Abstract
Cleavage stimulation factor 64 kDa (CstF64) is an essential pre-mRNA 3′ processing factor and an important regulator of alternative polyadenylation (APA). Here we characterized CstF64–RNA interactions in vivo at the transcriptome level and investigated the role of CstF64 in global APA regulation through individual nucleotide resolution UV crosslinking and immunoprecipitation sequencing and direct RNA sequencing analyses. We observed highly specific CstF64–RNA interactions at poly(A) sites (PASs), and we provide evidence that such interactions are widely variable in affinity and may be differentially required for PAS recognition. Depletion of CstF64 by RNAi has a relatively small effect on the global APA profile, but codepletion of the CstF64 paralog CstF64τ leads to greater APA changes, most of which are characterized by the increased relative use of distal PASs. Finally, we found that CstF64 binds to thousands of dormant intronic PASs that are suppressed, at least in part, by U1 small nuclear ribonucleoproteins. Taken together, our findings provide insight into the mechanisms of PAS recognition and identify CstF64 as an important global regulator of APA.
The 3′ ends of almost all eukaryotic mRNAs are formed cotranscriptionally by an endonucleolytic cleavage and the subsequent addition of a poly(A) tail (1–5). More than half of human genes produce alternatively polyadenylated mRNAs, and alternative polyadenylation (APA) has been increasingly recognized as a critical mechanism for eukaryotic gene regulation (6–11; reviewed in ref. 12). How poly(A) sites (PASs) are recognized and how APA is regulated remain poorly understood, however. Here PAS refers to the region including the cleavage site (CS) and all of the major cis elements required for recruiting the 3′ processing machinery to this site.
PAS recognition is mediated by protein–RNA interactions (3, 4). For example, the two critical cis elements found in the majority of mammalian PASs, the AAUAAA hexamer and the downstream U/GU-rich element, are recognized by the essential pre-mRNA 3′ processing factors cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF), respectively (3, 4). In vitro studies have shown that CPSF and CstF bind to RNAs in a synergistic manner (13–15). CstF is composed of three subunits, CstF77, CstF50, and CstF64 (16). CstF64 binds directly to RNA via its RNA recognition motif (RRM) (17). CstF64τ is a paralog of CstF64, and the two proteins have similar domain structures (18). CstF64τ has been isolated as a part of the CstF complex (19), but its functions in mRNA 3′ processing remain poorly understood. Although the AAUAAA hexamer is highly conserved, the downstream U/GU-rich elements are much more heterogeneous, and how CstF64 recognizes such divergent sequences is unclear (3, 4). CstF64 is also an important regulator of APA. For example, an increase in CstF64 protein levels during B-cell differentiation leads to a switch from the distal PAS containing a strong CstF64 binding sequence to a weaker proximal PAS in the IgM pre-mRNAs, resulting in the switch of IgM protein products from a membrane-bound form to a secreted form (20). A similar mechanism has been proposed to control the APA of nuclear factor of activated T-cell c (NF-ATc) mRNAs in effector T cells (21). How CstF64 regulates APA globally remains unknown, however.
To comprehensively characterize the functions of CstF64 in vivo, we mapped the CstF64–RNA interactions in human cells at the transcriptome level. In addition, we characterized CstF64-mediated global APA regulation by quantitative RNA polyadenylation profiling of CstF64-expressing and CstF64-depleted cells. Taken together, these data provide significant insight into the mechanisms of PAS recognition and APA regulation.
Results
Mapping CstF64–RNA Interactions in Vivo at Single Nucleotide Resolution by Individual Nucleotide Resolution UV Crosslinking and Immunoprecipitation Sequencing.
We set out to map the CstF64–RNA interactions in vivo by individual nucleotide resolution UV crosslinking and immunoprecipitation sequencing (iCLIP-seq) (22). CstF64 was efficiently crosslinked to RNAs in vivo by UV irradiation, and CstF64–RNA complexes were specifically immunoprecipitated by anti-CstF64 antibodies (Fig. S1A, lane 2). When the cell lysate was treated with RNase I before immunoprecipitation, a sharper band of ∼70 kDa was observed (Fig. S1A, lane 1), corresponding to CstF64 crosslinked to short RNAs. When UV irradiation was omitted (−UV) or when immunoprecipitation was carried out with protein A beads alone (with no Ab), no CstF64–RNA complex signal was detected (Fig. S1A, lanes 3–8). In addition, when CstF64 iCLIP-seq was performed using cells in which CstF64 had been depleted by RNAi, the signals for CstF64–RNA complexes were significantly diminished (Fig. S1A, lanes 9 and 10). Taken together, these results demonstrate that our iCLIP-seq procedure was highly specific. To further ensure data quality, three independent replicate iCLIP-seq libraries were prepared and sequenced.
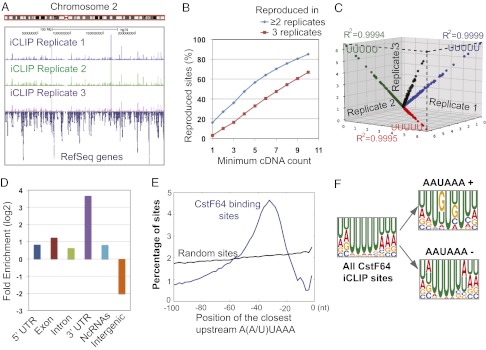
The three replicate datasets of CstF64 iCLIP-seq were highly consistent (Fig. 1A). To quantitatively assess the reproducibility of our data, we carried out the following analyses. We first calculated the percentage of CstF64 crosslinking nucleotides detected in multiple replicate experiments using different minimum cDNA count (i.e, the number of crosslinking events) thresholds. Higher percentages of crosslinking sites were detected in two or more replicates when higher cDNA count thresholds were used (Fig. 1B). For CstF64 crosslinking positions with cDNA counts of 10 or more, 85% of these sites were detected in at least two replicates, and 67% were detected in all three replicates. We next compared the frequency of all possible pentamers overlapping the crosslinking nucleotides, as described previously (22), and found extremely high correlations among all three replicates (R2 = 0.9994–0.9999; Fig. 1C). These results demonstrate that our iCLIP-seq datasets were of high quality. For subsequent analyses, we focused on the 264,522 crosslinking sites that were detected in all three replicates.
Fig. 1.
iCLIP-seq mapping of CstF64-RNA interactions. (A) iCLIP mapping results. The three replicates are labeled. (B) Reproducibility of CstF64 crosslinking sites. The x-axis is the minimum cDNA count threshold, and the y-axis is the corresponding percentage of crosslinking sites reproduced in at least two (blue) or three (red) replicates. (C) Reproducibility of nucleotide composition at crosslinking sites. Frequencies of all possible pentamers overlapping the crosslinking sites are compared among three replicates. R2 values and the top pentamer for each comparison are shown. The numbers on the axis represent the percentage of crosslinking sites that overlap with a specific pentamer. (D) Enrichment of CstF64 binding sites in different genomic regions. The log2 ratios between the observed and expected frequencies of reads in specified genomic regions are shown. (E) Position of the closest upstream A(A/U)UAAA for all CstF64 crosslinking sites (blue line) or random sites (black). Position 0 on the x-axis represents the CstF64 crosslinking site. (F) Overrepresented motifs at all CstF64 crosslinking sites or AAUAAA+ and AAUAAA− crosslinking sites.
Global CstF64–RNA Interaction Landscape.
CstF64 iCLIP-seq mapping results are summarized in Fig. S1B. Comparing the observed read distributions and the expected frequencies based on the genomic compositions revealed that CstF64 RNA binding sites are highly enriched in the extended 3′ UTRs (annotated 3′ UTRs in Refseq genes plus 200 nt of downstream sequences) (Fig. 1D), consistent with the known functions of CstF64 in mRNA 3′ processing. Based on previous in vitro studies (14, 15), we hypothesized that CstF and CPSF bind to RNA together as a complex. Given that CPSF specifically recognizes the A(A/U)UAAA hexamer, we searched for this hexamer upstream of CstF64 crosslinking sites. Strikingly, A(A/U)UAAA was highly enriched in a discrete region 20∼50 nt upstream of CstF64 binding sites (Fig. 1E). This observation strongly suggests that CPSF and CstF bind to many RNA sequences in vivo as a heterodimer.
We next characterized the CstF64 RNA-binding sequence specificity. When sequences of all nonoverlapping CstF64 crosslinking sites and 10 nt on either side were aligned, U was the most frequently seen nucleotide at almost all positions (Fig. S1C). In keeping with this, when we searched for overrepresented hexamers in the same 21-nt regions based on z-scores, the most enriched hexamers were again highly U-rich (Fig. 1F). These results demonstrate that CstF64 binds to U-rich sequences in vivo. As mentioned earlier, CPSF and CstF bind to many RNA sequences together (Fig. 1E). To further examine the binding specificity of CstF64, we divided CstF64 crosslinking sites into two groups, those with A(A/U)UAAA within 40 nt upstream (AAUAAA+) and those without A(A/U)UAAA within 40 nt upstream (AAUAAA−). Interestingly, both z-score and Multiple Em for Motif Elicitation (MEME) analyses showed that GU-rich motifs were enriched in the AAUAAA+ CstF64 binding sites, but U-rich sequences were overrepresented in the AAUAAA− sites (Fig. 1F and Fig. S1D). These data suggest that CstF64 binds to U- or GU-rich sequences globally, and that its binding specificity in vivo is likely influenced by CPSF.
CstF64–RNA Interactions and PAS Recognition.
Our comparison of conservation levels of CstF64 binding sites in 3′ UTRs and the neighboring sequences with random 3′ UTR sequences revealed that sequences 30–100 nt upstream of the CstF64 crosslinking sites were significantly more conserved than CstF64 crosslinking sites themselves and the downstream regions (Fig. S1E). This pattern is consistent with the fact that CstF64 binding sites are fairly variable, and provides further evidence that upstream sequences, which most likely represent CPSF binding sites, play an important role in recruiting CstF64 to RNAs.
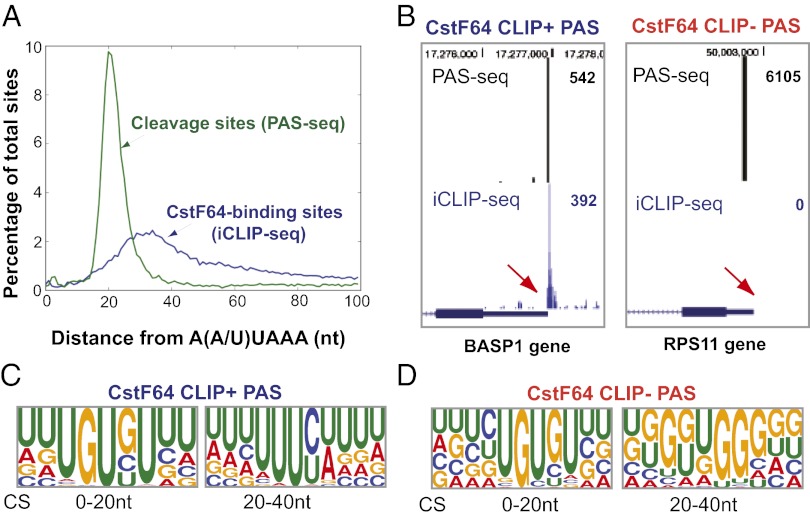
To better understand the role of CstF64 in PAS recognition in vivo, we characterized the distribution of CSs and CstF64 binding sites relative to the upstream A(A/U)UAAA hexamers in 3′ UTRs. For global analyses of the CSs, we used our recently published PAS-seq analysis of HeLa transcriptome in which we mapped the CSs of more than 9,000 actively expressed genes (7). Remarkably, the CSs displayed a sharp peak ∼20 nt downstream of the A(A/U)UAAA hexamers, whereas CstF64 binding sites were more broadly distributed, centered at ∼30 nt downstream of the A(A/U)UAAA hexamer (Fig. 2A). These data suggest that CSs are generally located between CPSF and CstF RNA-binding sites at a relatively fixed distance (∼20 nt) from the A(A/U)UAAA hexamers. These results are consistent with previous in vitro studies and confirm the role of CstF64–RNA interactions in PAS recognition in vivo (3, 4).
Fig. 2.
Differential CstF64–RNA interactions at PASs. (A) Distribution of CSs (green line, based on PAS-seq data) and CstF64 binding sites (blue line, based on iCLIP-seq data) relative to the closest upstream A(A/U)UAAA. Position 0 represents the 5′ end of AAUAAA. (B) PAS-seq and CstF64 iCLIP-seq results for BASP1 and RPS11 genes. The red arrows indicate the same regions downstream of the CSs. The read counts for the PAS-seq and iCLIP-seq peaks shown are listed on the right. (C and D) Web logos of the top 20 most greatly enriched motifs in the 0- to 20-nt and 20- to 40-nt regions downstream of the CSs of CstF64 CLIP+ and CLIP− PASs.
Consistent with the current model for PAS recognition, CstF64 binds RNA downstream of the CS at many PASs. For example, a cluster of CstF64 iCLIP tags was detected immediately downstream of the CS at the BASP1 PAS (Fig. 2B, Left and Fig. S2A). Hereinafter, these PASs are referred to as CstF64 CLIP-positive (CLIP+) PASs. Surprisingly, however, more than 50% of all active PASs in HeLa cells had no reproducible CstF64 iCLIP tags detected within 60 nt downstream of the CS (Fig. 2B, Right and Fig. S2B); we call these PASs CstF64 CLIP-negative (CLIP−) PASs. The lack of CstF64–RNA crosslinking signals at CstF64 CLIP− PASs could be related to several factors, including the transient nature of CstF64–RNA interactions and/or rapid degradation of CstF64-bound RNAs during 3′ processing. It is also possible that the lack of CstF64–RNA crosslinking signals at some of the CLIP− PASs might reflect weak interactions. We have obtained multiple lines of evidence suggesting that the CstF64 affinities for different PASs vary widely, and that the recognition of some CstF64 CLIP− PASs may be less dependent on stable CstF64–RNA interactions. First, we compared the sequences of the top 1,000 (ranked by PAS-seq read counts) CstF64 CLIP+ and CLIP− PASs between –100 nt and +100 nt relative to the CS. Over the entire region, CstF64 CLIP+ PASs were significantly more A/U-rich than CLIP− PASs (Fig. S3A). We next focused on the 40-nt sequence downstream of the CSs, the region in which most CstF64–RNA interactions occur (Fig. 2A). Within this region, CstF64 CLIP+ PASs generally are more U-rich, whereas the CLIP− sites have a higher G content (Fig. S3B). Interestingly, different subregions within this fragment seem to have distinct sequence features. Within the first 20 nt downstream of the CS, GU-rich motifs are enriched in both CstF64 CLIP+ and CLIP− PASs; however, in the 20- to 40-nt region, CstF64 CLIP+ PASs are generally U-rich, whereas G-rich motifs are enriched within the same region in the CLIP− PASs (Fig. 2 C and D). These results indicate that CstF64–RNA crosslinking at PASs is strongly influenced by RNA sequences, and that a U-rich sequence context promotes stable CstF64–RNA interactions.
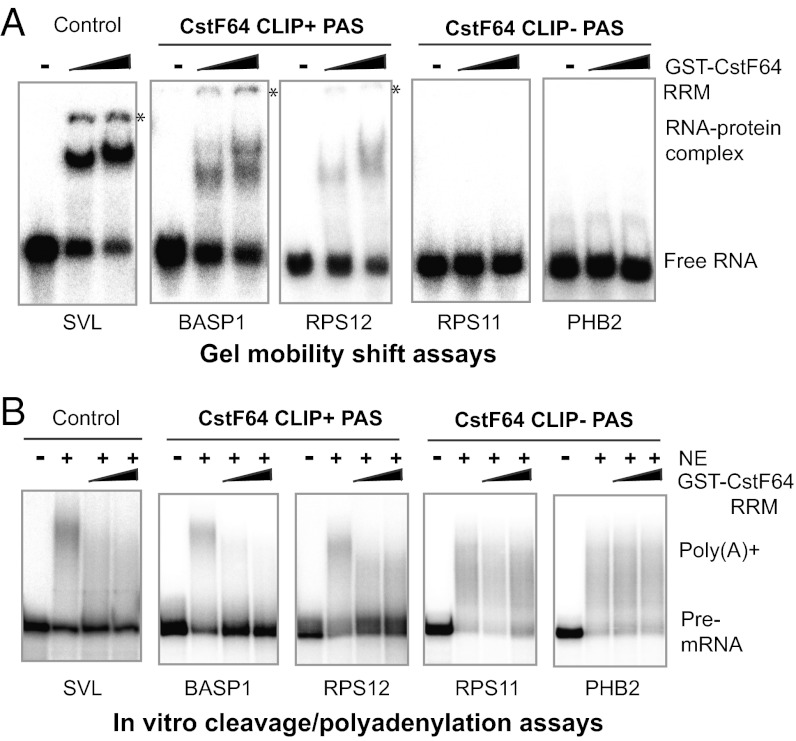
We next directly compared the affinities of CstF64 for the CLIP+ and CLIP− PASs in vitro. RNAs corresponding to the 60-nt sequences downstream of the CSs of two CLIP+ PASs (BASP1 and RPS12) (Fig. S2A) and two CLIP− PASs (PHB and RPS11) (Fig. S2B; sequences shown in Fig. S2C) were synthesized. We used these RNAs in gel mobility shift assays with the recombinant CstF64 RRM domain (GST-CstF64-RRM), with SVL, a commonly used viral PAS substrate known to bind CstF64 (17), as a positive control. CstF64-RRM was seen to bind to SVL as well as to BASP1 and RPS12; in contrast, no interaction was detected between CstF64-RRM and the two CLIP− PASs (Fig. 3A). These results are consistent with our iCLIP-seq data and demonstrate that the CstF64 has lower affinity for CLIP− PAS sequences. Finally, we examined whether these two types of PASs have different requirements for CstF64–RNA interactions for recognition. Toward this end, we carried out in vitro cleavage/polyadenylation assays with HeLa nuclear extract in the presence of recombinant CstF64-RRM. We reasoned that if CstF64 interacts with RNA and such interactions are required for PAS recognition, then excess CstF64-RRM will act as a dominant negative factor by competing with the endogenous CstF64 for RNA binding, thereby blocking 3′ processing. In contrast, if CstF64 does not bind the PAS and/or if PAS recognition is less dependent on CstF64–RNA interactions, then adding excess CstF64-RRM will have little or no effect. Indeed, the addition of CstF64-RRM blocked the cleavage/polyadenylation of all three CstF64 CLIP+ PASs but had little if any effect on the CstF64 CLIP− PASs (Fig. 3B). We also examined the role of CstF64 in the recognition of CstF64 CLIP+ and CLIP− PASs in vivo using the pPASPORT bicistronic reporter (Fig. S3C), which follows a similar design principle as a previously described reporter (23). In this system, the Renilla luciferase (Rluc)/Firefly luciferase (Fluc) ratio provides a quantitative measurement of the “PAS strength” of the tested sequences. Unexpectedly, the PAS activities (Rluc/Fluc) of both BASP1 (CLIP+) and PHB2 (CLIP−) PASs increased after CstF64 depletion by RNAi (Fig. S3D), likely due to the up-regulation of the redundant factor CstF64τ (Fig. 4A), as discussed in more detail below. After double knockdown of CstF64 and CstF64τ, the activity of both BASP1 and PHB2 PASs decreased. However, because CstF64τ was still present in the double-knockdown cells (Fig. 4A), our reporter assay results are inconclusive. Nonetheless, our iCLIP-seq and in vitro data demonstrate that CstF64–RNA interactions at PASs are highly variable in affinity, and evidence suggests that recognition of some CstF64 CLIP− PASs may be less dependent on stable CstF64–RNA interactions.
Fig. 3.
CstF64–RNA interactions and PAS recognition. (A) Gel mobility shift assays using GST-CstF64-RRM (0, 25, and 50 μM) and the 60-nt sequences downstream of the CSs of the listed genes. SVL RNA was used as a positive control. (B) In vitro cleavage/polyadenylation assays in the presence of GST-CstF64-RRM (0, 30, and 60 μM). SVL was used as a control.
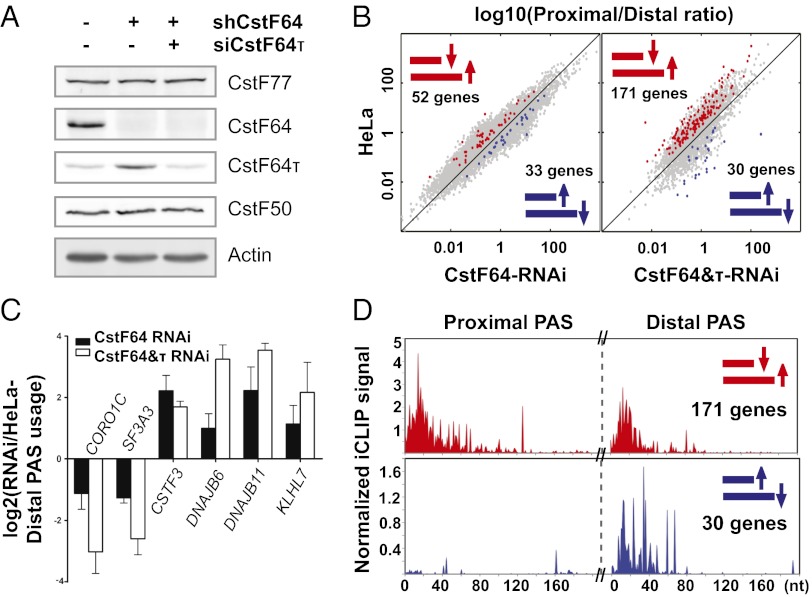
Fig. 4.
CstF64-mediated global APA regulation. (A) Western blot analysis of control HeLa, CstF64-RNAi, and CstF64&τ-RNAi cells. (B) Pairwise comparison of PAS use in HeLa, CstF64-RNAi, and CstF64&τ-RNAi cells. The y-axis shows log10(proximal/distal)–HeLa. The x-axis shows log10(proximal/distal)–CstF64-RNAi (Left) or –CstF64&τ-RNAi (Right). PAS pairs with statistically significant differences in use are highlighted in blue (higher use of proximal PAS in RNAi cells) or red (higher use of distal PAS in RNAi cells). (C) qRT-PCR verification of the APA changes in six genes. The y-axis shows the log2 ratio of RNAi/HeLa (extended/common). (D) Total normalized iCLIP signals for proximal-to-distal shift (red) and distal-to-proximal shift (blue) PAS pairs (the same highlighted PAS pairs as in B).
Global Analyses of CstF64-Mediated APA Regulation.
To characterize the role of CstF64 in global APA regulation, we generated HeLa cell lines (CstF64-RNAi cells) that stably expressed specific shRNAs against CstF64 mRNAs. CstF64 was efficiently depleted in these cells, whereas CstF77 and CstF50 levels were not significantly affected (Fig. 4A). Interestingly, these cells exhibited no apparent growth defects. We then isolated total RNAs from control HeLa cells and CstF64-RNAi cells and performed direct RNA sequencing (DRS) using the Helicos platform to quantitatively map RNA APA profiles. Because RNAs are directly sequenced without reverse-transcription and PCR amplification, DRS is a highly quantitative method (8). Our comparison of the APA profiles in control HeLa cells and CstF64-RNAi cells identified 327 PASs with significantly different uses (Dataset S1). We identified 85 genes as high-confidence targets because they contained two alternative PASs that showed significant difference in use. Fifty-two of these 85 genes exhibited an increase in the relative use of the distal PAS in CstF64-RNAi cells (proximal-to-distal shift), whereas the other 33 genes demonstrated changes in the opposite direction (distal-to-proximal shift) (Fig. 4B, Left and Dataset S1).
Given the known function of CstF64 as an essential mRNA 3′ processing factor, the finding that depletion of CstF64 had a relatively small effect on the global APA profile was unexpected. Interestingly, we found significantly higher CstF64τ protein levels in CstF64-RNAi cells (Fig. 4A). We next compared the RNA-binding specificity of CstF64 and CstF64τ using gel shift assays with purified GST-CstF64 or CstF64τ-RRM and the aforementioned CstF64 CLIP+ (SVL, BASP1, RPS11) and CLIP− (RPS12 and PHB2) PASs. For all RNAs tested, the affinities of CstF64 and CstF64τ were almost indistinguishable (Fig. S4). These results suggest that CstF64τ and CstF64 have overlapping RNA-binding specificities and may play redundant roles in mRNA 3′ processing. Thus, the enhanced levels of CstF64τ in CstF64-RNAi cells may compensate, at least partially, for the loss of CstF64.
To assess the specific role of CstF64 in global APA regulation, we knocked down CstF64τ in CstF64-RNAi cells to a level similar to that in control HeLa cells through transient transfection of siRNAs against CstF64τ (Fig. 4A, right lane). Our DRS analysis of the CstF64 and CstF64τ double-knockdown (CstF64&τ-RNAi) cells revealed two interesting findings. First, we identified 873 PASs with significantly different use in the CstF64&τ-RNAi cells and control HeLa cells (Dataset S1), a significantly higher number than found in the CstF64-RNAi cells. We identified 201 genes as high-confidence targets with two PASs that displayed significantly different use (Fig. 4B, Right). There was significant overlap between the genes with significant APA changes in CstF64-RNAi cells and in CstF64&τ-RNAi cells (Fig. S5A), and the regulated PASs in the two datasets also shared some sequence features (Fig. S5B). Second, of the identified genes with APA changes, the majority (171 genes; 85%) exhibited a proximal-to-distal shift, whereas only 30 genes (15%) had changes in the opposite direction (Fig. 4B). We validated our APA analyses results on six selected target genes through quantitative RT-PCR (qRT-PCR) using primer sets targeting the common regions shared by both APA isoforms or the extended regions found only in the longer isoforms. For all six genes tested, the directionality of APA changes detected by the DRS analysis was confirmed by qRT-PCR (Fig. 4C), suggesting that our DRS analyses of APA were highly reliable. In most cases, the magnitude of the APA changes was greater in CstF64&τ-RNAi cells than in CstF64-RNAi cells.
To understand the role of CstF64–RNA interactions in APA regulation, we compared CstF64 iCLIP signals in the PASs regulated by CstF64. We divided the genes with significant APA changes in CstF64&τ RNAi cells into “proximal-to-distal shift” and “distal-to-proximal shift” groups. For all of the genes within each APA group, we then plotted the total normalized iCLIP signals at the proximal and distal sites (see SI Materials and Methods for details). We detected similar levels of iCLIP signals at both proximal and distal PASs for genes in the proximal-to-distal group (Fig. 4D, Upper). In contrast, the distal PASs had significantly higher CstF64 iCLIP signals compared with the proximal PASs for genes in the distal-to-proximal group (Fig. 4D, Lower).
To gain mechanistic insight into CstF64-mediated APA regulation, we performed further analyses of the DNAJB6 and CSTF3 genes. Both demonstrated proximal-to-distal APA changes in CstF64&τ-RNAi cells (Fig. 4C and Fig. S6A). We first compared the CstF64–RNA interactions at the proximal and distal PASs. We detected greater CstF64 iCLIP signals at the proximal PAS than at the distal PAS for both genes (Fig. S6A). Consistent with the iCLIP data, in vitro gel mobility shift assays using GST-CstF64-RRM and RNAs from the two DNAJB6 PASs revealed that CstF64 has greater affinity for the proximal PAS (Fig. S6B). Second, we measured the activities of the proximal and distal PASs of DNAJB6 and CSTF3 using reporter assays. Both distal PASs were significantly more active than the proximal PASs (Fig. S6C). In addition, for all PASs except the CSTF3 distal site, their activities decreased after CstF64&τ knockdown. We incorporated these results into a mechanistic model of CstF64-mediated APA regulation (Discussion).
Characterization of Intronic CstF64 Binding Sites.
Almost half of the CstF64 binding sites are in introns (Fig. S1B). Several lines of evidence suggest that many of these intronic CstF64 binding sites constitute parts of functional PASs. First, there is a significant enrichment of A(A/U)UAAA within 20∼50 nt upstream of intronic CstF64 crosslinking sites (Fig. S7A), suggesting that CstF and most likely CPSF are recruited to these intronic sites. Second, intronic CstF64 binding sites are more conserved than random intronic sequences (Fig. S7B). We next directly tested whether intronic CstF64 binding regions can function as PASs using reporter assays. We tested three CstF64-bound intronic regions from the BASP1, CMIP, and NR3C1 genes (Fig. S7C), each spanning a cluster of strong CstF64 crosslinking sites and the closest upstream A(A/U)UAAA hexamer. We used the strong viral PAS, SVL, as a positive control. The Rluc/Fluc ratio for the tested intronic CstF64 binding sites ranges from ∼2% (for BASP1) to >15% (for CMIP and NR3C1) of the SVL PAS (Fig. S7C). Finally, our in vitro cleavage/polyadenylation assays further confirmed that the intronic CstF64 binding sites from the CMIP and NR3C1 genes can support 3′ processing in HeLa nuclear extract (Fig S7D). Taken together, these results suggest that there is a large number of potentially functional PASs in introns, and that these intronic PASs are bound by CPSF–CstF complexes.
Using a stringent standard (cDNA counts of ≥5 and reproduced in all three replicates), we identified 9,584 high-confidence intronic CstF64 binding sites. However, our PAS-seq analyses showed that <2% of active PASs are found in introns (7). How are most intronic PASs suppressed? Based on previous studies (24, 25), we hypothesized that CstF64-bound intronic PASs are suppressed by U1 small nuclear ribonucleoproteins. To test this hypothesis, we took advantage of the recently published Affymetrix GeneChip human tiling array analyses of total RNAs isolated from HeLa cells treated with control or U1 antisense morpholino oligos, which blocks U1 binding to RNAs (25). For example, their analyses detected an abrupt decrease in RNA signals in the introns of the CMIP, BASP1, NR3C1, and HSPA4 genes, indicating premature cleavage/polyadenylation at these sites (Fig. S8 A–D). Strikingly, we found that each of these premature termination sites corresponds precisely to a prominent CstF64 binding site (Fig. S8 A–D, lower tracks), suggesting that these CstF64-bound intronic PASs are activated when U1 is inhibited. To determine whether this phenomenon is general, we plotted the average tiling array signals surrounding all intronic CstF64 crosslinking sites. Significantly, we observed a sharp decrease in the U1 antisense morpholino oligo microarray signals upstream of CstF64 binding sites, but not at random intronic sites (Fig. S8E). The cleavage/polyadenylation, as indicated by a transition from positive values to negative values in microarray signals, occurs 50∼100 nt upstream of the CstF64 crosslinking sites (Fig. S8E). Taken together, these findings strongly suggest that U1 small nuclear ribonucleoprotein suppression of CstF64-bound intronic PASs is a widespread phenomenon, and that U1 inhibits 3′ processing at intronic PASs in a step after the recruitment of CPSF and CstF.
Discussion
A central question in mRNA 3′ processing is how PAS recognition is regulated. The first step in addressing this question is to comprehensively characterize the protein–RNA interactions for the core pre-mRNA 3′ processing factors. In this study, we generated a single-nucleotide resolution protein–RNA map for the essential pre-mRNA 3′ processing factor CstF64. In addition, we characterized CstF64-mediated global APA regulation. Integration of these data provides insight into the mechanisms of PAS recognition and APA regulation.
Consistent with previous in vitro studies, we have shown that CstF64 recognizes many PASs in vivo through direct interactions with both U-rich and GU-rich sequences downstream of their CSs (Fig. 2). Unexpectedly, however, our iCLIP-seq and in vitro biochemical data revealed that CstF64 interactions with PAS RNAs are of highly variable affinity (Fig. 3A). Furthermore, our in vitro 3′ processing assays provided supportive evidence that the recognition of PASs with low CstF64 affinities may be less dependent on stable CstF64–RNA interactions (Fig. 3B). However, our results do not rule out the possibility that CstF64 is physically present in the 3′ processing complex assembled on CstF64 CLIP− PASs. These results provide further evidence that mammalian PASs with different sequence features may rely on different protein–RNA interactions for their recognition. For example, the CPSF–AAUAAA interaction is believed to be essential for recognition of the majority of mammalian PASs (3, 4). Approximately 30% of human PASs do not have the AAUAAA hexamer, however (26). For some of these “noncanonical” PASs, cleavage factor I (CF Im) binding to UGUA motifs has been proposed to play a major role in recognizing these PASs (27). Similarly, for CstF64 CLIP− PASs, different proteins may be responsible for recognizing the downstream sequences. Given that G-rich motifs are enriched at these PASs (Fig. 2D), a possible candidate is heterogenous nuclear ribonucleoprotein H, which has been shown to bind to the G-rich sequences in SVL PAS and contribute to efficient PAS recognition (28). Alternatively, CPSF and CF Im interactions with upstream sequences may be sufficient for the recognition of CstF64 CLIP− PASs. Thus, these results suggest the possible existence of diverse mechanisms for PAS recognition in mammalian systems.
The present study provides some important insight into the role of CstF64 in global APA regulation. First, our data reveal that CstF64 and its paralog CstF64τ have overlapping RNA-binding specificities and play redundant roles in APA regulation. The functional redundancy between the two proteins provides an explanation for our observations that CstF64 depletion had little effect on cell growth or the global APA profile, and that codepletion of both CstF64 and CstF64τ led to greater APA changes. Given that CstF64τ was still present in our CstF64/τ-RNAi cells (Fig. 4A), the actual number of APA events regulated by CstF64 and CstF64τ may exceed those identified in this study. Second, our data suggest that CstF64 is an important global regulator of APA and in most cases promotes the use of proximal PASs (Fig. 4B). We propose the following model for CstF64-mediated APA regulation. When CstF64 is abundant, it promotes efficient recognition of the proximal and weaker PASs through direct protein–RNA interactions. The 3′ processing at proximal PASs prevents the transcription and use of the distal PASs. In the presence of limited CstF64, however, recognition of the proximal PASs becomes less efficient, which allows the distal and stronger PASs to be transcribed and recognized by the 3′ processing machinery. Our results are consistent with those of previous studies showing that higher levels of CstF64 led to increased use of the proximal PASs in the IgM and NF-ATc mRNAs (20, 21). Third, although CstF64 is believed to be a general 3′ processing factor, our results suggest that CstF64 depletion affects the APA of a specific subset of genes. Interestingly, a similar phenomenon has been reported in splicing, where changes in the concentration of core spliceosomal components regulate specific alternative splicing events (29). This may be a common theme for the regulation of mRNA processing. Finally, a number of recent studies have reported widespread and systematic APA changes under varying physiological and pathological conditions (12). Interestingly, systematic APA shifts to the distal PASs during stem cell differentiation and development are accompanied by a decrease in the mRNA levels of many core 3′ processing factors, including CstF64 and CstF64τ (7, 9, 30). Our study provides direct evidence that a decrease in the protein levels of a general 3′ processing factor leads to largely unidirectional APA changes. It is important for future studies to examine whether, and if so, how, the protein levels of CstF64/τ and other core 3′ processing factors are regulated under different physiological conditions, and how these changes contribute to global APA regulation.
Materials and Methods
The iCLIP-seq analyses were carried out as described previously (22). Direct RNA sequencing was performed by Helicos BioSciences as described previously (8). All sequencing data have been submitted to the National Center for Biotechnology Information’s Gene Expression Omnibus database (accession no. GSE40859). CstF64-RNAi cell lines were generated by transfecting HeLa cells with pSuperior-puro constructs and selecting stably transfected cells with puromycine. Single colonies were expanded and used for this study. GST-CstF64/τ-RRM was expressed in Escherichia coli and purified with glutathione-conjugated beads in accordance with the manufacturer’s instructions (GE Healthcare Life Sciences). Gel mobility shift assays, in vitro cleavage/polyadenylation assays, and bioinformatic analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Bin Tian for providing the scripts for motif analysis and Dr. Anne-Catherine Prats for providing reagents. This study was supported by National Institutes of Health Grants R01 GM090056 (to Y.S) and R01 GM088342 (to Y.X.), American Cancer Society Grant RSG-12-186 (to Y.S), National Science Foundation Grant DBI-084621 (to X.X.), and a junior faculty grant from the Edward Mallinckrodt Jr. Foundation (to Y.X.). J.B. was partially supported by National Institutes of Health/National Library of Medicine Bioinformatics Training Grant T15LM07443.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40859).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211101109/-/DCSupplemental.
References
- 1.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38(9):2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J. 2008;27(3):482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11(21):2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 5.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2(3):321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17(4):761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143(6):1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rüegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271(11):6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 14.Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267(21):14804–14811. [PubMed] [Google Scholar]
- 15.Gilmartin GM, Nevins JR. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3(12B):2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 16.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4(12A):2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 17.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14(10):6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace AM, et al. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci USA. 1999;96(12):6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87(5):941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 21.Chuvpilo S, et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10(2):261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- 22.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Z, et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1(2):255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 25.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468(7324):664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30(8):1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101(45):15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4(12):e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.