Abstract
Chromatin remodeling is essential for controlling the expression of genes during development. The histone-modifying enzyme G9a/KMT1C can act both as a coactivator and a corepressor of transcription. Here, we show that the dual function of G9a as a coactivator vs. a corepressor entails its association within two distinct protein complexes, one containing the coactivator Mediator and one containing the corepressor Jarid1a/KDM5A. Functionally, G9a is important in stabilizing the Mediator complex for gene activation, whereas its repressive function entails a coordinate action with the histone H3 lysine 4 (H3K4) demethylase Jarid1a for the maintenance of gene repression. The essential nature of cross-talk between the histone methyltransferase G9a and the demethylase Jarid1a is demonstrated on the embryonic Ey-globin gene, where the concurrent introduction of repressive histone marks (dimethylated H3K9 and dimethylated H3K27) and removal of activating histone mark (trimethylated H3K4) is required for maintenance of gene silencing. Taken together with our previous demonstration of cross-talk between UTX and MLL2 to mediate activation of the adult βmaj-globin gene, these data suggest a model where “active” and “repressive” cross-talk between histone-modifying enzymes coexist on the same multigene locus and play a crucial role in the precise control of developmentally regulated gene expression.
Keywords: beta-globin locus, epigenetics, erythroid differentiation, hemoglobin
Histones are subjected to a number of posttranslational modifications that play important roles in regulating diverse cellular processes. These modifications are highly dynamic and are established through a competition between “writer” enzymes that introduce the modifications and “eraser” enzymes that remove them (1). Proper coordination between histone-modifying enzymes is critical for the regulation of gene expression (2, 3). For example, it has been shown that the histone H3 lysine 4 (H3K4) methyltransferases MLL3/4 (also called KMT2C/B) and the H3K27 demethylase UTX (also called KDM6A) are part of the same protein complex and coordinate their histone-modifying functions to activate the expression of specific genes through the concurrent introduction of the active histone mark trimethylated H3K4 (H3K4me3) and removal of the repressive histone mark H3K27me3 (4–6). These results underline the importance of cross-talk between histone methyltransferase and demethylase enzymes to activate transcription.
G9a (also called Ehmt2 or KMT1C) and its homolog GLP (also called Ehmt1 or KMT1D) have been identified as major euchromatic methyltransferases. These enzymes work as heterodimers to introduce monomethyl and dimethyl modifications on histone H3 at Lys-9 [single- or dimethylated H3K9 (H3K9me1/me2] (7–10). In addition, several studies have shown that G9a and GLP mediate dimethylation of H3K27, both in vitro (11) and in vivo (12–14).
It has been well established that G9a can repress transcription by the introduction of repressive histone modifications (H3K9me2 and H3K27me2) and/or the recruitment of DNA methyltransferases (reviewed in 14, 15). In addition, G9a can activate transcription of specific genes through a mechanism that does not require its methyltransferase activity (12, 16–19). It is currently unclear how G9a can act to mediate both the activation and repression of transcription. In addition, we do not know whether the G9a-GLP heterodimer interplays with other histone methylating/demethylating enzymes.
Results
G9a and GLP Associate with the Coactivator Complex Mediator and the Corepressor Protein Jarid1a.
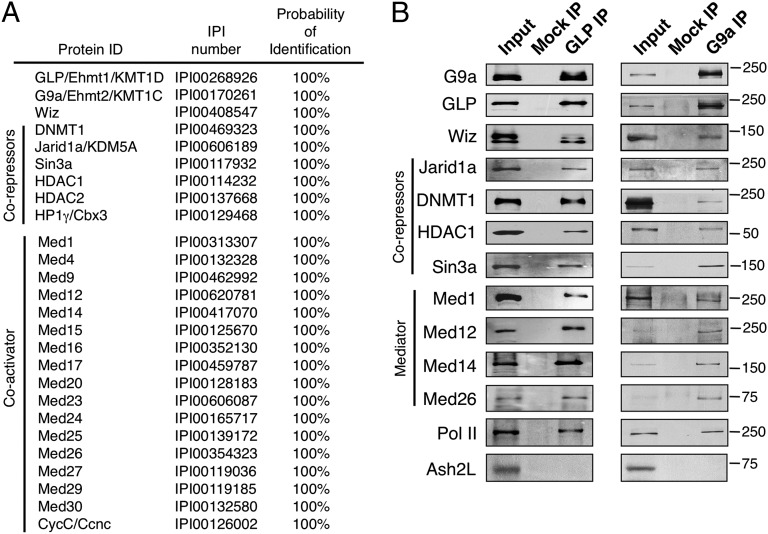
To identify proteins that interact with the G9a-GLP heterodimer, we performed immunoprecipitation (IP) experiments using anti-GLP Abs and normal IgG as a negative control. Endogenous proteins were immunoprecipitated from nuclear extracts of murine erythroid (MEL) cells and identified by mass spectrometry (MS) (Fig. 1A and Dataset S1). Validating our approach, G9a and GLP themselves were identified, as well as a number of previously known G9a-interacting partners, including Wiz (20), DNMT1 (21), and HP1γ (22, 23). In addition, several previously unknown G9a-GLP interacting proteins, including both coactivators and corepressors, were identified, which is consistent with the dual role of G9a in regulating transcription (Fig. 1).
Fig. 1.
Identification of Jarid1a and the Mediator complex as previously undescribed G9a- and GLP-interacting proteins. (A) MS analysis of a GLP immunoprecipitate. The probability of identification was determined using ProteinProphet (44). IPI, International Protein Index. (B) Confirmation of the interaction of GLP and G9a with Jarid1a and the Mediator complex. Proteins immunoprecipitated via Abs against GLP and G9a were analyzed by Western blot. A mock IP with normal IgG was used as a negative control. Abs used for Western blot (Left) and molecular masses (Right; in kilodaltons) are indicated.
Among the coactivators that interact with G9a-GLP, 17 subunits of the Mediator complex, which is a fundamental component of the RNA polymerase II (Pol II) transcriptional machinery (24), were identified (Fig. 1). This result is consistent with our previous finding that G9a interacts with Pol II (12) and with the previously shown interaction between G9a and Med12 (25). Furthermore, we confirmed the association of G9a-GLP with the Mediator complex by both independent G9a and GLP IPs revealed by Western blot (Fig. 1B) and by a reciprocal Med1 IP in which Med12, G9a, and GLP were detected (Fig. 2B, Right).
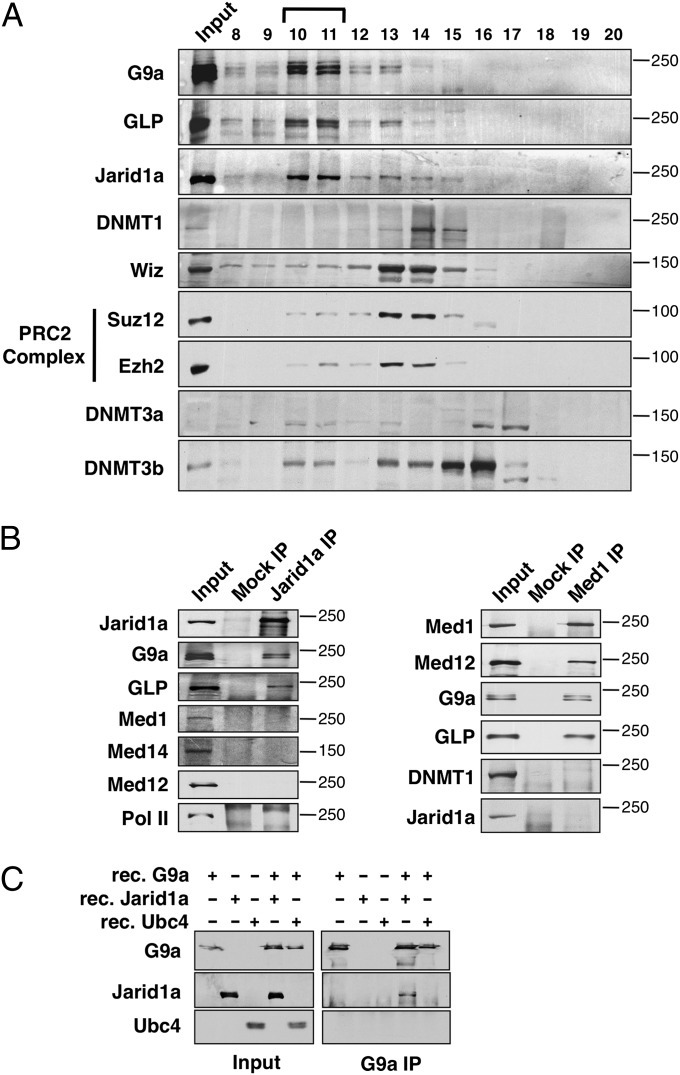
Fig. 2.
Jarid1a is tightly associated with the G9a-GLP heterodimer. (A) G9a, GLP, and Jarid1a coelute by size exclusion chromatography. A nuclear extract was separated on a Superose-6 column, and the presence of specific proteins in different fractions revealed by immunoblotting with Abs indicated on the left. Molecular masses (in kilodaltons) are indicated on the right. (B) G9a-GLP heterodimer associates with Jarid1a and the Mediator complex in a mutually exclusive manner. Proteins immunoprecipitated via Abs against Jarid1a and Med1 were analyzed by Western blot. Mock IPs with normal IgG were used as negative controls. Abs used for Western blot (Left) and molecular masses (Right; in kilodaltons) are indicated. (C) Jarid1a interacts directly with G9a. Rec purified Jarid1a or Ubc4 was incubated with rec purified G9a before IP with G9a Abs. Immunoprecipitated proteins were revealed by immunoblotting with the Abs indicated on the left.
In addition to the coactivator complex Mediator, several corepressor proteins were identified as G9a-GLP–associated partners, including the DNA methyltransferase DNMT1, the histone deacetylases HDAC1 and HDAC2, and HP1γ. Furthermore, the H3K4me3 demethylase Jarid1a (also called KDM5A or Rbp2) (26–28) was identified as a previously unknown G9a-GLP–interacting protein (Fig. 1).
In an attempt to separate the various corepressors that associate with G9a and GLP, we performed size exclusion chromatography on nuclear extracts. Consistent with G9a and GLP being part of a heterodimer (10), these two proteins coelute in the same two gel filtration fractions. Strikingly, the main peak of Jarid1a comigrates with the main peak of G9a and GLP (Fig. 2A, fractions 10 and 11). This is in contrast to other G9a-interacting proteins, such as DNMT1 (21), DNMT3a/b (29), and Wiz (20), or Jarid1a-interacting proteins Ezh2 and Suz12 (30, 31), which coelute with G9a and GLP but are present predominantly in lower molecular mass fractions (Fig. 2A, fractions 13–16). This result suggests that Jarid1a and the G9a-GLP heterodimer might be tightly associated in the cell. To address this possibility further, we performed a reciprocal IP using Abs against endogenous Jarid1a. We observed both G9a and GLP in the Jarid1a IP fraction, which confirms the interaction between these proteins (Fig. 2B). To determine whether the interaction between G9a and Jarid1a is direct, we incubated recombinant (rec) purified G9a and Jarid1a proteins and performed a G9a IP. As shown in Fig. 2C, rec G9a specifically pulled down rec Jarid1a (but not the control rec Ubc4), demonstrating a direct interaction between these proteins. Taken together, these results indicate that G9a, GLP, and Jarid1a are tightly associated within the same protein complex that is distinct from the core PRC2 complex containing Ezh2 and Suz12 (32).
Because these results were obtained using a mouse MEL cell line, we wanted to determine whether the interaction between G9a-GLP and Jarid1a is conserved in other cell types. To address this question, the G9a, GLP, and Jarid1a IPs, as well as the size exclusion chromatography experiments, were repeated with nuclear extract prepared from undifferentiated murine embryonic stem (mES) cells. The association between these proteins was reproduced in mES cells (Fig. S1), suggesting that the interaction between the G9a-GLP heterodimer and Jarid1a is conserved in cell types of different cell fate potential.
Cross-Talk Between G9a and Jarid1a Is Important for the Maintenance of Gene Repression.
The interaction between G9a and Jarid1a is particularly interesting in light of the cross-talk between histone methyltransferase and demethylase that has been implicated in the regulation of transcription. Indeed, it has been shown previously that the interaction between the H3K4 methyltransferases MLL3/4 and the H3K27 demethylase UTX is important for the activation of transcription (4–6). In this context, the detected interaction between G9a and Jarid1a suggests that cross-talk between methyltransferase and demethylase might also be important to maintain gene repression through a mechanism that involves the concurrent introduction of repressive histone marks (H3K9me2 and H3K27me2) by G9a and removal of active histone mark (H3K4me3) by Jarid1a. To test this possibility, we studied the mechanism of gene repression on the β-globin locus. This locus comprises several β-globin genes that are differentially regulated during development, including the embryonic gene Ey, which is repressed, and the adult gene βmaj, which is actively transcribed in differentiated MEL cells (33–36). Furthermore, it was shown previously that G9a maintains the Ey gene in a repressed state in a methyltransferase-dependent manner (12).
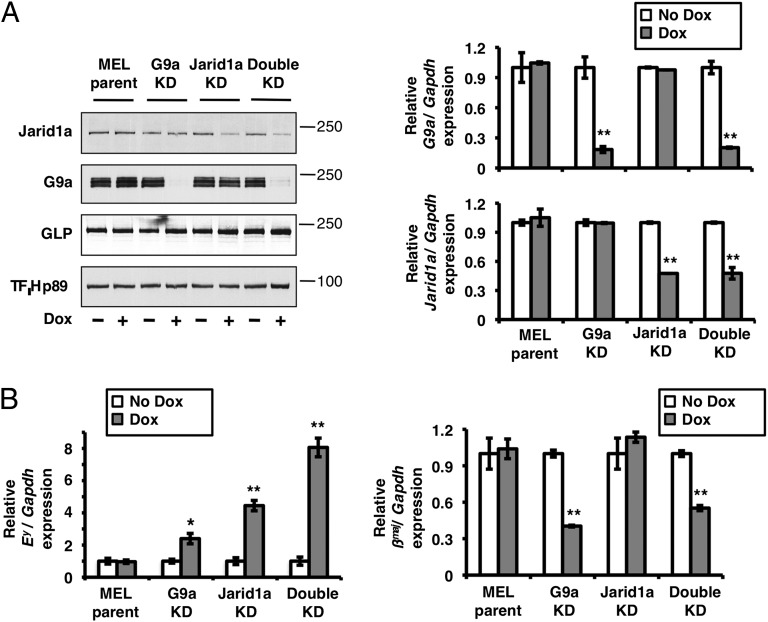
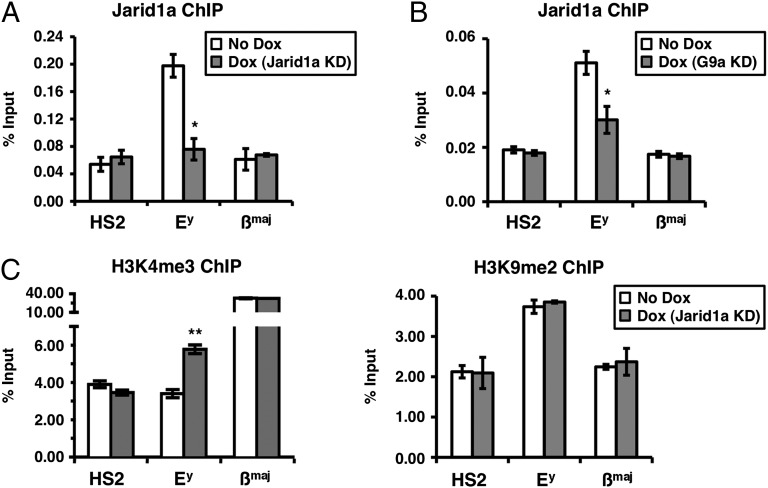
To determine whether Jarid1a is also involved in maintaining the Ey gene in a repressed state, stable MEL clones were generated in which the shRNA-mediated knockdown (KD) of Jarid1a can be induced on doxycycline (Dox) treatment. We found that although the KD of Jarid1a (Fig. 3A) does not affect the proliferation of cells during erythroid differentiation (Fig. S2), it leads to derepression of the embryonic Ey globin gene (Fig. 3B, Left), suggesting that Jarid1a is involved in maintaining this gene in a silenced state. In contrast, Jarid1a KD has no effect on transcription of the adult βmaj globin gene (Fig. 3B, Right). To determine whether Jarid1a directly represses Ey, the binding of Jarid1a to the promoter of this gene was analyzed by ChIP. Jarid1a was found to bind to the Ey gene promoter but not to the βmaj promoter or the distal enhancer HS2 in differentiated erythroid cells (Fig. 4A). In addition, Jarid1a binding was lost on Jarid1a KD (Fig. 4A), which validates the specificity of the Jarid1a Ab. Interestingly, Jarid1a binding to the Ey promoter is also decreased on the KD of G9a (Fig. 4B), emphasizing the importance of the interaction between G9a and Jarid1a for the binding of Jarid1a to chromatin. We note, however, that in the absence of G9a, Jarid1a binding to the Ey promoter is not completely lost (compare Fig. 4 A and B). This residual weak binding could be due to the association of Jarid1a with other chromatin-associated proteins, such as GLP, which continues to interact with Jarid1a even in the absence of G9a (Fig. S3). Importantly, control experiments demonstrate that the KD of Jarid1a does not affect the amount of G9a (at the transcript and protein levels) and, similarly, that the KD of G9a does not affect the amount of Jarid1a in the cell (Fig. 3A). To determine whether Jarid1a actively demethylates H3K4me3 on the repressed Ey gene, we performed an H3K4me3 ChIP experiment. Depletion of Jarid1a results in increased levels of H3K4me3 on the Ey promoter (but not on the βmaj promoter), whereas the H3K9me2 mark is not affected (Fig. 4C). We note that the level of increase in H3K4me3 observed on Ey on Jarid1a KD is equivalent to the H3K4me3 increase measured on the Sdf1 gene in Jarid1a−/− mouse embryonic fibroblasts (MEFs) (26). Taken together, these results indicate that Jarid1a is involved in actively demethylating H3K4me3 on the Ey gene’s promoter.
Fig. 3.
Jarid1a and G9a cooperate in the maintenance of gene repression. (A) Establishment of stable MEL cell clones in which the single KDs of Jarid1a or G9a and the double KD of Jarid1a plus G9a are induced by Dox-mediated expression of specific shRNAs. This figure shows the results using Jarid1a shRNA1 (SI Materials and Methods). The levels of Jarid1a and G9a in Dox-treated (Dox) vs. untreated (No Dox) cells were analyzed at the protein level by Western blot of nuclear extracts using the indicated Abs (Left) and at the mRNA level by real-time RT-qPCR (Right). MEL parent represents a control cell line with no induction of shRNA on Dox treatment. (Left) Molecular masses of proteins (in kilodaltons) are indicated on the right. (Right) Transcript levels are normalized to GAPDH with the ratio observed in the absence of Dox set to 1. (B) Additive effect of the double Jarid1a plus G9a KD on derepression of the Ey globin gene. Transcripts levels were measured by real-time RT-qPCR in Dox-treated/untreated cells. Transcript levels are normalized to GAPDH with the ratio observed in the absence of Dox set to 1. (A and B) Average values from triplicate experiments are represented with error bars corresponding to SDs. *P < 0.05; **P < 0.01.
Fig. 4.
Jarid1a maintains the globin gene Ey in a repressed state through demethylation of H3K4me3. (A) Jarid1a binds to the Ey promoter in erythroid cells. Jarid1a ChIPs were performed in the presence of normal (No Dox) or reduced (Dox) levels of Jarid1a. (B) G9a stabilizes the binding of Jarid1a to the Ey promoter. Jarid1a ChIPs were performed in the presence of normal (No Dox) or reduced (Dox) levels of G9a. (C) KD of Jarid1a leads to an increase in H3K4me3 on the Ey promoter. Native ChIPs were used to measure the levels of H3K4me3 and H3K9me2 in the presence of normal (No Dox) or reduced (Dox) levels of Jarid1a. (A–C) ChIPs were revealed by real-time qPCR using Taqman probes located at the HS2 site of the locus control region and at the promoters of the Ey and βmaj genes. Error bars represent SDs calculated from triplicate experiments. *P < 0.05; **P < 0.01.
Finally, to determine whether G9a and Jarid1a work together to maintain the Ey gene in a repressed state, we generated clones in which double KD of G9a and Jarid1a is induced on Dox treatment. Study of these clones revealed that the G9a/Jarid1a double KD has an additive effect on the derepression of Ey compared with either single KD on its own (Fig. 3; confirmation of this result in a clone expressing a shRNA targeting a different region of the Jarid1a transcript is shown in Fig. S4), indicating that G9a and Jarid1a work in combination to maintain the Ey gene in a transcriptionally silenced state. Taken together, these results show that cross-talk between the methyltransferase G9a and the demethylase Jarid1a occurs on the Ey globin gene and is important to maintain this gene in a repressed state.
G9a-Dependent Binding of the Mediator Complex.
We have shown previously that G9a participates in the activation of adult βmaj globin gene transcription in a methyltransferase-independent manner (12). In contrast, Jarid1a does not appear to regulate transcription of the βmaj gene (Fig. 3B), suggesting that G9a could be present in distinct coactivator and corepressor complexes. To test this possibility, we performed IPs of both Jarid1a and Mediator. As shown in Fig. 2B, G9a, but not the Mediator complex (Med1, Med12, and Med14 subunits), coimmunoprecipitates with Jarid1a. Reciprocally, Jarid1a is not detected in the G9a complex immunoprecipitated with a Med1 Ab (Fig. 2B). This shows that G9a interacts with separate coactivator and corepressor complexes containing Mediator and Jarid1a, respectively.
Because the G9a–Jarid1a repressive complex is restricted to the transcriptionally silent Ey gene (Fig. 4A), we next examined the localization of the G9a–Mediator complex on the β-globin locus that comprises both the silent Ey globin and the active βmaj globin genes by ChIP. This experiment showed that the Mediator complex subunits Med1, Med12, Med14, and Med17 are bound to the active βmaj gene but not to the repressed Ey gene (Fig. S5). Furthermore, G9a is important for binding of the Mediator complex to the βmaj gene because Mediator ChIP signals are decreased on G9a KD (Fig. S5). Taken together, these results show that G9a is important for the assembly and/or stability of the Mediator complex to the promoter of the active βmaj gene promoter but not to the repressed Ey gene.
Discussion
Cross-Talk Between a Histone Methyltransferase and a Histone Demethylase in the Maintenance of Gene Silencing.
The establishment of tissue-specific gene expression programs during differentiation and development is influenced by changes in histone modifications. Gene activation encompasses the coordinated removal of histone marks that are refractory to the transcriptional process and introduction of histone marks that are permissive to transcription (37). Coordination of this process is permitted by several mechanisms of cross-talk between histone modifications, including the association of different histone-modifying enzymes within the same protein complex (2, 3). The prime example of cross-talk facilitating gene activation is the association between UTX and MLL3/4, which work together within the same protein complex to remove the repressive mark H3K27me3 and introduce the active mark H3K4me3, respectively (4–6). In this paper, we have identified previously undescribed cross-talk between two histone-modifying enzymes that play major roles in the maintenance of gene silencing: the H3K9/K27 methyltransferase G9a (14, 15) and the H3K4 demethylase Jarid1a (38). We show that these proteins interact directly and are part of the same protein complex in both erythroid cells and mES cells. Furthermore, our results show that the G9a–GLP–Jarid1a complex is distinct from the core PRC2 repressive complex that contains Ezh2 and Suz12, consistent with the previous finding that Jarid1a and the PRC2 complex bind to largely nonoverlapping genomic regions (31). Functionally, the results show that G9a stabilizes the binding of Jarid1a to chromatin and that these two enzymes work together to maintain gene repression through introduction of the repressive histone marks H3K9/27me2 and removal of the active histone mark H3K4me3. Thus, the maintenance of gene silencing is an active process permitted by cross-talk between the histone methyltransferase G9a and the demethylase Jarid1a. Furthermore, this mechanism may be generalized to additional Jarid proteins, because G9a is also present in a Jarid1c-containing complex (39).
Cross-Talk Between Histone-Modifying Enzymes Differentially Regulates Gene Expression Within a Multigene Locus.
G9a plays a crucial role as a “gatekeeper” of cell identity by protecting cells from spurious expression of genes from other lineages (15). For example, G9a prevents the expression of neuronal genes in nonneuronal cell types (40) and the expression of pluripotency genes in differentiated cells (29). Similarly, G9a controls the developmental timing of gene expression within specific lineages, as exemplified by the derepression of neuronal progenitor genes in G9a-ablated adult neurons (40). In addition, G9a has a dual role in regulating transcription on the β-globin locus, where it is involved in both repressing the embryonic gene Ey (through the introduction of repressive histone marks H3K9me2 and H3K27me2) and activating the adult gene βmaj (in a methyltransferase-independent manner) (12). However, it was not clear how G9a acts as both an activator and a repressor. The present study shows that the dual function of G9a entails an association with two physically distinct complexes: the coactivator Mediator and the corepressor Jarid1a. The existence of these distinct repressor and activator complexes is recapitulated on the β-globin locus, where the G9a–Mediator complex is bound to the active βmaj gene, whereas the G9a–Jarid1a complex is bound to the repressed Ey gene (Fig. 5). On the βmaj promoter, G9a stabilizes binding of the Mediator complex (Fig. S5) to enable formation of the preinitiation complex and maximal transactivation of this gene (12) (Fig. 3B). In contrast, on the Ey promoter, G9a is required for stable binding of Jarid1a (Fig. 4B) and for the maintenance of the Ey gene in a repressed state through cross-talk with Jarid1a (Fig. 5). Future studies are required to determine what regulates the association of G9a with Jarid1a vs. Mediator.
Fig. 5.
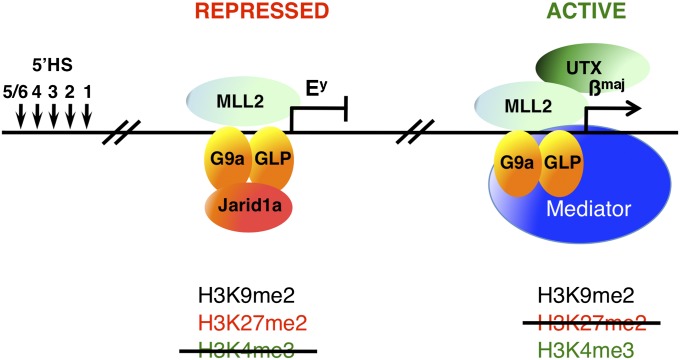
Model of the regulation of transcription on the β-globin locus through cross-talk between histone methyltransferases and demethylases. In adult erythroid cells, the embryonic globin gene Ey and the adult globin gene βmaj are both bound by the histone methyltransferases G9a-GLP, which introduce the repressive marks H3K9me2 and H3K27me2, and MLL2, which introduces the active mark H3K4me3. What differentiates these two genes is the presence of specific demethylases. Indeed, on the Ey gene, the coordinate actions of the H3K4me3 demethylase Jarid1a and the H3K9/K27 methyltransferase G9a-GLP allow for the maintenance of a transcriptionally repressed state. In contrast, on the βmaj gene, the demethylase UTX actively removes the repressive mark H3K27me2, whereas the methyltransferase MLL2 trimethylates H3K4, allowing full activation of this gene in erythroid cells. 5′HS1–6 represents the distal β-globin locus control region.
Previously, it was shown that the H3K4 methyltransferase MLL2/KMT2D is bound to both the silenced gene Ey and the active gene βmaj in differentiated erythroid cells (41). However, the MLL2-dependent H3K4me3 mark was detected on the βmaj gene but not on the Ey gene. At this point, it was not clear whether MLL2 is enzymatically inactive on the Ey gene or whether the H3K4me3 mark is dynamically removed. The finding here that Jarid1a demethylates H3K4me3 on the Ey gene promoter suggests that MLL2 could be enzymatically active at this location but that the H3K4me3 mark is removed by Jarid1a. In contrast, the adult βmaj gene, which is highly enriched for H3K4me3, is not bound by Jarid1a. Instead, this gene is bound by another demethylase (i.e., UTX), which actively removes the G9a-dependent repressive mark H3K27me2, thereby allowing activation of the βmaj gene (12). The presence of both MLL2 and UTX on the adult βmaj gene suggests that cross-talk between these histone-modifying enzymes is mediating activation of this gene. Therefore, there are at least two different examples of cross-talking between histone methyltransferases and demethylases that take place on the β-globin locus, one that activates transcription and one that represses transcription (Fig. 5, model).
Finally, it is interesting to note that although the repressed Ey gene and the active βmaj gene are bound by both the histone methyltransferases MLL2 and G9a, the active vs. repressed status of these genes correlates with the presence of the histone demethylases (UTX vs. Jarid1a).
Taken together, these results reveal that “active” and “repressive” cross-talk of histone-modifying enzymes coexists on the same multigene locus and plays a crucial role in the precise control of developmentally regulated gene expression.
Materials and Methods
Abs.
Abs used in this study are described in SI Materials and Methods.
Nuclear Extraction and IPs.
Nuclear extraction and IPs were performed as described (41), except that proteins were eluted at 37 °C by 30 min of vortexing at 200 × g in a 6-M urea buffer containing 0.05% SDS, 50 mM Tris (pH 8.3), and 5 mM EDTA. Eluted proteins were analyzed by Western blot and/or digested with trypsin and analyzed by liquid chromatography (LC)-tandem MS (MS/MS). Before performing IPs for Western blot analysis, ethidium bromide was added to the nuclear extract at a final concentration of 100 μg/mL.
LC-MS/MS.
LC-MS/MS was performed on a linear trap quadrupole (LTQ) mass spectrometer with a nanospray ion source and Surveyor HPLC (Thermo). Data analysis was performed using the Trans-Proteomics Pipeline (TPP), version 4.0 (42), with SEQUEST and the International Protein Index mouse database (43). A probability of identification for each protein was determined using the ProteinProphet algorithm (44) integrated in the TPP.
Size Exclusion Chromatography.
Nuclear extracts from MEL and mES cells were resolved by size exclusion chromatography using a Superose-6 10/300 GL column on an AKTA Explorer FPLC system following the instructions of the manufacturer (Amersham). Fractions of 0.5 mL were collected, precipitated with trichloroacetic acid (TCA), and analyzed by Western blot.
MEL Clones with Inducible KD of Jarid1a and G9a.
MEL clones with Dox-inducible G9a KD were described previously (12). MEL clones with an inducible KD of Jarid1a and clones with inducible double KD of G9a plus Jarid1a were established as previously described (12). Details are provided in SI Materials and Methods. KD was induced by treating cells with 5 μg/mL Dox for 4 d.
ChIPs.
Histone ChIPs were performed using a native ChIP protocol (45). X-ChIPs were performed as described (41). Real-time quantitative PCR (qPCR) was done on a Rotorgene 6000 (Corbett Research) using TaqMan probes and primers. ChIP qPCR signals are presented as a percentage of input. Statistical significance was determined using a Student t test. Abs used for ChIP and primers/probes used for qPCR are discussed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J.-F. Couture for discussions, S. Pradhan for the gift of rec G9a, J. Wu for technical help, and E. Pranckeviciene and G. Palidwor for help with MS data analysis. MS was done at the Ottawa Hospital Research Institute Mass Spectrometry Core Facility. This project was funded by Canadian Institutes of Health Research Grant MOP-89834 (to M.B.), Grant MOP-77778 (to F.J.D.), and Grant MOP-89910 (to W.L.S.). C.-P.C. was funded, in part, by a fellowship from the Canadian Institutes of Health Research/Thalassemia Foundation of Canada. C.-P.C and R.L.C. were funded, in part, by the Government of Ontario Ministry of Economic Development and Innovation for the Ontario Research Fund supporting the International Regulome Consortium. M.B., F.J.D., and W.L.S. hold Canadian Research Chairs in the Regulation of Gene Expression, Epigenetic Regulation of Transcription, and Integrative Stem Cell Biology, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213951109/-/DCSupplemental.
References
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135(4):604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 3.Verrier L, Vandromme M, Trouche D. Histone demethylases in chromatin cross-talks. Biol Cell. 2011;103(8):381–401. doi: 10.1042/BC20110028. [DOI] [PubMed] [Google Scholar]
- 4.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 6.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12(6):1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 9.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 10.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19(7):815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276(27):25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 12.Chaturvedi CP, et al. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci USA. 2009;106(43):18303–18308. doi: 10.1073/pnas.0906769106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, et al. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res. 2011;21(2):365–367. doi: 10.1038/cr.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25(8):781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins R, Cheng X. A case study in cross-talk: The histone lysine methyltransferases G9a and GLP. Nucleic Acids Res. 2010;38(11):3503–3511. doi: 10.1093/nar/gkq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281(13):8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehnertz B, et al. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J Exp Med. 2010;207(5):915–922. doi: 10.1084/jem.20100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell DJ, Jeong KW, Bittencourt D, Gerke DS, Stallcup MR. A distinct mechanism for coactivator versus corepressor function by histone methyltransferase G9a in transcriptional regulation. J Biol Chem. 2011;286(49):41963–41971. doi: 10.1074/jbc.M111.298463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell DJ, et al. Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription. J Cell Biochem. 2012;113(7):2406–2414. doi: 10.1002/jcb.24114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueda J, Tachibana M, Ikura T, Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem. 2006;281(29):20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 21.Estève PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20(22):3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin HG, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35(21):7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath SC, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27(4):596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 24.Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011;22(7):729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31(3):347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klose RJ, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128(5):889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128(6):1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128(6):1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasini D, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22(10):1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand M, Dilworth FJ. Modulation of Developmentally Regulated Gene Expression Programs Through Targeting of Polycomb and Trithorax Group Proteins. Chichester, UK: Wiley; 2012. [Google Scholar]
- 33.Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci USA. 2001;98(18):10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand M, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 35.Hosey AM, Chaturvedi CP, Brand M. Crosstalk between histone modifications maintains the developmental pattern of gene expression on a tissue-specific locus. Epigenetics. 2010;5(4):273–281. doi: 10.4161/epi.5.4.11522. [DOI] [PubMed] [Google Scholar]
- 36.Kiefer CM, Hou C, Little JA, Dean A. Epigenetics of beta-globin gene regulation. Mutat Res. 2008;647(1-2):68–76. doi: 10.1016/j.mrfmmm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 38.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10(3):413–424. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahiliani M, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447(7144):601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64(5):678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demers C, et al. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27(4):573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kersey PJ, et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4(7):1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 44.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 45.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3(3):398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.