Abstract
Stroke is a major cause of mortality and morbidity worldwide. Extracellular glutamate accumulation leading to overstimulation of the ionotropic glutamate receptors mediates neuronal injury in stroke and in neurodegenerative disorders. Here we show that miR-223 controls the response to neuronal injury by regulating the functional expression of the glutamate receptor subunits GluR2 and NR2B in brain. Overexpression of miR-223 lowers the levels of GluR2 and NR2B by targeting 3′-UTR target sites (TSs) in GluR2 and NR2B, inhibits NMDA-induced calcium influx in hippocampal neurons, and protects the brain from neuronal cell death following transient global ischemia and excitotoxic injury. MiR-223 deficiency results in higher levels of NR2B and GluR2, enhanced NMDA-induced calcium influx, and increased miniature excitatory postsynaptic currents in hippocampal neurons. In addition, the absence of MiR-223 leads to contextual, but not cued memory deficits and increased neuronal cell death following transient global ischemia and excitotoxicity. These data identify miR-223 as a major regulator of the expression of GluR2 and NR2B, and suggest a therapeutic role for miR-223 in stroke and other excitotoxic neuronal disorders.
MicroRNAs (miRNAs) are small noncoding endogenous RNA molecules that repress their target mRNA through complementary binding in the message 3′-UTR (1). MiRNAs play important roles in multiple physiological processes such as cell death and survival, cellular response to stress, stem cell division, and pluripotency (2). MiRNAs also play important roles in disease processes including cancer (3), cardiovascular disease (4), and neurodegenerative diseases (5). Due to their small size, relative ease of delivery, and sequence specificity in recognizing their targets, miRNAs are promising therapeutic targets (6).
Stroke is the second major killer and the leading cause of disability worldwide (7). Overstimulation of the glutamate receptor (glutamate excitotoxicity) is a major mechanism for neuronal cell death during stroke, central nervous system (CNS) trauma, and chronic neurodegenerative disorders. Excessive calcium influx through the N-methyl-d-aspartate receptors (NMDARs) results in abnormally high intracellular calcium concentrations leading to lethal consequences. This calcium influx through the NMDAR requires membrane depolarization induced by sodium influx through 2-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs) (8). Whereas accumulating evidence indicates that phosphorylation and trafficking play important roles in regulation of glutamate receptor signaling, the molecular mechanisms regulating glutamate receptor expression levels remain unexplored. Recent work suggests that the miRNA pathway regulates the AMPAR subunit GluR2 expression (9, 10). In hippocampal neurons, MiR-125b has been shown to regulate the NMDAR subunit NR2A (11). However, miRNA regulation of glutamate receptor expression remains poorly characterized. Identifying miRNAs that could regulate the glutamate receptor provide the opportunity for treating stroke and chronic neurodegenerative diseases. Recent success in therapeutic targeting of small RNAs in animal models and in humans emphasizes such treatment strategies (6).
MiR-223 is highly expressed in bone marrow and neutrophils where it plays an important role in regulating granulopoeisis and neutrophil function (12, 13). MiR-223 is deregulated in acute myeloid leukemia (14, 15). We find that miR-223 is also expressed in the nervous system and we demonstrate that miR-223 controls the expression and function of GluR2 and NR2B subunits of the glutamate receptor. Using in vitro and in vivo models of ischemic reperfusion brain injury and excitotoxic neuronal death we show that miR-223 is a neuroprotective microRNA.
Results
MiR-223 Expression in the Brain and Potential CNS Targets.
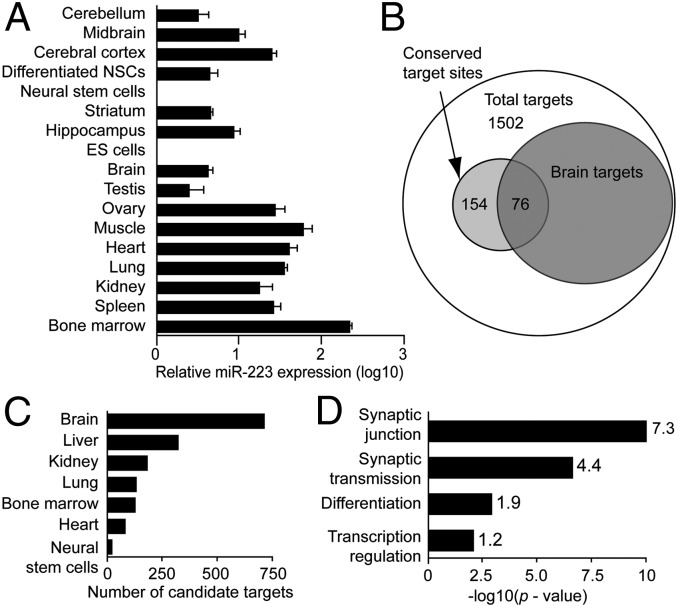
To characterize the expression of miR-223 in the brain, quantitative PCR (qPCR) for miR-223 relative to U6 snRNA was performed in different organs compared with brain and subregions of the brain. Whereas miR-223 is highly expressed in the bone marrow as previously described (16, 17), it is also widely expressed in other organs (Fig. 1A). Notably, miR-223 up-regulation is detected upon differentiation of adult mouse neural progenitors. Whereas miR-223 expression in the brain was low compared with other organs, hippocampus, midbrain, and cortex demonstrate relative enrichment of miR-223 expression compared with whole brain (Fig. 1A).
Fig. 1.
MiR-223 expression in the brain and peripheral tissue. (A) MiR-223 real-time PCR in different organs normalized to U6 snRNA. (B–D) DAVID bioinformatic (v6.7) analysis of TargetScan (release 5.1) total predicted miR-223 targets. (B) Venn diagram of total miR-223 predicted targets with the subset expressed in the brain. (C) Select tissue distribution comparison for miR-223 predicted targets (see Dataset S1 for full comparison). (D) Functional annotation analysis for brain-expressed miR-223 predicted targets. The x axis represents the EASE score P values for gene-enrichment annotation clusters. Enrichment scores are indicated next to columns (modified Fisher’s exact test).
To explore potential brain targets for miR-223, the online miRNA target prediction algorithm TargetScan was interrogated (18). There are 1,769 potential targets, with a total of 154 conserved sites and 1,881 poorly conserved sites (Dataset S1). Functions of potential miR-223 targets were evaluated using the National Institutes of Health Database for Annotation, Visualization, and Integrated Discovery (DAVID). Surprisingly, out of the 1,502 DAVID annotated genes (from the total 1,769 potential targets), 709 (∼47%) are brain-expressed genes, whereas only 124 (8%) are bone-marrow–expressed genes (Fig. 1 B and C and Dataset S1). This brain target gene expression enrichment suggested a functional role for miR-223 in the brain. Using the DAVID functional annotation tool to annotate the 709 brain-expressed potential miR-223 targets, there is a 7.34 enrichment score for synapse-related genes (P = 9.35 × 10−11) and a 4.43 enrichment score for synaptic-transmission–related genes (P = 2.64 × 10−07) (Fig. 1D and Dataset S1). Using a different miRNA target prediction algorithm, miRanda predicted targets for miR-223; the results are similar to those observed using the TargetScan algorithm-predicted targets, further supporting a functional role for miR-223 in the brain (Fig. S1 A–C and Dataset S2). These results indicate that miR-223 is expressed in the adult brain and suggest that miR-223 may be involved in the regulation of synaptic function.
MiR-223 Targets AMPA and NMDA Receptor Isotypes.
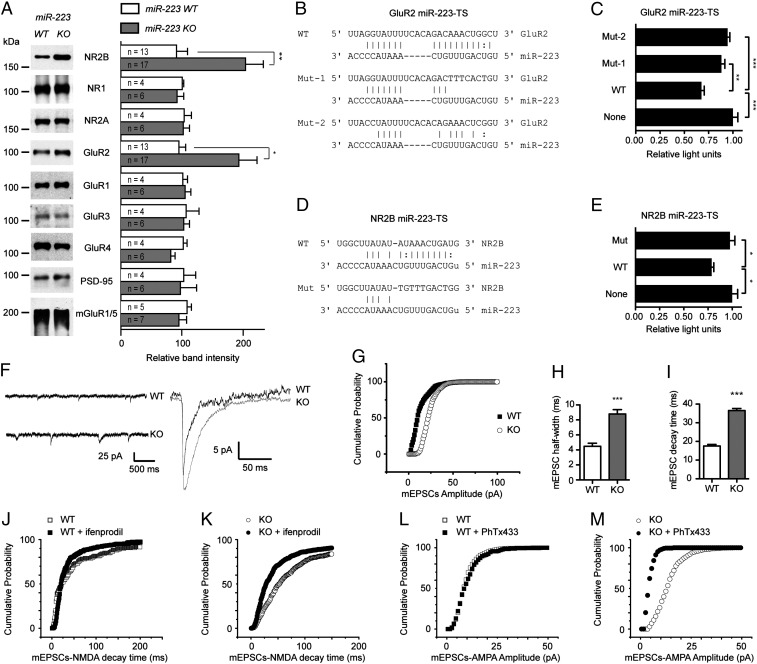
Among the potential synaptic junction targets identified, there are miR-223 binding sites in AMPAR subunit GluR2 and NMDAR subunit NR2B 3′-UTR. Performing multiple sequence alignment using ClustalW (19) demonstrates that the identified miR-223 target site (TS) in GluR2 3′-UTR is conserved among mammals (Fig. S2A). The miR-223 target site in NR2B 3′-UTR is shared between mouse and rat. However, there is a miR-223 target site in the NMDAR subunit NR3A 3′-UTR conserved among human, chimpanzee, and rhesus monkey (Fig. S2B). Computational modeling of miR-223 binding to potential GluR2 and NR2B 3′-UTR target sites using RNAhybrid algorithm (20) demonstrates extensive base pairing of 18 and 17 of 22 nucleotides, respectively. The binding minimum free energy is −23.3 and −18.2 kcal/mol, respectively, suggesting the possibility for strong binding (Fig. S2 C and D). To determine whether miR-223 regulates the expression of GluR2 and NR2B, Western blot analysis in hippocampal lysates from miR-223 wild-type (WT) and knockout (KO) mice was performed. A significant increase in GluR2 and NR2B protein levels in KO mice relative to the WT mice is observed (Fig. 2A). The protein expression levels of NR1, NR2A, GluR1, GluR3, GluR4, mGluR1/5, and PSD-95 are not significantly different in either WT or KO mice (Fig. 2A). These results suggest that miR-223 selectively represses the expression of GluR2 and NR2B subunits. To test whether miR-223 regulates GluR2 and NR2B messages through binding to their respective 3′-UTR candidate target sites, a dual Firefly/Renilla luciferase reporter assay was conducted. Candidate GluR2 and NR2B TSs for miR-223 were cloned into the Renilla luciferase 3′-UTR region, whereas Firefly luciferase served as a loading control in a dual-luciferase vector and was expressed in the presence of miR-223. Wild-type GluR2 miR-223–TS significantly inhibited luciferase activity, which was rescued by mutating the entire target site seed sequence (Mut-1) and point mutant of the seed sequence (Mut-2) (Fig. 2 B and C). Similarly, WT NR2B miR-223–TS significantly inhibited luciferase activity. This inhibition was abolished by mutating the entire target site seed sequence in Mut (Fig. 2 D and E). These studies confirm that the 3′-UTR of GluR2 and NR2B harbor functional miR-223 target sites that are sufficient to confer miR-223–mediated repression. Taken together, these data demonstrate that miR-223 regulates both GluR2 and NR2B protein expression.
Fig. 2.
MiR-223 targets GluR2 and NR2B. (A) Effect of miR-223 deficiency on the protein expression levels of indicated proteins in miR-223 wild-type (WT) and knockout (KO) mice hippocampus. Representative Western blots shown. Graph on Right shows quantification of band intensities normalized to actin. Means ± SEM are shown (n = number of mice analyzed). *P = 0.01, **P = 0.005 (two-tailed t test). (B) MiR-223 target site in GluR2. WT, wild type; Mut-1, entire seed sequence mutant; Mut-2, seed sequence point mutant. (C) Dual-luciferase assay for GluR2 target site illustrated (B) in HEK 293 cells stably expressing miR-223. (D) MiR-223 target site in NR2B. Mut, entire seed sequence mutant. (E) Dual-luciferase assay for NR2B target site constructs illustrated in D. Luciferase assay graphs show Renilla/Firefly luciferase activity measured 48 h posttransfection. Means ± SEM of representative experiments performed in six replicates are shown. (F) Representative mEPSC traces of recordings from hippocampal slices from WT or miR-223 KO mice. (G) Cumulative probability distribution of mEPSC amplitude from WT and miR-223 KO mice (P < 0.05, Kolmogov–Smirnov test). (H) Half-width of and (I) decay time for mEPSCs from WT and miR-223 KO hippocampal slices. For G–I, n = 3 animals, 10–12 cells per group. For H and I, 1,029 and 1,405 mEPSCs were analyzed for WT and KO, respectively. ***P < 0.001, **P < 0.01, *P = < 0.05 (two-tailed t test). (J and K) Decay time for NMDA mEPSCs in primary hippocampal neurons (PHNs) derived from miR-223 WT (J) and KO (K) mice, ± the specific NR2B inhibitor ifenprodil (P < 0.05, Kolmogov–Smirnov test). (L and M) mEPSCs-AMPA amplitude in PHNs derived from miR-223 WT (L) and KO (M) mice, ± PhTx433 (a calcium-permeable AMPAR inhibitor). (M) (P < 0.05, Kolmogov–Smirnov test). (J–M) n = 10 in each group.
To determine whether miR-223–induced changes in GluR2 and NR2B alter the physiologically relevant pool of AMPA and NMDA receptors, we recorded miniature excitatory postsynaptic currents (mEPSCs) in hippocampal slices from miR-223 KO and WT littermates (Fig. 2F). The amplitude of mEPSCs in miR-223 KO mice was significantly increased (Fig. 2G and Fig. S2E; P < 0.05, Kolmogov–Smirnov test), consistent with an increase in functional synaptic AMPARs. As measured by both mEPSC half-width (Fig. 2H) and decay time (Fig. 2I), miR-223 KO mice demonstrate a profile consistent with increased NMDA receptor current (t test, P < 10−8). Together, these data indicate that genetic deletion of miR-223 enhances the function of NMDA and AMPA receptors in the hippocampus.
To investigate miR-223 regulation of the NMDAR phenotype, mEPSCs-NMDA decay time was assessed in primary hippocampal neurons derived from miR-223 WT and KO mice, in the presence or absence of the specific NR2B inhibitor ifenprodil. A longer decay time was observed in miR-223 KO neurons consistent with up-regulation of NMDAR containing the NR2B subunit (Fig. 2 J and K). MiR-223 regulation of the AMPAR phenotype was investigated. High sensitivity to philanthotoxin-433 (PhTX-433) treatment (Fig. 2 L and M) and inward rectification of synaptic currents (Fig. S2F) were observed, a signature of calcium-permeable AMPARs. These findings suggest that miR-223 regulates the synaptic phenotypes of NMDARs and AMPARs.
MiR-223 Regulates NMDA-Induced Calcium Influx in Hippocampal Neurons.
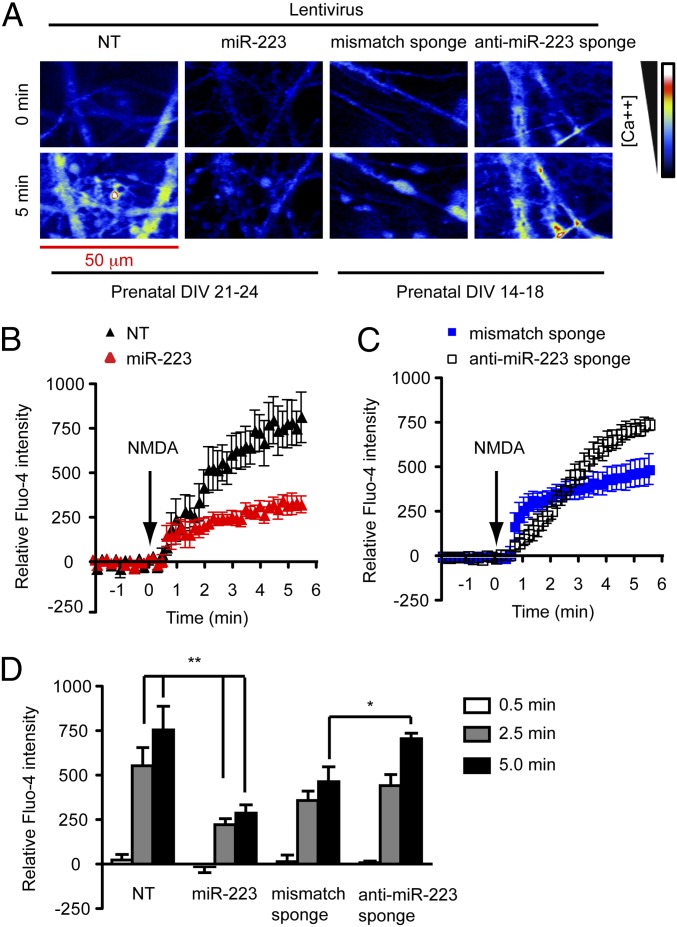
To determine whether miR-223 acts cell autonomously to regulate excitability, Fluo-4 calcium mobilization assays in primary hippocampal neurons accompanied by miR-223 gain or loss of function were performed. Calcium influx was triggered by a sublethal 100-μM NMDA stimulation, which was inhibited by NMDAR antagonist (5S,10R)-(+)-5-Methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine (MK801) (10 μM) and AMPAR antagonist 6,7-Dinitroquinoxaline-2,3(1H,4H)-dione (DNQX) (10 μM) (Fig. S3 A and B). This might be attributed to the previously reported DNQX affinity for the glycine site of the NMDAR (21). For gain of function (GOF), miR-223 or a nontargeting (NT) miRNA control were overexpressed. For loss of function (LOF), an anti–miR-223 sponge or a mismatch sponge control was used. The activity of the miR-223 sponge in inhibiting miR-223 function was verified using a 3′-UTR luciferase assay (Fig. S3G). Calcium influx by confocal microscopy live imaging was determined over 5 min after establishing a baseline. These conditions did not induce neural cell death throughout the live imaging duration (Fig. S3F). GOF experiments demonstrate that miR-233 but not NT-miRNA inhibited NMDA-induced calcium influx in 21–24 d in vitro (DIV) primary hippocampal neurons (Fig. 3 A, B, and D). On the other hand, the anti–miR-223 sponge but not the sponge-mismatch control enhances NMDA-induced calcium influx in DIV 14–18 primary hippocampal neurons (Fig. 3 A, C, and D). DIV 14–18 hippocampal neurons were used because anti–miR-223 sponge-expressing neurons’ viability decreased by DIV 21–24. In addition, postnatal hippocampal neurons isolated from miR-223 KO mice demonstrated enhanced NMDA-induced calcium influx compared with their WT littermates (Fig. S3 C–E). These findings demonstrate that miR-223 regulates NMDA-induced calcium influx in hippocampal neurons.
Fig. 3.
MiR-223 regulates NMDA-induced calcium influx in hippocampal neurons using Fluo-4 calcium mobilization assay. (A) Confocal images of Fluo-4 loaded hippocampal neurons transduced with lentiviral vectors encoding nontargeting miRNA (NT) (n = 7), miR-223 (n = 9), mismatch sponge (n = 9), or anti–miR-223 sponge (n = 6) before (0 min) and after (5 min) 100 μM NMDA stimulation. (B–D) Quantification of regions of interest demonstrating effect of miR-223 on increased fluorescence over time in response to NMDA administered at 140 s. Data represent mean ± SEM. **P < 0.01, *P < 0.05 two-tailed t test.
MiR-223 Protects Against in Vitro and in Vivo NMDA-Induced Excitotoxicity.
To determine whether miR-223 could reduce neuronal cell death in vitro, an excitotoxicity assay was performed in primary hippocampal neurons. Overexpression of miR-223 but not NT-miRNA decreases neuronal loss following a lethal (500 μM) NMDA insult (Fig. S4 A and B). These findings indicate that miR-223 confers neuroprotection against excitotoxicity in primary hippocampal neurons.
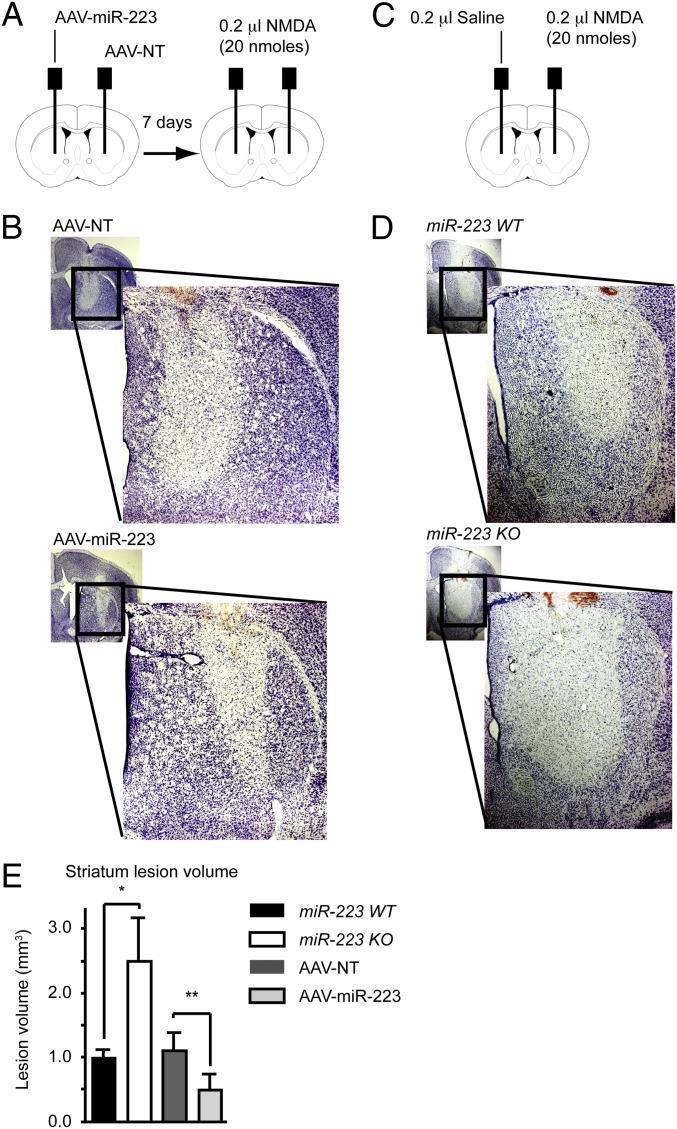
To activate excitotoxicity in vivo, 20 nmols of NMDA were injected into the striatum. The extent of neuronal damage was assessed 48 h following the injections by Nissl staining. To test the neuroprotective effect of miR-223 in vivo, an adeno-associated viral vector (AAV) was used to overexpress miR-223 (AAV–miR-223) or a NT-miRNA control (AAV-NT) in the striatum (Fig. 4A). MiR-223 overexpression in the striatum is associated with decreased NMDA-induced lesion volume compared with NT-miRNA overexpression (Fig. 4 B and E and Fig. S4C). On the other hand, miR-223 KO mice have nearly twice the striatal lesion volume compared with WT littermate mice (Fig. 4 D and E and Fig. S4 D and E), whereas the saline-injected mice exhibited a negligible lesion along the needle track in both miR-223 KO and WT littermate mice. In this context, it is important to note that AAV showed strong preference to neurons in the striatum (Fig. S4F). AAV-mediated overexpression of miR-223 was robust in primary neurons. In vivo overexpression of miR-223 showed a modest but significant increase in its expression (Fig. S4G). Taken together, these findings support the notion that miR-223 is a neuroprotective miRNA.
Fig. 4.
MiR-223 protects against excitotoxic damage in vivo. (A) Schematic diagram of AAV viral vector then NMDA striatum stereotaxic injections. (B) Nissl stain demonstrating damage in striatum following NMDA injection as indicated by reduced staining areas. (C) Schematic diagram of NMDA striatum stereotaxic injections. (D) Nissl stain demonstrating damage in striatum following NMDA injection as indicated by reduced staining areas. (E) Striatum lesion volumes quantification. Means ± SEM are shown (*P = 0.041 two-tailed t test), WT (n = 5), KO (n = 4), (*P = 0.033 paired t test), AAV-NT (n = 5), AAV-miR-223 (n = 5).
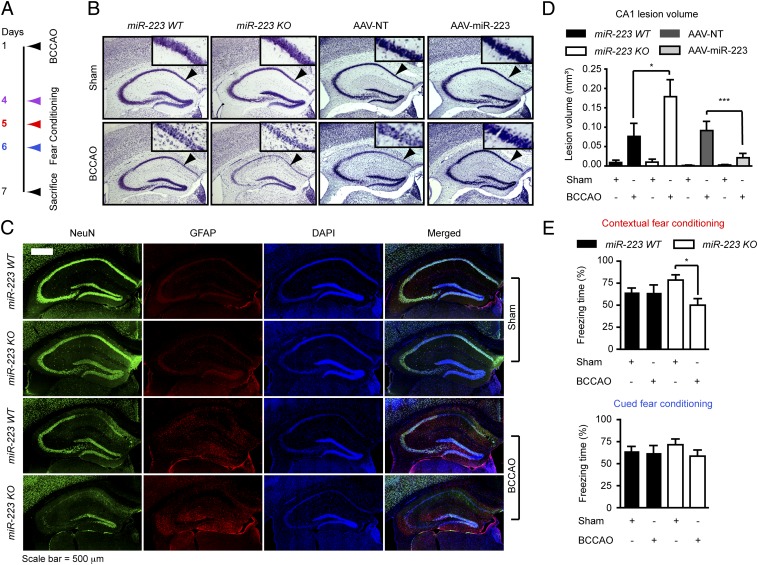
MiR-223 Modulates Contextual Memory and Delayed Hippocampal Cornu Ammonis 1 (CA1) Neuronal Cell Death Following Transient Global Ischemia.
To test the physiologic relevance of miR-223 regulation of glutamate receptors in vivo, transient global ischemia by temporary bilateral common carotid artery occlusion (BCCAO) for 20 min followed by reperfusion was performed. Selective delayed CA1 neuronal death in the hippocampus and memory retention was evaluated (Fig. 5A). The extent of neuronal loss in the hippocampus CA1 region was assessed by Nissl staining. MiR-223 KO mice had more severe CA1 neuronal loss after BCCAO compared with WT mice as determined by loss of Nissl staining (Fig. 5B) and the loss of antineuronal nuclei marker (anti-NeuN) immunostaining (Fig. 5C and Fig. S5A). On the other hand, mice overexpressing miR-223 (AAV–miR-223) in the CA1 area demonstrate less severe CA1 neuronal loss after BCCAO compared with mice overexpressing NT-miRNA (AAV-NT) (Fig. 5B and Fig. S5B). Quantification of Nissl staining loss in the CA1 region revealed that the lesion volume in KO mice was twice that observed in WT mice, whereas the lesion in the AAV–miR-223 group is decreased compared with the AAV-NT control group (Fig. 5D). Memory retention was assayed by a fear-conditioning test (Fig. S5C). There was no difference between miR-223 WT mice and KO mice in footpad-shock–induced pain threshold (Fig. S5D) excluding differences in pain perception as a confounder. Linear regression analysis reveals a strong correlation between the CA1 lesion volume and hippocampal-dependent contextual fear conditioning memory deficit but not the amygdala-dependent cued memory (Fig. S5 E and F). There is no significant difference in memory retention between WT and KO mice subjected to sham surgery (Fig. 5E). Whereas WT mice exhibit no difference in fear conditioning memory retention after BCCAO, miR-223 KO mice exhibit significant contextual but not cued fear conditioning memory deficit (Fig. 5E). On the other hand there was no difference in fear conditioning memory retention after BCCAO in both AAV–miR-223 and AAV-NT groups (Fig. S5 G and H). It is worth noting that AAV had a strong preference to neurons in the CA1 region (Fig. S5I). Overexpression of miR-223 in the CA1 region was modest but significant, which is likely due to a dilution effect because the assay was done on the whole hippocampus (Fig. S4G). Taken together, these data suggest a neuroprotective role for miR-223 against ischemia–reperfusion-induced neuronal cell death following transient global ischemia in vivo.
Fig. 5.
MiR-223 modulates contextual memory and delayed CA1 neuronal cell death following transient global ischemia. (A) Schematic diagram of experimental time line. BCCAO, bilateral common carotid artery occlusion. (B) Nissl stain demonstrating damage in CA1 region (arrowheads) of the hippocampus following BCCAO as indicated by reduced staining areas. (C) Immunostaining for neuronal nuclei marker (NeuN), GFAP, and DAPI showing neuronal loss in the CA1 hippocampal region following BCCAO in miR-223 WT and KO mice. (D) Lesion volumes of the CA1 quantification. Means ± SEM are shown (*P < 0.05, ***P < 0.001, one-way ANOVA followed by Newman–Keuls multiple comparison test). WT sham (n = 7), WT BCCAO (n = 6), KO sham (n = 4), KO BCCAO (n = 5). AAV-NT sham (n = 9), AAV-NT BCCAO (n = 7), AAV–miR-223 sham (n = 6), AAV–miR-223 BCCAO (n = 8). (E) Fear conditioning memory test, 24 h following training exposure to one cue (30-s tone) ending by a 2-s shock. Data points represent mean ± SEM (contextual fear conditioning one-way ANOVA P < 0.05, *P < 0.05 Tukey's multiple comparison post hoc test). WT sham (n = 17), WT BCCAO (n = 15), KO sham (n = 13), KO BCCAO (n = 13).
Discussion
Our findings reveal a functional role for miR-223 in the expression of the glutamate receptor subunits, GluR2 and NR2B. We show that genetic deletion of miR-223 results in selective increased expression of GluR2 and NR2B that has functional consequences. The amplitude of mEPSCs is increased consistent with an increase in functional AMPA receptors at the synapse and NMDA-mediated calcium influx is increased in miR-223–deficient neurons. Synaptic calcium-permeable AMPARs are increased, suggesting a change in APMAR phenotype. The NR2B subunit was found previously to enhance the NMDAR excitotoxicity (22). This explains the neuroprotective effect of miR-223 overexpression and the enhanced vulnerability of the hippocampal CA1 neurons in miR-223 KO mice to excitotoxicity and BCCAO compared with WT mice. Unlike the calcium-permeable NMDARs, calcium permeability of AMPARs is regulated by GluR2 pre-mRNA editing at Q/R site 607, rendering AMPARs containing the GluR2 subunit calcium impermeable. AMPARs lacking GluR2 or containing unedited GluR2 are permeable to calcium and promote cell death following ischemia–reperfusion injury. GluR2 607 Q/R editing is disrupted in CA1 pyramidal neurons following forebrain ischemia, resulting in high calcium permeability of AMPARs and increased cell death (23). Cumulative evidence indicates that the increase in AMPAR excitotoxicity results from down-regulation of GluR2 expression in CA1 neurons following ischemia (24, 25). On the other hand, in the absence of GluR2, AMPARs are less efficiently assembled at the synapse in hippocampal neurons (26). This might help explain the neuroprotective effect of miR-223 overexpression in the CA1 region following BCCAO. However, in other areas of the brain, we cannot exclude the possibility that miR-223 overexpression might increase the proportion of calcium-permeable AMPARs, which could promote ischemia-induced cell death. Taken together, these data suggest that the NMDAR and AMPAR composition and function are regulated by miR-223. It is well established that NMDAR-mediated calcium influx increases nitric oxide production (27). It is therefore likely that miR-223 KO mice’s neural vulnerability to cell death by excitotoxicity or ischemia–reperfusion injury is mediated at least in part by enhanced nitric oxide neurotoxicity. Thus, miR-223 is a major regulator of glutamate receptor expression and function and it plays a major role in neuronal survival.
Because miR-223 deficiency results in an expanded hypersensitive granulocytic compartment and an increased oxygen burst by phagocytic cells (13), this raises the possibility that inflammation plays a role in the enhanced neural cell death observed in miR-223 KO mice following excitotoxicity or ischemia–reperfusion injury. However, our local miR-223 overexpression experiments in the striatum and CA1 region of the hippocampus argue against a significant primary role for infiltrating inflammatory cells in the observed difference in neural cell death in vivo. In addition, the selective preference of AAV for neurons (Figs. S4F and S5I) suggests that the neuroprotective effect of miR-223 is cell autonomous. However, further studies are needed to determine whether a local brain immune response is under the regulation of miR-223 and whether it plays a role in its neuroprotective effect.
Functional target sites for miR-223 in GluR2 and NR2B mRNAs 3′-UTR were identified, verified, and are consistent with bioinformatics predictions. However, the expression of miR-223 might not be stoichiometrically matched to GluR2, NR2B, and other potential neuronal target mRNAs. Mammalian miRNAs, which are low-abundance miRNAs such as miR-223, can use extensive 3′-supplementary pairing in addition to the seed pairing to limit the amount of miRNA to a small fraction of target sites (1). We find extensive 3′-base pairing between miR-223 and its GluR2 and NR2B 3′-UTR target sites (Fig. S2 C and D). In addition, previous studies in miR-223 KO mice demonstrated that lowered mRNA levels account for 84% or more of decreased protein levels, implicating mRNA destabilization as the major mechanism for repression (28, 29). Thus, a catalytic model for miR-223 regulation of its targets is more likely than a stoichiometric model.
In summary, these data reveal a unique function for miR-223 in the CNS by targeting the AMPAR subunit GluR2 and the NMDAR subunit NR2B, which control neuronal excitability in response to glutamate. Small molecule modulators of miR-223 levels in neurons or direct delivery of miR-223 could be a potentially useful strategy to protect against neuronal cell death in stroke and other excitotoxic neuronal disorders.
Materials and Methods
Real-Time PCR.
MicroRNA quantitative real-time PCR was done using the Taqman real-time probes. The U6 snRNA level was used to normalize miR-223 values.
Hippocampal Slice Electrophysiological Recording.
Horizontal hippocampal slices (300 μm) were prepared from mice (postnatal day 15–20) using standard methods and as described in SI Materials and Methods.
Fluo-4 Calcium Mobilization Assay.
Confocal laser scanning microscope was used to monitor the Fluo-4 calcium indicator dye in real-time following stimulation with 100 μM NMDA in HBSS + 2 mM calcium chloride + 10 μM glycine.
Transient Global Ischemia Model.
Male mice (age: 8–12 wk) were anesthetized with 2–3% (vol/vol) isoflurane for induction and 1–1.5% for maintenance. BCCAO was performed for 20 min followed by reperfusion. Arterial blood pH, PO2, glucose, hemoglobin, and hematocrit did not show differences between groups.
Fear Conditioning (FC).
Mice were placed individually in a fear-conditioning apparatus chamber. Freezing (immobility) was traced and analyzed using a video-based tracking system (Fig. S5C).
Stereotaxic Injection.
Coordinates used for striatum: from bregma rostral, −0.5 mm; lateral, ±2.0 mm, and ventral, −4.0 mm. Coordinates used for CA1 area: from bregma caudal, 2.0 mm; lateral, ±1.6 mm, and 1.5 mm ventral.
Statistical Analysis.
Unless otherwise indicated, multiple group comparisons were evaluated using one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison post hoc test. Two group comparisons were evaluated using a two-tailed t test. Data were expressed as mean ± SEM. Experiments for quantification were performed in a blinded fashion.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute on Drug Abuse Grant DA000266 and a Maryland Stem Cell Research Fund fellowship (to M.M.H.). T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121288109/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 3.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr Opin Cardiol. 2011;26(3):181–189. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- 5.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10(12):837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambari R, et al. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82(10):1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371(9624):1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 8.Lee JM, Zipfel GJ, Choi DW. The changing landscape of ischaemic brain injury mechanisms. Nature. 1999;399(6738)(Suppl):A7–A14. doi: 10.1038/399a007. [DOI] [PubMed] [Google Scholar]
- 9.Karr J, et al. Regulation of glutamate receptor subunit availability by microRNAs. J Cell Biol. 2009;185(4):685–697. doi: 10.1083/jcb.200902062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saba R, et al. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32(3):619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65(3):373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 14.Fazi F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Pulikkan JA, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 17.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler M, Baudry M, Lynch G. Quinoxaline derivatives are high-affinity antagonists of the NMDA receptor-associated glycine sites. Brain Res. 1989;489(2):377–382. doi: 10.1016/0006-8993(89)90875-5. [DOI] [PubMed] [Google Scholar]
- 22.Martel MA, et al. The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron. 2012;74(3):543–556. doi: 10.1016/j.neuron.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng PL, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49(5):719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Kwak S, Weiss JH. Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr Opin Neurobiol. 2006;16(3):281–287. doi: 10.1016/j.conb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci. 2007;30(3):126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Sans N, et al. Aberrant formation of glutamate receptor complexes in hippocampal neurons of mice lacking the GluR2 AMPA receptor subunit. J Neurosci. 2003;23(28):9367–9373. doi: 10.1523/JNEUROSCI.23-28-09367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sattler R, et al. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284(5421):1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- 28.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.