbc.M111.240044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci USA. 2004;101(31):11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereverzev YV, Prezhdo OV, Forero M, Sokurenko EV, Thomas WE. The two-pathway model for the catch-slip transition in biological adhesion. Biophys J. 2005;89(3):1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barsegov V, Thirumalai D. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc Natl Acad Sci USA. 2005;102(6):1835–1839. doi: 10.1073/pnas.0406938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favre M, et al. Force-clamp spectroscopy with a small dithering of AFM tip, and its application to explore the energy landscape of single avidin-biotin complex. Ultramicroscopy. 2007;107(10-11):882–886. doi: 10.1016/j.ultramic.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64(7):1868–1873. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.