Abstract
Ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is a crucial enzyme in carbon fixation and the most abundant protein on earth. It has been studied extensively by biochemical and structural methods; however, the most essential activation step has not yet been described. Here, we describe the mechanistic details of Lys carbamylation that leads to RuBisCO activation by atmospheric CO2. We report two crystal structures of nitrosylated RuBisCO from the red algae Galdieria sulphuraria with O2 and CO2 bound at the active site. G. sulphuraria RuBisCO is inhibited by cysteine nitrosylation that results in trapping of these gaseous ligands. The structure with CO2 defines an elusive, preactivation complex that contains a metal cation Mg2+ surrounded by three H2O/OH molecules. Both structures suggest the mechanism for discriminating gaseous ligands by their quadrupole electric moments. We describe conformational changes that allow for intermittent binding of the metal ion required for activation. On the basis of these structures we propose the individual steps of the activation mechanism. Knowledge of all these elements is indispensable for engineering RuBisCO into a more efficient enzyme for crop enhancement or as a remedy to global warming.
Keywords: nitrosylation in algae, activity control, intermediate trapping
Ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (EC 4.1.1.39) is a dominant contributor to conversion of gaseous CO2 into biomass (1). Aerobic life forms, including heterotrophs, primarily derive their carbon through an authotrophic/photosynthetic route using CO2 as a carbon source. RuBisCO accomplishes this task by incorporating CO2 into a phospho-sugar, ribulose 1,5-bisphosphate (RuBP). Incorporation of CO2 into RuBP generates two molecules of 3-phosphoglycerate. This simple compound is subsequently used to build other organic molecules of life. Synthesized RuBisCO does not have a fully functional active site (2–4). It needs to be activated by a CO2 molecule that carbamylates its catalytic Lys to bind Mg2+ that completes the activation process (5, 6). The mechanism of RuBisCO activation by the CO2 molecule is presented in this work. This knowledge will contribute to acceleration of the progress in remodeling the RuBisCO for practical uses (7–10).
RuBisCO is present in many photosynthetic organisms including bacteria, algae, plants (2–4), and archaea (11). It accounts for roughly a half of the soluble protein mass in C3 plant leaves (1). However, RuBisCO is an inefficient enzyme with a low, 1–10/s, turnover rate (12). A side reaction with O2 reduces its functional activity even further. The side reaction happens when a promiscuous enodiolate intermediate of RuBP is attacked by O2 (versus CO2) and produces 2-phosphoglycolate (13). This reaction drains a pool of RuBP and decreases the efficiency of carbon fixation by up to 50%. The efficiency of RuBisCO is thus dependent on the ability to discriminate between CO2 and O2 or on the specificity ratio. The specificity ratio S is defined as: S = VcKo/VoKc, where Vc and Vo are maximal velocity for carboxylation and oxygenation, and Kc and Ko are the relative Michaelis constants for CO2 and O2, respectively. The specificity ratio has implications for crop yield, nitrogen and water utilization in plants, and the global carbon cycle. We propose the principles of substrate selectivity and suggest how to improve it.
Mutational studies produced a limited success in improving the catalytic efficiency of RuBisCO (9, 14, 15). This limited success stems from an incomplete understanding of the complex mechanistic principles of RuBisCO activation and catalysis (16). The inefficiency of the enzyme, however, does not affect its cellular function; therefore, it has been recently postulated that RuBisCO has evolved as an inefficient enzyme (17, 18), which is compensated by its abundance. Our work delineates the fundamental mechanism of RuBisCO activation by carbamylation and suggests the principles of gaseous ligand discrimination (O2 versus CO2) by the enzyme. As a result, we provide a road map for the potential improvement of the enzyme’s selectivity and catalytic efficiency.
Forms of RuBisCO.
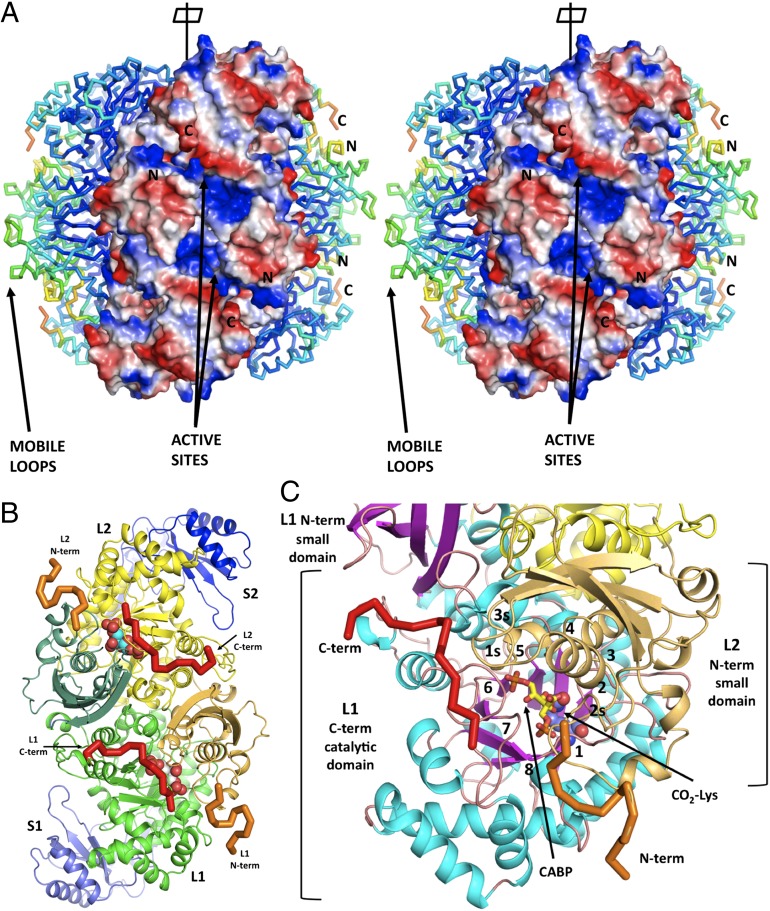
The RuBisCO family represents a diverse group of enzymes. There are four organizational forms of RuBisCO: I, II, III, and IV (13, 19). Representative X-ray structures of these forms (19) are now available. The minimal functional enzyme is a homodimer, composed of two large subunits (L2). An individual subunit contains a catalytic, TIM-like barrel and a small C-terminal α+β domain. Frequently, RuBisCO contains two large subunits (L, 50–55 kDa) that are associated with two small subunits (S, 12–18 kDa). The most common form I is a L8S8 hexadecamer (Fig. 1). This form is present mostly in chemoautotrophic bacteria, cyanobacteria, red and brown algae, and all higher plants. Form I RuBisCO has four subtypes: A and B from cyanobacteria, eukaryotic algae, and higher plants and C and D from nongreen algae and phototropic bacteria (red-type enzymes). The form II enzyme is an L2 dimer of large subunits only. Form III is diverse in composition (L2, L8, and L10) but does not contain small subunits. Form IV encompasses RuBisCO-like proteins from organisms that do not use CO2 as a major carbon source (20). The focus of this study is the L8S8 hexadecameric form ID of RuBisCO with a molecular weight of ∼0.6 MDa from red algae Galdieria sulphuraria (21).
Fig. 1.
(A) General view of hexadecameric structure G. sulphuraria RuBisCO with three L2S2 dimers visible. The dimers on both sides are in a Cα representation colored by the temperature factors. The front-facing dimer is depicted in a surface representation colored by the polarity of the electrostatic field. Two large, positively charged patches (in blue) indicate the entry to the active site cavity that is lined with highly mobile loops with higher temperature factors coming from the β-barrel catalytic domains and the N-terminal domain (visible in dimers on both sides). These positively charged patches are visible because the disordered N- and C-terminal fragments were not included in our model (not visible in ED). They are responsible for sealing off the active sites during the catalytic cycle. (B) A L2S2 dimer of G. partita RuBisCO in the same orientation as in A with the highlighted N-terminal (orange) and C-terminal (red) regions that are disordered in G. sulphuraria structure. (C) Close-up of the L1 subunit with N- and C-terminal tails marked and the loops numbered. Both termini play a crucial role in stabilizing close conformation of the catalytically competent enzyme. The C-terminal tail stabilizes loops 6, 7, and 5 whereas the N-terminal region stabilizes loop 1 (containing the KPK motif) and loops 2s and 3s of the small domain.
Results and Discussion
Structure.
The enzyme was crystallized in the presence of ammonium sulfate (∼2 M) and 2 mM Mg2+. The data were collected on lens-shaped crystals (21), characterized by the I422 space group, to ∼1.9 Å resolution. The structure was solved by molecular replacement using the tobacco structure (3RUB) (22) as a template. The asymmetric unit contained a dimer of the small subunit (138 aa) and the 21–475 residues of a large subunit (493 aa) with the N and C termini disordered (L1S1). This constituted 1/8 of the biological hexadecamer (L8S8) (Fig. 1), which is reconstituted by the crystal symmetry of the lattice.
The final model was similar to that of Galdieria partita RuBisCO (23). The catalytic domain of the large subunit was an α/β (TIM) barrel. The structure showed the unmodified Lys-210, implying that the enzyme was not activated (22). The smaller dimerization domain of the large catalytic subunit was well defined. The small subunit contained an extension forming a β-hairpin. Four hairpins formed an eight-stranded β-barrel around the fourfold symmetry axis. This β-barrel, absent in other forms of RuBisCO (Fig. S1), was first described in the crystal structure of RuBisCO from green bacteria (24).
The structure of the WT enzyme had several unusual features: (i) an electron density (ED) radiating from the sulfur of Cys460 identified as NO (Fig. 2), (ii) a large clathrate of water molecules organized around the fourfold axis that organizes the β-barrel of the small subunits (Fig. S1), and (iii) the large ED identified as dioxygen at the center of the active site (Fig. 3A). Crystallographic measures such as low-temperature factors compatible with surrounding atoms, clear ED combined with minimal residual difference ED, and bonding distances indicated the presence of O2 rather than a single water molecule (Fig. 3A). The site was located within the region of positive electrostatic potential at the center of the β-barrel. We are confident that our interpretation is correct based on the high quality of the resulting model that included alternative conformations for 15 residues and ∼350 water molecules. Additionally, when we tried to activate the enzyme by placing crystals in a CO2 atmosphere and adding of Mg2+, the resulting structure showed disappearance of the oxygen molecule that was replaced by a water molecule. The same experiment performed in higher temperature showed a replacement of a water molecule by CO2 (see below and Table S1).
Fig. 2.
The bond-stick model of the nitrosylated Cys460 covered with the omit map electron density (3 σ).
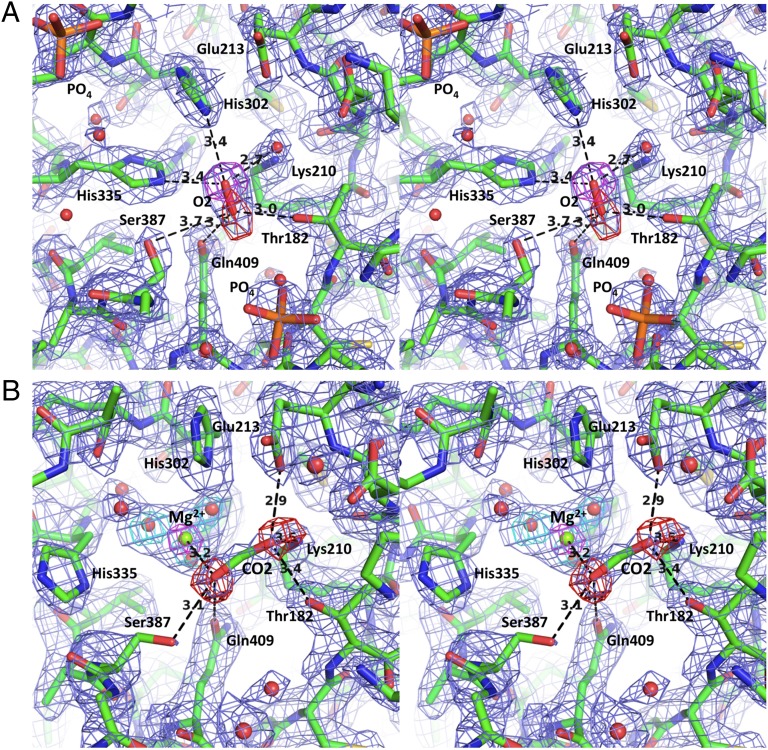
Fig. 3.
The stereo representation of the active site of the enzyme (in bond-stick). (A) The apo-enzyme with O2 bound at the C-terminal end of the inner β-barrel constituting the active site. The 2Fo-Fc electron density contoured at the 1 σ level is in slate blue, whereas the two omit maps for individual oxygen atoms in the O2 molecule are contoured at the 4 σ level (purple and red). (B) The active site of the preactivation complex with Mg2+ surrounded by three water molecules and the CO2 molecule. The 2Fo-Fc ED map (contoured at the 1 σ level) is in slate blue; the omit maps, calculated with oxygen atoms omitted from the CO2 molecule (in red), are contoured at 4 σ; the omit map for Mg2+ (in green) is contoured at the 6 σ level; and the omit maps for the surrounding water molecules are contoured at 3.5 σ (in cyan).
The presence of nitrosylated Cys residues in G. sulphuraria RuBisCO was highly unexpected. Only recently has evidence emerged that plant RuBisCO is regulated by NO and that nitrosylation leads to enzyme inhibition (25). Two NO modifications were refined at Cys460 and Cys181, albeit, with higher temperature factors compared with Cys460, thus suggesting partial disorder. Both Cys residues have been implicated in the inactivation of RuBisCO by oxidation (26). The sequence 175-GRPLLGCTIKPKLGLS-190 is highly homologous in G. sulphuraria and green plants RuBisCOs. This sequence includes the active site Thr182 and the highly conserved cis-Pro185 (in KPK motif, in boldface type above). This sequence also contains highly conserved Cys181, the presence of which correlates with inhibition by nitrosylation in several RuBisCOs investigated so far. The nitrosylation of Cys181 that is adjacent to Thr182 must cause a steric disturbance and marked change in the electrostatic signature of the active site. These changes must influence in a detrimental manner the unstable character of the activation complex making the carbamylation inoperable. Nitrosylation of Cys-460 was also implicated in this inhibition (26). Nitrosylation of Cys residues is a frequent posttranslational modification in higher eukaryotes; however, this modification has not been reported in algae (27), and thus the presence of nitrosylated Cys in G. sulphuraria RuBisCO is a unique finding.
The active site showed an increased disorder (mobility) that included the solvent as well as the protein (3) (Fig. 1 and Figs. S2–S4). Despite the increased mobility (higher Bs) in the active site region, we detected two sizable patches of difference ED that were identified and refined as phosphate/sulfate anions (∼60% occupied). Both phosphate positions correlate well with phosphate groups of substrates or analogs that were found in the other RuBisCO structures (28) (Fig. 3A).
Activation.
Subsequently, to investigate the active enzyme, we attempted to activate the G. sulphuraria RuBisCO in both soluble and crystal forms. We were unable to activate the enzyme using up to 20 mM Mg2+ and excess CO2. Nevertheless, when the crystals were incubated at ∼40 °C in activating solutions, a positive difference ED emerged near His335 (Fig. S2A) and the oxygen position. A proper identification of these and remaining difference ED features became possible only after reinterpreting the WT structure (Fig. S2B), and by modeling Mg2+ surrounded by water molecules and the CO2 molecule (Fig. 3B and Fig. S2B).
A detailed comparison of ED in the WT and the activation complex structures shows numerous and significant conformational changes at the active site and mobile loops that are critical to activity. These changes are presented in Figs. S3 and S4. At the active site of the activation complex, the side chain of His335 rotates away from the position attained in the WT structure to an alternative conformation. The triangular difference density that emerged after the move of His335 has been modeled by an Mg2+ cation coordinated by water molecules (Fig. S2B). The refined model of the activation complex shows a pseudotetrahedral arrangement of the Mg2+ cation to three water molecules (H2O/OH) that are coordinated directly by three His residues (300, 302, and 335). The fourth tetrahedral position is occupied by CO2.
Our refinement of the activation complex structure identified the CO2 molecule in analogous position to that of the O2 molecule in the WT structure. Both gaseous ligands are bound at the center of the TIM barrel of the catalytic domain and have B-factors fully compatible with the mobility of the surrounding atoms (Fig. S3 A and B). A good convergence (R and Rfree), an excellent fit to the ED, and chemically acceptable contacts (Fig. 3B) allowed us to uniquely identify the ligands in this transient structure. An additional support to this conclusion comes from the only known high-resolution protein structure (PDB ID code 2VVA) that has gaseous CO2 bound to a metal ion (Fig. S2C). The active site residues present in both enzymes are similar (His, Gln, Glu, Thr, Lys) and have remarkably similar spatial arrangements. The RuBisCO and carbonic anhydrase (CA) have similar distances between a metal ion and carbon dioxide (∼3.2Å), as well as the planar angle subtended between the CO2 molecule and the metal ion (∼106°). Moreover, the general direction of the CO2 molecule in our structure agrees well with that of the carboxy group observed in the complex of RuBisCO (1IR1) with a transition state analog, 2-carboxyarabinitol 1,5-bisphosphate, as well as the direction of the carbamyl group of Lys210. The character of the CO2-binding site suggests that the directionality of CO2 in the activation complex and in fully active enzyme with the substrate bound might be preserved as dictated by the protein electrostatic field. The bonding geometry of the metal ion is additionally supported by our Quantum Mechanics calculations and by the experimental results of others (29). These results support the interpretation of the immediate surroundings of the metal ion, including the suggestion that two water molecules are in an OH form.
A comparison of the relative positions of the gaseous ligands—O2 (in the WT structure) and CO2 (in the activation complex) of the active site of RuBisCO (Fig. 4)—suggests the mechanism of substrate selectivity of G. sulphuraria RuBisCO (CO2:O2, 222:1) (30). Both ligands bind in a semiorthogonal manner to the same region of RuBisCO, which has a high electrostatic field gradient (closeness of blue and red surfaces in Fig. 4). CO2 and O2 are neither charged nor have dipole moments, which, combined with a very strong electric field at the active site, suggests that the quadrupole moments of both ligands provide the only discriminating factor for binding to RuBisCO. Additionally, the CO2 quadrupole moment is 15-fold higher than that of O2 (Fig. S5), which, combined with relative solubility of both gases in water, provides a semiquantitative explanation for selectivity toward CO2. The high gradient of the electric field at the active site favors CO2 binding by all RuBisCOs. Selectivity is elevated in G. sulphuraria RuBisCO because of several factors such as its high activation temperature (increased mobility), a stronger electrostatic field, and the directionality of the bound ligands. All these factors provide an explanation for a higher affinity and turnover rate for CO2 of G. sulphuraria RuBisCO that exhibits a specificity factor of 222 (30). The role of activation temperature associated with protein’s mobility in increasing the level of specificity, as highlighted by the kinetics of RuBisCO (from Thermococcus kodakarensis) that exhibits S of ∼310 at 90 °C but only S of ∼6 at 25 °C (31), needs to be further examined.
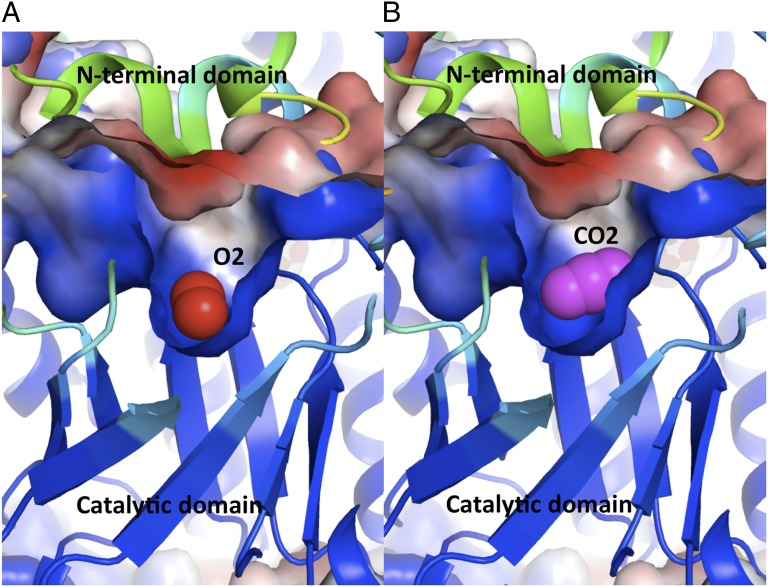
Fig. 4.
The ribbon model of the central catalytic domain with bound gaseous ligands at the active site and the accessible surface colored by the electrostatic potential. The ligands are embedded in a positively charged cavity (blue) located at the C-terminal end of the β-barrel of the catalytic domain closed from the top with a negatively charged lid (red) formed by the N-terminal domain of the L subunit of the opposite dimer. Both of these features are responsible for a strong gradient of the electrostatic field across the active site that interacts with quadrupole moments of the gaseous ligands. The bound ligands are (A) dioxygen (in red) and (B) carbon dioxide (in purple).
The quadrupole moments of charge-less and dipole-less ligands interact only with the high electrostatic field gradient that provides a sufficient guiding force for discrimination and binding at the active site. This apparent need for a high gradient at the active site region explains why the minimal active form of RuBisCO is a dimer of large subunits. Unlike monomers, only dimers produce sufficiently strong electric field gradient across the active site by combining highly and oppositely charged domains that are required for the interaction of RuBisCO with the gaseous substrates. The dimeric form recombines the monomers that have negatively and positively charged elements separated into the N-terminal (dimerization) and the C-terminal (catalytic) domains to avoid misfolding. This conclusion is partially supported by the use of chaperones in the larger oligomer assembly (L8S8) (32).
This modular design of RuBisCO also suggests a function for many flexible loops present at the active site. During catalysis, the enzyme closes the C- and N-terminal segments (33) to increase the strength of the electric field by decreasing the dielectric constant and simultaneously sealing off the active site from solvent. Closing of the active site loops has a high entropic cost; therefore, it is rare and results in a low turnover rate. These conclusions agree well with claims that the specificity is mostly imparted by the mobile loops of the enzyme (16), but also agree with modest improvement in the catalytic efficiency and selectivity of RuBisCO achieved in the mutational studies (9, 15).
Mechanism of Carbamylation.
The structures described above suggest the mechanism of RuBisCO activation by Lys210 carbamylation. Initially, the rotation of His335 to its alternative conformation opens up the active site and creates a transient binding site for the Mg2+ ion. When two water molecules bound by the Mg2+ ion are transiently converted to OH, which results in partial neutralization, the hydrated metal binds at the active site defined by three His (300, 302, 335). His residues (Fig. 3 and Fig. S6) coordinate the metal cation indirectly through the water molecules, which results in a weak and unstable complex. This environment favors binding of Mg2+ in a rare pseudotetrahedral coordination with three H2O/OH molecules and one CO2 molecule bound at the solvent-exposed tetrahedral site. The metal-bound hydroxides drive the deprotonation of the Nε of Lys210 that results in a conformational change in which the Nε of Lys210 rotates by 120° to the trans conformer. This rotation significantly decreases the distance between the Nε of Lys and the C atom of CO2. Next, the CO2 forms a covalent bond with the Nε atom of Lys210 and creates a negatively charged carbamyl group that facilitates metal-ion binding. As a result of CO2 covalent attachment to Lys210, the destabilized Mg2+ ion relocates to a new binding site, created by the carbamyl group of Lys210 and the carboxyl groups of Glu-213 and Asp212. At the final step, His335 that undergoes thermal fluctuation returns to its original conformation, thus finalizing the activation and the formation of the mature active site (Fig. S6) and thus enabling a full closure of loop 6 (Fig. S2) upon binding of the substrates.
The RuBisCO activation mechanism has a chemical precedent in CA. Its reverse reaction (to primary dehydration) mirrors the proposed mechanism (albeit resulting in a different product) (34). In the CA complex with CO2, the position of the activated water is similar to that of the alternative position of the Lys210 Nε, compared with a model of the activation mechanism (Fig. S6). The carbonic acid reaction of CA therefore provides a prototype for the carbamylation reaction in RuBisCO (Fig. S2 B and C).
Conclusions
In this paper, I propose the mechanism of activation of RuBisCO. This work provides an insight into the elusive preactivation complex with Mg2+ and CO2 and defines the principles for the gaseous ligand differential binding. I conclude that the trapping of the intermediate and gaseous ligands becomes possible because the enzyme is inactivated by nitrosylation. I believe that nitrosylation is a common mechanism for controlling RuBisCO activity that has not received sufficient attention.
All of the factors described in this paper, i.e., existence of a strong electric field gradient that interacts with quadrupole moments of the substrates, temperature activation associated with protein mobility, and finally directional binding of the ligands, affect the efficiency of the enzyme, and thus they all should be considered in reengineering of the enzyme to improve its catalytic efficiency and the substrate specificity. The methods for proper accounting for changes in the electrostatic field interactions with the quadrupole moment of the ligands as well as the mobility factors in the mutant proteins must be first developed to fully use the findings of this report. In light of the results presented in this study, I propose that the dimeric forms of RuBisCO are much more amenable material for genetic manipulations and future successful reengineering of the enzyme for practical applications.
Materials and Methods
Protein production and crystallization methods have been described before (21). The native enzyme was inactive. The activation process was carried out by treating the enzyme with 20 mM MgSO4 under CO2 atmosphere for an extended period (up to 24 h). We did not manage to activate the purified enzyme. The activation process of the protein crystals was attempted at room temperature as well as at the elevated temperature (∼40 °C). The crystals were usually harvested after ∼2 h in activating conditions. The crystals of the wild-type enzyme subjected to treatment with Mg2+ and CO2 were immediately frozen in liquid nitrogen upon harvesting from the crystallization wells. The data were collected at low temperature (100 K) for the preactivation complexes whereas the data for the WT native crystals were collected at room temperature. The data were collected on the Rigaku Microfocus 007 and Raxis IV++ imaging plate detector. We refined the structures using Shelx93 and Refmac. The use of both programs was instrumental in identifying gaseous ligands. The ligands were identified by several criteria: (i) elimination of residual difference densities, (ii) comparable level of B-factors to surrounding atoms, and (iii) proper hydrogen-bonding patterns. Their identity was confirmed by omit maps. The refinements converged with excellent stereochemistry, and the final model fit to the ED maps. The R (Rfree) of the refined models was 0.166 (0.22) and 0.181 (0.241) for the native and preactivation complex with CO2 and 0.163 (0.234) for the activation complex without gaseous ligands. For a summary of the refinement statistics, see Table S1.
Supplementary Material
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4F0H, 4F0K, and 4F0M).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210754109/-/DCSupplemental.
References
- 1.Ellis RJ. Most abundant protein in the world. Trends Biochem Sci. 1979;4(11):241–244. [Google Scholar]
- 2.Andersson I. Catalysis and regulation in Rubisco. J Exp Bot. 2008;59(7):1555–1568. doi: 10.1093/jxb/ern091. [DOI] [PubMed] [Google Scholar]
- 3.Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiol Biochem. 2008;46(3):275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Ashida H, et al. RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: Functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO. J Exp Bot. 2008;59(7):1543–1554. doi: 10.1093/jxb/ern104. [DOI] [PubMed] [Google Scholar]
- 5.Lorimer GH, Badger MR, Andrews TJ. The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry. 1976;15(3):529–536. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- 6.Lorimer GH, Miziorko HM. Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+ Biochemistry. 1980;19(23):5321–5328. doi: 10.1021/bi00564a027. [DOI] [PubMed] [Google Scholar]
- 7.Spreitzer RJ, Salvucci ME. Rubisco: Structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- 8.Parry MA, Andralojc PJ, Mitchell RA, Madgwick PJ, Keys AJ. Manipulation of Rubisco: The amount, activity, function and regulation. J Exp Bot. 2003;54(386):1321–1333. doi: 10.1093/jxb/erg141. [DOI] [PubMed] [Google Scholar]
- 9.John Andrews T, Whitney SM. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch Biochem Biophys. 2003;414(2):159–169. doi: 10.1016/s0003-9861(03)00100-0. [DOI] [PubMed] [Google Scholar]
- 10.Farazdaghi H. 2009. Modeling the kinetics of activation and reaction of Rubisco from gas exchange. Photosynthesis in Silico: Understanding Complexity from Molecules to Ecosystems, eds Laisk A, Nedbal L, Govindjee; Advances in Photosynthesis and Respiration 29 (Springer, Berlin), pp 275–294.
- 11.Mueller-Cajar O, Badger MR. New roads lead to Rubisco in archaebacteria. Bioessays. 2007;29(8):722–724. doi: 10.1002/bies.20616. [DOI] [PubMed] [Google Scholar]
- 12.Sage RF. Variation in the k(cat) of Rubisco in C(3) and C(4) plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot. 2002;53(369):609–620. doi: 10.1093/jexbot/53.369.609. [DOI] [PubMed] [Google Scholar]
- 13.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59(7):1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 14.Mueller-Cajar O, Whitney SM. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem J. 2008;414(2):205–214. doi: 10.1042/BJ20080668. [DOI] [PubMed] [Google Scholar]
- 15.Spreitzer RJ, Peddi SR, Satagopan S. Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc Natl Acad Sci USA. 2005;102(47):17225–17230. doi: 10.1073/pnas.0508042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlitter J, Wildner GF. The kinetics of conformation change as determinant of Rubisco’s specificity. Photosynth Res. 2000;65(1):7–13. doi: 10.1023/A:1006425607995. [DOI] [PubMed] [Google Scholar]
- 17.Tcherkez GG, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103(19):7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savir Y, Noor E, Milo R, Tlusty T. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci USA. 2010;107(8):3475–3480. doi: 10.1073/pnas.0911663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson I, Taylor TC. Structural framework for catalysis and regulation in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys. 2003;414(2):130–140. doi: 10.1016/s0003-9861(03)00164-4. [DOI] [PubMed] [Google Scholar]
- 20.Watson GM, Yu JP, Tabita FR. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J Bacteriol. 1999;181(5):1569–1575. doi: 10.1128/jb.181.5.1569-1575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranowski M, Stec B. Crystallization and characterization of Galdieria sulphuraria RUBISCO in two crystal forms: Structural phase transition observed in P21 crystal form. Int J Mol Sci. 2007;8(10):1039–1051. [Google Scholar]
- 22.Curmi PM, Cascio D, Sweet RM, Eisenberg D, Schreuder H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution. J Biol Chem. 1992;267(24):16980–16989. [PubMed] [Google Scholar]
- 23.Sugawara H, et al. Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita. J Biol Chem. 1999;274(22):15655–15661. doi: 10.1074/jbc.274.22.15655. [DOI] [PubMed] [Google Scholar]
- 24.Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I. First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J Biol Chem. 2001;276(51):48159–48164. doi: 10.1074/jbc.M107765200. [DOI] [PubMed] [Google Scholar]
- 25.Moreno J, García-Murria MJ, Marín-Navarro J. Redox modulation of Rubisco conformation and activity through its cysteine residues. J Exp Bot. 2008;59(7):1605–1614. doi: 10.1093/jxb/erm310. [DOI] [PubMed] [Google Scholar]
- 26.Sakihama Y, Nakamura S, Yamasaki H. Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol. 2002;43(3):290–297. doi: 10.1093/pcp/pcf034. [DOI] [PubMed] [Google Scholar]
- 27.Abat JK, Deswal R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: Change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics. 2009;9(18):4368–4380. doi: 10.1002/pmic.200800985. [DOI] [PubMed] [Google Scholar]
- 28.Mizohata E, et al. Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate. J Mol Biol. 2002;316(3):679–691. doi: 10.1006/jmbi.2001.5381. [DOI] [PubMed] [Google Scholar]
- 29.Kluge S, Weston J. Can a hydroxide ligand trigger a change in the coordination number of magnesium ions in biological systems? Biochemistry. 2005;44(12):4877–4885. doi: 10.1021/bi047454j. [DOI] [PubMed] [Google Scholar]
- 30.Uemura K, Anwaruzzaman , Miyachi S, Yokota A. Ribulose-1,5-bisphosphate carboxylase/oxygenase from thermophilic red algae with a strong specificity for CO2 fixation. Biochem Biophys Res Commun. 1997;233(2):568–571. doi: 10.1006/bbrc.1997.6497. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida S, Atomi H, Imanaka T. Engineering of a type III rubisco from a hyperthermophilic archaeon in order to enhance catalytic performance in mesophilic host cells. Appl Environ Microbiol. 2007;73(19):6254–6261. doi: 10.1128/AEM.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature. 2010;463(7278):197–202. doi: 10.1038/nature08651. [DOI] [PubMed] [Google Scholar]
- 33.Satagopan S, Spreitzer RJ. Plant-like substitutions in the large-subunit carboxy terminus of Chlamydomonas Rubisco increase CO2/O2 specificity. BMC Plant Biol. 2008;8:85. doi: 10.1186/1471-2229-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjöblom B, Polentarutti M, Djinovic-Carugo K. Structural study of X-ray induced activation of carbonic anhydrase. Proc Natl Acad Sci USA. 2009;106(26):10609–10613. doi: 10.1073/pnas.0904184106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.